Abstract
Background
Variants in FTO (fat mass and obesity associated) gene are associated with obesity and type 2 diabetes (T2D) in white Europeans. These associations are not consistent in Asians and there are few reports in South Asian Indians who develop T2D at a much lower body mass index (BMI) than that in the white Europeans.
Aims and hypothesis
We studied the association of FTO variants with T2D and measures of obesity in South Asian Indians in Pune, India.
Methods
We genotyped by sequencing, two SNPs rs9939609 and rs7191344, in the FTO gene in 1453 type 2 diabetes patients and 1361 controls and a further 961 population based individuals from India .
Results
We observed a strong association of the minor allele A at rs9939609 with T2D (OR per allele =1.26 [95% CI, 1.13-1.40], P=3×10-5). The variant was also associated with BMI but this association appeared to be weaker (0.06SDs; 95%CIs:0.01-0.10, p=0.017) than the previously reported effect in Europeans (0.10SDs 95%CIs:0.09-0.12). Unlike in the Europeans, the association with T2D remained when adjusting for BMI (OR per allele for T2D=1.21 (95% CI, 1.06-1.37); P=4.0 × 10-3). Similar results were obtained when using waist circumference and other anthropometric parameters.
Conclusions
Our study replicates the strong association of FTO variants with type 2 diabetes in South Asian Indians but suggests that the association of FTO with T2D in them might operate through mechanisms other than obesity. This could imply a fundamental difference between Indians and Europeans in the mechanisms linking body size with T2D.
Keywords: FTO, type 2 diabetes mellitus, polymorphisms, ethnicity, body mass index
Introduction
Common variations in the FTO (fat mass and obesity associated) gene have been found to be strongly associated with body mass index (BMI), obesity and type 2 diabetes (T2D) in a study of over 41,000 white European adults and children [1,2]; the association with T2D was entirely explained by the association with BMI. The association of FTO variants with T2D and BMI has been replicated in several white European populations [3-7] but there are only a few reports in non-European populations, and the findings are not consistent [8,9]. Thus, in a population based study in Chinese subjects, FTO variants were neither associated with BMI nor with type 2 diabetes [8] while in a case-control study in Japanese subjects, there was no association with T2D but a weak association with BMI [9]. Another study in Japanese subjects observed an association of FTO variants with severe obesity [10], while recently, Ng et al found association of FTO variants with both T2D and BMI in East Asians but the FTO association with BMI was weaker in Asians as compared to the European studies [11]. This raises a possibility that the association of FTO variants with BMI and type 2 diabetes might be different in Asian populations. India has the largest number of T2D patients in any single country and this number will increase to over 79 million by the year 2030, when it is estimated that one in five diabetic patients in the world will be an Indian [12]. Even though obesity is a major risk factor for T2D in all populations, Indians develop diabetes at a much lower BMI than that in the white Europeans, and for any given BMI Indians have a higher prevalence of diabetes [13]. This increased risk at lower BMI is thought to be due to higher central obesity (waist-hip ratio) as well as higher adiposity (body fat percent) for any given BMI in Indians compared to white Europeans [13]. Therefore, studying the association of FTO variants with T2D and different measures of obesity in Indians might provide interesting insight in these relationships.
Materials and Methods
Patients
The data and analysis reported in this study included 1453 type 2 diabetes mellitus patients of Indo-European ethnicity, diagnosed before 45 years of age from in and around Pune in western India. These patients were attending the Diabetes Clinic of the King Edward Memorial Hospital and Research Centre (KEMHRC), Pune. They were diagnosed and classified according to WHO 1999 criteria [14], based on response to oral hypoglycemic treatment and perceived lack of insulin dependence. All the cases with probable diagnosis of type 1 diabetes or other rare forms of diabetes were excluded. The phenotypic details were collected through an investigator administered questionnaire; anthropometric measurements were done as per standardized protocols and ethnicity established by their mother tongue and geographic origin [15]. Fasting and postprandial blood was collected for biochemical tests and DNA studies. Biochemical measurements included plasma glucose, insulin, cholesterol and triglyceride concentrations using standard laboratory assays [16].
Controls
Primary cohort
The control subjects in this study are 1361 Indo-European, non-diabetic subjects (75g oral glucose tolerance test, WHO 1999 criteria) resident in and around Pune and belonged to different cohorts being studied at the Diabetes Unit of the KEMHRC, Pune. These include parents of the children in the village based Pune Maternal Nutrition Study (PMNS) which is a study of the relationship between maternal nutrition, fetal growth and future risk of T2D [17], parents of children in the city based Pune Children Study (PCS) which is a study of the relationship between child’s birth weight and its risk for T2D [18], and the Coronary Risk of Insulin Sensitivity in Indian Subjects (CRISIS) study which is a rural-urban comparison of adiposity and risk factors for T2D and coronary artery disease [19]. All subjects have been extensively phenotyped for different biochemical-metabolic and anthropometric parameters.
Dravidian Cohort
To increase power, we also analyzed another cohort of non-diabetic subjects (n=960) of Dravidian origin from Mysore in South India. These are the parents of children in the Parthenon Study, which studied the relationship between maternal glucose tolerance, fetal growth and future risk of T2D [20]. These subjects are also extensively phenotyped for biochemical and anthropometric parameters.
Genotyping and replication analysis
All the samples were genotyped at Genome Research Group, Centre for Cellular and Molecular Biology, Hyderabad. Using the online software (http://www.primer3.org), we designed primers flanking the 2 SNPs rs9939609 and rs7191344 in the FTO gene for sequence analysis as well as for tetra primer analysis. The purified PCR products were sequenced using ABI3730 Genetic Analyzer and the sequences were analyzed using Autoassembler software after aligning the sequences with the reference sequence from NCBI (NT_010498.15). Genotypes for about ~10% samples (n=516) were validated by re-genotyping them using other genotyping platforms, the tetra primer and the Taqman methods.
Statistical analysis
We calculated the allele and genotype frequency for both variants in patients and control groups and analyzed deviation from Hardy Weinberg Equilibrium using a contingency table of observed and expected genotype frequencies by Markov simulation based on goodness of fit test [21]. The distributions of body mass index (BMI), waist circumference, triglycerides, insulin concentration, insulin resistance, and 120-min plasma glucose concentrations were skewed. These values were log10 transformed to satisfy assumptions of normality. Homeostasis model assessment of insulin resistance (HOMA-R) was calculated using the formula ‘fasting plasma glucose’ × ‘fasting plasma insulin/22.5’. We used logistic regression to analyze the association between type 2 diabetes and FTO genotype (coded as 0, 1 or 2 risk alleles) in patients and non-diabetic controls of Indo-European origin. To test whether this association was mediated through BMI or other anthropometric measures, we included each measure as a covariate in the logistic regression calculation. We used linear regression to analyze the association between FTO genotype and BMI or other anthropometric measures. These analyses were stratified by ethnicity and type 2 diabetes case/control status (Indo-European cases, Indo-European controls and Dravidian controls). We used meta-analysis to combine the linear regression coefficients from within the three cohorts. The distributions of the anthropometric measurements differed among the three cohorts and hence, study- and gender-specific Z-scores were used in these analyses. Meta-analysis statistics and plots were generated using the inverse variance method, assuming fixed effects, as implemented in the METAN module, written for Stata [22]. All statistical analyses were carried out using Stata (version 9; Stata Corporation, College Station, TX, USA) and inter-study heterogeneity was estimated using Cochran’s Q-test and the I2 statistic [23]. Power calculations were performed using Quanto v.1. 2. [24].
The Indo-European case control study had >92% power to detect the association between FTO genotype and type 2 diabetes observed in the European populations (OR = 1.27, ref Frayling et al; log-additive model) at α = 0.01, given the minor allele frequency in Indians (0.30). Although the Indo-European case (n=1448) and control (n=1355) and Dravidian (n=960) studies individually had limited power to detect the 0.1SD per allele association with BMI seen in Europeans at α = 0.01 (power = 46%, 43% and 29%, respectively), the meta-analysis (n=3763) gave >85% power.
Results
We compared the distribution of genotypes in 1453 T2D patients of Indo-European ethnicity and 1361 geographically and ethnically matched non-diabetic controls [15] (table 1). The minor allele A at rs9939609 in the FTO gene had a frequency of 0.30 in controls and genotypes were in Hardy Weinberg equilibrium (HWE). The A allele was strongly associated with T2D (OR per allele =1.26 [95% CI, 1.13-1.40], P=3×10-5) (Table 2), but not with BMI either in patients (P=0.29) or in controls (P=0.82) and not with other anthropometric parameters (Table 3). The association of FTO genotype with T2D remained significant after adjusting for BMI [OR per allele for T2D=1.21 (95% CI, 1.06-1.37); P=4×10-3], for waist circumference [OR per allele =1.20, 95% CI, 1.04-1.37; P=0.01)], for hip circumference [OR per allele =1.24, 95% CI, 1.10-1.40; P= 6×10-4] or for WHR (OR per allele =1.22, 95% CI, 1.08-1.39; P=2×10-3]. Similar results were obtained on analysis of 960 non-diabetic controls from the Parthenon study [20] (Table 1). This suggests that the increased risk of developing T2D in those with FTO variants is not entirely mediated through its effect on BMI or central obesity in our subjects. Another SNP rs7193144 {highly correlated with rs9939609 (r2=1.0) in the Europeans [1, 2], and in our study (r2=0.99)} was also in HWE and showed similar association with T2D [OR=1.31, (1.12-1.54), P=8.2×10-4] (Table 2) but was not associated with BMI or other anthropometric parameters (data not presented).
Table 1.
Basic characteristics of patients and control groups
| Patients |
Controls |
||
|---|---|---|---|
| Pune | Parthenon | ||
| N | 1453 | 1361 | 961 |
| Sex: Male (%) | 818 (56.3) | 729 (53.6) | 446 (46.4) |
| Female (%) | 635 (43.7) | 632 (46.4) | 515 (53.6) |
| Present age (years) | 46.6 (9.3) | 34.5 (6.1) | 32.4 (5.9) |
| Age at diagnosis (years) | 37.0 (16.4) | -- | -- |
| Ethnicity | Indo-European | Indo-European | Dravidian |
| Systolic Blood pressure (mm Hg) | 128 (16) | 116 (14) | 112 (13) |
| Diastolic blood pressure (mm Hg) | 80 (9) | 69 (10) | 69 (10) |
| Height (cm) | 160.9 (9.1) | 159.7 (8.5) | 160.4 (8.7) |
| Weight (kg) | 67.8 (12.0) | 55.0 (11.9) | 61.4 (12.5) |
| Waist circumference (cm) | 94.1 (10.6) | 77.0 (12.4) | 84.0 (11.4) |
| Hip circumference (cm) | 99.9 (9.3) | 91.3 (8.4) | 92.7 (8.1) |
| BMI (kg/m2) | |||
| Male | 25.4 (4.0) | 21.9 (3.6) | 23.9 (3.6) |
| Female | 27.2 (4.3) | 20.9 (4.1) | 23.6 (4.5) |
| Waist (cm) | |||
| Male | 95.4 (10.7) | 83.2 (10.5) | 86.3 (10.3) |
| Female | 92.4 (10.3) | 69.9 (10.4) | 82.1 (11.9) |
| Hip (cm) | |||
| Male | 97.3 (7.4) | 91.6 (7.5) | 92.8 (7.4) |
| Female | 103.2 (10.2) | 90.9 (9.3) | 92.5 (8.7) |
| WHR | |||
| Male | 0.98 (0.06) | 0.91 (0.06) | 0.93 (0.06) |
| Female | 0.89 (0.06) | 0.77 (0.06) | 0.88 (0.07) |
| Fat mass percent by DEXA | -- | -- | |
| Male | -- | 17.7 (8.4) | -- |
| Female | -- | 27.3 (8.7) | |
| FPG (mmol/l) | 8.50 (6.89-11.28) | 5.06 (4.61-5.56) | 5.49 (5.11-5.88) |
| 2-h PG (mmol/l) | 13.17 (10.72-16.39) | 5.50 (4.56-6.72) | 5.94 (5.09-6.74) |
| FPI (pmol/l) | -- | 32.8 (20.6-48.7) | 44.8 (30.1-69.1) |
| 2-h PI (pmol/l) | -- | 173.3 (97.9-319.9) | 246.9 (160.6-378.3) |
| TC (mmol/l) | 4.19 (0.98) | 3.91 (0.86) | 4.30 (0.91) |
| TG (mmol/l) | 1.66 (1.12) | 1.09 (0.81) | 1.57 (1.17) |
| HDLC (mmol/l) | 1.06 (0.25) | 1.07 (0.32) | 1.06 (0.22) |
| HOMA-R | -- | 1.85 (1.89) | 2.17 (1.54) |
n, number of individuals; BMI, body mass index; WHR, waist-hip ratio; DEXA, dual energy x-ray absorptiometry; FPG, fasting plasma glucose 2-h PG, 2-hour plasma glucose; FPI, fasting plasma insulin; 2-h PI, 2-hour plasma insulin; TC, plasma total cholesterol; TG, plasma triglycerides HDLC, plasma HDL cholesterol; HOMA-R, Homeostasis model assessment of insulin resistance
Values are mean (SD) for all except for FPG, 2-h PG, FPI and 2-h PI, which are median (Inter Quartile Range)
Table 2.
Allelic and genotypic frequencies and estimates of relative risks for the FTO variants in type 2 diabetes patients & control subjects (Indo-Europeans)
| SNP | Position (NCBI 36.1)a |
Allele | Patients (n=1453) |
Controls (n=1361) |
Genotype | Patients (n=1453) |
Controls (n=1361) |
Allele OR (95% CI) |
P |
|---|---|---|---|---|---|---|---|---|---|
| T | 0.646 | 0.700 | TT | 633 (43.6) | 678 (49.8) | ||||
| rs9939609 | 52378028 | TA | 613 (42.2) | 550 (40.4) | |||||
| A | 0.354 | 0.300 | AA | 207 (14.2) | 133 (9.8) | 1.26 (1.13-1.40) | 3.0×10-5 | ||
| T | 0.646 | 0.704 | TT | 559 (43.6) | 243 (49.8) | ||||
| rs7193144* | 52368187 | TC | 538 (41.9) | 202 (41.4) | |||||
| C | 0.354 | 0.296 | CC | 186 (14.5) | 43 (8.8) | 1.31 (1.12-1.54) | 8.2×10-4 |
SNP, single nucleotide polymorphism; n, number of individuals; figures in parentheses indicate percentage, OR, odds ratio; P, p-value; 95% CI; 95% confidence interval
National Centre for Biotechnology Information, Build 35.1
on analysis of 1283 patients and 488 controls
Table 3.
Correlation of FTO rs9939609 genotype with measures of obesity (body size characteristics) in patients and non-diabetic control subjects
| Patients |
Controls |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Indo- Europeans |
Indo- Europeans |
Dravidians |
|||||||||||||
| Genotype |
p | p1 | Genotype |
p | p1 | Genotype |
p | p1 | |||||||
| TT | TA | AA | TT | TA | AA | TT | TA | AA | |||||||
| N | 633 | 613 | 207 | 678 | 550 | 133 | 435 | 419 | 107 | ||||||
| Weight (kg) | 67.6 (11.9) | 67.8 (12.5) | 68.8 (11.1) | 0.26 | 0.38 | 55.2 (11.7) | 54.7 (12.2) | 55.6 (11.9) | 0.88 | 0.98 | 60.2 (12.0) | 62.4 (12.9) | 61.8 (12.0) | 0.04 | 0.05 |
| Height (cm) | 160.9 (9.2) | 160.9 (9.1) | 161.1 (8.6) | 0.92 | 0.84 | 160.1 (8.6) | 159.4 (8.5) | 159.4 (8.6) | 0.17 | 0.49 | 160.1 (8.5) | 160.8 (9.1) | 160.6 (7.9) | 0.33 | 0.61 |
| BMI (kg/m2) | 26.1 (4.0) | 26.2 (4.6) | 26.5 (3.8) | 0.20 | 0.29 | 21.5 (3.8) | 21.4 (3.9) | 21.8 (4.0) | 0.64 | 0.82 | 23.4 (3.9) | 24.0 (4.3) | 23.9 (3.9) | 0.07 | 0.09 |
| Waist circumference (cm) |
93.9 (10.6) | 94.1 (10.9) | 95.2 (9.6) | 0.11 | 0.27 | 77.2 (12.0) | 76.7 (12.7) | 77.7 (12.8) | 0.94 | 0.95 | 83.4 (11.4) | 84.7 (11.6) | 84.1 (10.3) | 0.21 | 0.25 |
| Hip circumference (cm) |
99.7 (8.9) | 99.9 (9.6) | 100.4 (9.2) | 0.30 | 0.56 | 91.4 (8.1) | 91.0 (8.6) | 91.8 (9.0) | 0.99 | 0.89 | 91.9 (7.8) | 93.4 (8.5) | 92.9 (7.5) | 0.03 | 0.02 |
| WHR | 0.94 (0.07) | 0.94 (0.07) | 0.95 (0.08) | 0.17 | 0.23 | 0.84 (0.09) | 0.84 (0.09) | 0.84 (0.09) | 0.88 | 0.88 | 0.90 (0.07) | 0.91 (0.07) | 0.90 (0.07) | 0.95 | 0.98 |
| Fat mass % by DEXA | -- | -- | -- | -- | -- | 22.3 (9.7) | 22.1 (9.9) | 23.4 (10.0) | 0.58 | 0.59 | -- | -- | -- | -- | -- |
n, number of individuals; BMI, body mass index; WHR, waist-hip ratio; DEXA, dual energy x-ray absorptiometry; Values are mean (SD); p by ANOVA; p1, adjusted for age and gender
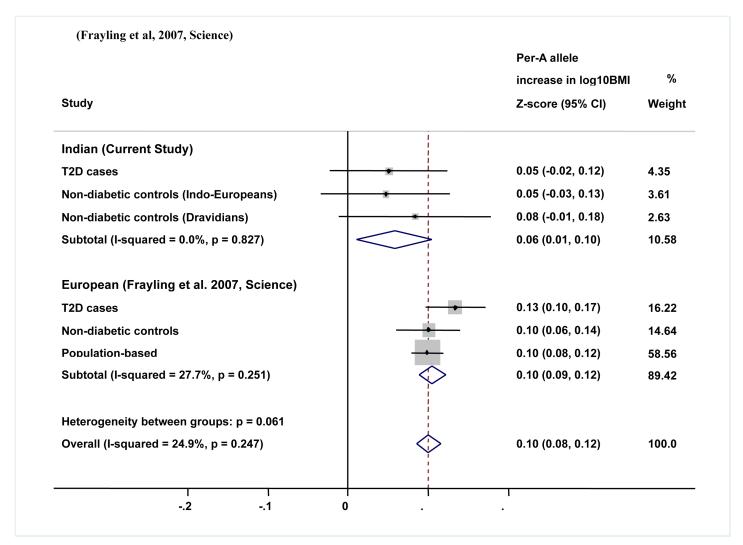
We performed a meta-analysis combining Indo-European cases, Indo-European controls and Dravidian controls (n= 3763) to assess the impact of FTO variants on BMI. There was a borderline association between FTO genotype and BMI (per allele SD change in log10BMI: 0.06 (95%CI, 0.01-0.10); P=0.017; I2=0%; Fig 1), waist circumference (per allele SD change in log10WC: 0.05 (95%CI, 0.01-0.10); P=0.030; I2=0%) and hip circumference (per allele SD change in hip circumference: 0.05 (95%CI, 0.01-0.10); P=0.027; I2=0%), but not waist-hip ratio (P = 0.42).(table 3).
Figure 1.
Meta-analysis plot for FTO variant rs9939609 and BMI association for the present study and comparison with the European Study
The FTO genotype at rs9939609 was not associated with traits associated with obesity including glycemia, insulin concentrations, insulin resistance (HOMA-R) or lipids in Indo-Europeans but was associated with fasting plasma insulin levels in Dravidians (Suppl table 1).
Discussion
Our results demonstrate for the first time in Asian Indians, that variants in the FTO gene predispose to type 2 diabetes, but unlike in Europeans does not appear to do this entirely through its influence on BMI, central obesity and adiposity [1, 2]. The strength of association of FTO variants with T2D was similar to that in Europeans, but the association with BMI and other body size parameters appears weaker (1, 3). Further studies in South Asians are needed to confirm this. The finding that the FTO variants are associated with T2D argues against a difference in linkage disequilibrium with a putative functional variant in these two populations, and our results suggest that there could be a fundamental difference in the way FTO gene works to influence risk of type 2 diabetes in Indians and in Europeans. Although population stratification may be a possible explanation that may confound the results, recent evidence suggests that despite the geographic and linguistic diversity, Indians as a whole display a low level of genetic heterogeneity [25, 26]. Two recent reports in Chinese and Japanese populations did not find an association of FTO variants with type 2 diabetes, and only a weak association with BMI in Japanese [8,9]. Another study reported that FTO variants might influence the risk of severe obesity in the Japanese [10], while a recent study also found borderline evidence that the FTO association with BMI was weaker in Asians as compared to the European studies, but the association with T2D was similar [11]. The frequency of the A allele at rs9939609 variant was lower in Chinese (0.12) and Japanese (0.18) compared to that in Europeans (0.45) and Indians (0.30), meaning studies in East Asians have less power [8-10]. It is well known that Indians (and Asians in general) have a lower BMI than the Europeans, and have a steeper relationship between obesity measures and risk of T2D [27], being more susceptible at a lower BMI. This has been ascribed to relatively higher central obesity (WHR) and adiposity (body fat percent) in Asians for a given BMI compared to that in the Europeans [17]. It is possible that there is an ethnic difference in the role, the FTO variants play in the pathogenesis of obesity and T2D.
The functional role of FTO gene is not yet clear, nor is it clear how the variants affect body size and predict the risk of T2D. It has recently been shown that physically inactive homozygous A-allele carriers have a relatively larger increase in BMI compared with that in non-carriers and those heterozygous for A-allele suggesting that low physical activity may accentuate the effect of FTO rs9939609 on body fat accumulation [6]. However, another study demonstrated lack of association of FTO variants with energy expenditure or physical activity [28]. The gene is expressed in different tissues, notably in hypothalamus, liver, muscle, adipose tissue and pancreatic B-cells [29]. Its expression has been shown to be higher in subcutaneous compared to visceral adipose tissue, though expression in only the latter is associated with BMI (but inversely) [30]. However, a recent study showed no association between the expression levels and the FTO genotype at rs9939609 [31]. In another study, individuals homozygous for the protective allele showed increased adipocyte lipolytic activity, both in vivo and in vitro, suggesting that they might be protected against fat deposition [31]. Based on sequence homology, FTO gene is predicted to code for a 2-oxoglutarate dependent demethylase enzyme [32] which influences nucleic acid demethylation and thus, may be important in epigenetic regulation. It is intriguing that we have demonstrated an association between intrauterine vitamin B12 and folate nutrition (the two major methyl donor vitamins) and adiposity and insulin resistance in Indian children in the Pune Maternal Nutrition Study [33]. Taken together, these results suggest that 1-C (methyl) metabolism could have important influences on development of obesity, adiposity and type 2 diabetes.
In conclusion our results demonstrate that in Indians, variants in the FTO gene predispose to T2D. Our results are consistent with a smaller effect of FTO variation on BMI in Indians compared to Europeans. These results reinforce our previous suggestion that the relationship between BMI and T2D may be different in Indians. and raise the possibility that comparative studies of FTO in South Asian and European populations, especially functional genomic studies (including epigenetics) may offer a novel opportunity to understand critical mechanisms in the pathogenesis of obesity and T2D.
Supplementary Material
Acknowledgements
We thank all the patients and control subjects for agreeing to participate in the study. We gratefully acknowledge the contributions made by many colleagues involved in sample collection, phenotyping, DNA extraction, data collection and data management. The WellGen study, PMNS study, PCS study and CRISIS study were supported by Wellcome Trust, London, UK. The Parthenon study was initiated by the former MRC Epidemiology Unit (Southampton) and was funded by the British Heart Foundation, UK. We thank Dr Sanjay Sirdeshpande, and Mrs Pallavi Yajnik for their support to these studies.
List of Abbreviations
- T2D
type 2 diabetes
- FTO
Fat mass and obesity associated
- SNP
Single nucleotide polymorphism
- OR
Odds ratio
- CI
Confidence interval
- BMI
Body mass index
- WHR
Waist hip ratio
- OR
Odds ratio
- PMNS
Pune maternal nutrition study
- HWE
Hardy-Weinberg equilibrium
- PCS
Pune Children study
- CRISIS
Coronary Risk of Insulin Sensitivity in Indian Subjects
- HOMA-R
Homeostasis model assessment of insulin resistance
Footnotes
Duality of interest None declared
References
- 1.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dina C, Meyre D, Gallina S, Durand E, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:706–707. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 4.Ron Do, Swneke D. Bailey, Katia Desbiens, et al. Genetic Variants of FTO Influence Adiposity, Insulin Sensitivity, Leptin Levels and Resting Metabolic Rate in the Quebec Family Study. Diabetes. 2008;57:1147–1150. doi: 10.2337/db07-1267. [DOI] [PubMed] [Google Scholar]
- 5.Hunt SC, Stone S, Xin Y, et al. Association of the FTO Gene with BMI. Obesity (Silver Spring) 2008;16:902–904. doi: 10.1038/oby.2007.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andreasen CH, Stender-Petersen KL, Mogensen MS, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57:95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- 7.Scuteri A, Sanna S, Chen WM, et al. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Wu Y, Loos RJ, Hu FB, Liu Y, Wang J, Yu Z, Lin X. Variants in the fat mass-and obesity-associated (FTO) gene are not associated with obesity in a Chinese Han population. Diabetes. 2008;57:264–268. doi: 10.2337/db07-1130. [DOI] [PubMed] [Google Scholar]
- 9.Omori S, Tanaka Y, Takahashi A, et al. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes. 2008;57:791–795. doi: 10.2337/db07-0979. [DOI] [PubMed] [Google Scholar]
- 10.Hotta K, Nakata Y, Matsuo T, et al. Variations in the FTO gene are associated with severe obesity in the Japanese. J Hum Genet. 2008 Apr 1; doi: 10.1007/s10038-008-0283-1. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng MC, Park KS, Oh B, et al. Implication of Genetic Variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2 and FTO in Type 2 Diabetes and Obesity in 6719 Asians. Diabetes. 2008 May 9; doi: 10.2337/db07-1583. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran A, Snehalatha C, Kapur A, et al. High prevalence of diabetes and impaired glucose tolerance in India: National Urban Diabetes Survey. Diabetologia. 2001;44:1094–1101. doi: 10.1007/s001250100627. [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran A, Snehalatha C, Viswanathan V, Viswanathan M, Haffner SM. Risk of non-insulin-dependent diabetes mellitus conferred by obesity and central obesity in different ethnic groups. A comparative analysis between Asian Indians, Mexican Americans and Whites. Diabetes Res Clin Pract. 1997;36:121–125. doi: 10.1016/s0168-8227(97)00040-5. [DOI] [PubMed] [Google Scholar]
- 14.Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 15.Chandak GR, Janipalli CS, Bhaskar S, et al. Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian population. Diabetologia. 2007;50:63–67. doi: 10.1007/s00125-006-0502-2. [DOI] [PubMed] [Google Scholar]
- 16.Yajnik CS, Joglekar CV, Lubree HG, et al. Adiposity, inflammation and hyperglycaemia in rural and urban Indian men: Coronary Risk of Insulin Sensitivity in Indian Subjects (CRISIS) Study. Diabetologia. 2008;51:39–46. doi: 10.1007/s00125-007-0847-1. [DOI] [PubMed] [Google Scholar]
- 17.Rao S, Yajnik CS, Kanade A, et al. Intake of micronutrient-rich foods in rural Indian mothers is associated with the size of their babies at birth: Pune Maternal Nutrition Study. J Nutr. 2001;131:1217–1224. doi: 10.1093/jn/131.4.1217. [DOI] [PubMed] [Google Scholar]
- 18.Bavdekar A, Yajnik CS, Fall CH, et al. Insulin resistance syndrome in 8-year-old Indian children: small at birth, big at 8 years, or both? Diabetes. 1999;48:2422–2429. doi: 10.2337/diabetes.48.12.2422. [DOI] [PubMed] [Google Scholar]
- 19.Bhat DS, Yajnik CS, Sayyad MG, et al. Body fat measurement in Indian men: comparison of three methods based on a two-compartment model. Int J Obes (Lond) 2005;29:842–848. doi: 10.1038/sj.ijo.0802953. [DOI] [PubMed] [Google Scholar]
- 20.Krishnaveni GV, Hill JC, Leary SD, et al. Anthropometry, glucose tolerance, and insulin concentrations in Indian children: relationships to maternal glucose and insulin concentrations during pregnancy. Diabetes Care. 2005;28:2919–2925. doi: 10.2337/diacare.28.12.2919. [DOI] [PubMed] [Google Scholar]
- 21.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris R, Bradburn M, Deeks J, et al. Statistical Software Components S456798. Boston College Department of Economics; 2006. METAN: Stata module for fixed and random effects meta-analysis. revised 19 Feb 2007. [Google Scholar]
- 23.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Br Med J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauderman WJ, Morrison JM. QUANTO 1.1: A computer program for power and sample size calculations for genetic epidemiology studies. 2006. http://hydra.usc.edu/gxe.
- 25.Rosenberg NA, Mahajan S, Gonzalez-Quevedo C, et al. Low levels of genetic divergence across geographically and linguistically diverse populations from India. PloS Genet. 2006;2(12):e215. doi: 10.1371/journal.pgen.0020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pemberton TJ, Mehta NU, Witonsky D, et al. Prevalence of common disease-associated variants in Asian Indians. BMC Genet. 2008 Feb 4;9:13. doi: 10.1186/1471-2156-9-13. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yajnik CS. Nutrition, growth, and body size in relation to insulin resistance and type 2 diabetes. Curr Diab Rep. 2003;3:108–114. doi: 10.1007/s11892-003-0033-x. [DOI] [PubMed] [Google Scholar]
- 28.Berentzen T, Kring SII, Holst C, et al. Lack of association of fatness-related FTO gene variants with energy expenditure or physical activity. J Clin Endocrinol Metab. 2008 Apr 29; doi: 10.1210/jc.2008-0007. 2008. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Stratigopoulos G, Padilla SL, Leduc CA, et al. Regulation of Fto/Ftm gene expression in mice and humans. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1185–1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klöting N, Schleinitz D, Ruschke K, et al. Inverse relationship between obesity and FTO gene expression in visceral adipose tissue in humans. Diabetologia. 2008;51:641–647. doi: 10.1007/s00125-008-0928-9. [DOI] [PubMed] [Google Scholar]
- 31.Wåhlén K, Sjölin E, Hoffstedt J. The common rs9939609 gene variant of the fat mass- and obesity-associated gene FTO is related to fat cell lipolysis. J Lipid Res. 2008;49:607–611. doi: 10.1194/jlr.M700448-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;30:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yajnik CS, Deshpande SS, Jackson AA, et al. Vitamin B12 and folate concentrations during pregnancy and insulin resistance in the offspring: the Pune Maternal Nutrition Study. Diabetologia. 2008;51:29–38. doi: 10.1007/s00125-007-0793-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.