Abstract
The RNA-dependent RNA-polymerase (RdRp) of human parainfluenza virus type 3 (HPIV3) is a large protein (L, 2233 amino acids), and along with the phosphoprotein (P, 603 amino acids) forms a hetero-complex that transcribes the genome RNA into mRNAs in vitro and in vivo that are 5′ capped and methylated and 3′- polyadenylated. The interaction of the P protein, an obligatory cofactor, imparts the RdRp activity of the L protein, which is otherwise inactive. The precise mechanism underlying this activation process remains unknown. Several recent reports suggested that the L proteins of paramyxoviruses, when expressed alone, self-associate to form an oligomeric structure. The presumptive oligomerization domain lies in the N-terminal part of the L protein (for HPIV3, 889 amino acids). Here, we demonstrate that a series of N-terminally deleted L proteins as well as several truncated proteins that span different regions of the L protein can also efficiently co-immunoprecipitate the full length L protein. In addition, by several biochemical parameters, the L-L interaction was shown to form aggregates rather than oligomers. In contrast, when the P protein was co-expressed with the L protein, the former bound to a domain spanning the N-terminal 1060 amino acids of the latter, which prevented L-L self-association, resulting in the formation of structurally competent and functionally active RdRp.
Keywords: Self-interaction of HPIV3 L protein, P-L interaction, prevention of L-L interaction
Introduction
The virion-associated RNA-dependent RNA-polymerase (RdRp), L protein, of human parainfluenza virus type 3 (HPIV3), like other members of non-segmented-negative sense-RNA (NNS) viruses, is a large (2233 amino acids) multifunctional protein (Banerjee, 1987; Banerjee et al, 1991; Karron and Collins, 2007; Lamb and Parks, 2007). It interacts with the phosphoprotein, P (603 amino acids) and carries out several enzymatic activities, both in vitro and in vivo, such as mRNA synthesis, 5′-capping and methylation and 3′-polyadenylation (Banerjee, 1987; Banerjee et al, 1991; Karron and Collins, 2007; Lamb and Parks, 2007). Alignment of amino acid sequences of L proteins derived from different members of the NNS virus family shows six highly conserved regions that are joined by variable length of spacer regions with relatively low sequence homology (Müller et al., 1994; Poch et al., 1990; Sidhu et al., 1993). Each conserved region is predicted to perform specific biochemical functions related to RNA synthesis. So far, conserved region III (the invariant GDNQ sequence) has been unequivocally assigned to be the polymerase active site (Sleat and Banerjee, 1993; Malur et al., 2002a). The conserved region VI (GXGXG motif) is linked to cap methylation activities (Grdzelishvili et al., 2005; Li et al., 2005; Li et al., 2006; Ogino et al., 2005) and recently, certain amino acids residues within the conserved region V were shown to be involved in mRNA capping (Li et al., 2008; Ogino and Banerjee, unpublished data). The precise functions of the other conserved modules remain unknown at present although several mutational analyses within blocks I, II and IV have been carried out for SeV, which gave rise to a spectrum of variable polymerase activity ranging from partial to complete inactivation of transcription and/or replication (Chandrika et al., 1995; Feller et al., 2000; Smallwood et al., 1999; Smallwood et al., 2002b).
The obligatory component of HPIV3 RdRp heterocomplex, the P protein, possibly a tetramer [in analogy to Sendai virus P (Tarbouriech et al., 2000)] interacts with the L protein to form a stable and functional RdRp holoenzyme (Lamb and Parks, 2007). Unlike the L protein of VSV, the paramyxoviral L proteins appear to be unstable and prone to degradation when expressed in cells, unless the cognate P protein is co-expressed (Horikami et al., 1992; Smallwood et al., 1994). The obligate association of P with L protein is further underscored by the fact that L protein is not found free in infected cells; it remains associated with the P protein. Therefore, the possibility exists that the P protein may act as a chaperone, required for the proper folding of L protein which provides stability to the RdRp complex. A similar chaperone-like function of P protein for the nucleocapsid (N) protein has previously been demonstrated; the N and P proteins form a soluble N0-P complex that prevents N protein from aggregation and non-specific binding to cellular RNAs (Masters and Banerjee, 1988a; Masters and Banerjee, 1988b, Majumder et al., 2001; Majumder et al., 2004). This complex presumably initiates encapsidation of the nascent viral RNA chains during replication of genome RNA (Majumder et al., 2001; Majumder et al., 2004; Masters and Banerjee, 1988a; Masters and Banerjee, 1988b; Chen et al., 2007; Davis et al., 1986). Also, for all NNS viruses, the biological function of P depends on oligomerization (Ding et al., 2006; Tarbouriech et al., 2000; Choudhary et al., 2002; Chen et al., 2006), deletion of which renders P protein incapable of supporting transcription (Choudhary et al., 2002; Chen et al., 2006).
It remains unclear, however, whether the L protein carries out its function as a monomer or needs to oligomerize to complex with P to be functionally active. Recently, it was suggested that in several paramyxoviruses including Sendai, HPIV3 and measles, L protein forms an oligomeric structure although the precise degree of oligomerization was not established. The presumptive oligomerization domain resides at the N-terminal, 174 amino acids, 889 amino acids* (see footnote) and 408 amino acids, respectively, for Sendai, HPIV3 and measles virus (Cevik et al., 2003; Cevik et al., 2004; Cevik et al., 2007; Smallwood and Moyer, 2004). Consistent with the observed putative oligomerization, the L protein of SeV appears to function in a modular fashion such that two inactive L mutants, when co-expressed (in the presence of P protein), restore viral RNA synthesis in vitro by intragenic complementation, albeit highly inefficiently (Smallwood et al, 2002a). The L protein of lymphocytic choriomeningitis virus (LCM) a member of the arenavirus family was also suggested to form such putatative oligomers (Sánchez and de la Torre, 2005).
Based on the fact that L protein does not remain free in the infected cells (remains bound to the P protein), and coupled with the above findings of putative oligomerization of L protein when expressed in cells, it was of interest to study the nature of such L-L complex formation and locate, if possible, the oligomerization domain, within the L protein. To address these questions we used a series of L protein mutants to study their interactions with the wild type L protein (Lwt). We were unable to locate any specific oligomerization domain within the L protein, since every deleted or truncated L protein mutants strongly interacted with the Lwt. However, when P protein was co-expressed with the L protein, L-L interaction did not occur, only (L-P) complex was formed, thus, underscoring the point that L-L interaction, in the absence of P protein, leads to possible aggregation of the L protein.
Results
L-L interaction as studied by co-immunoprecipitation
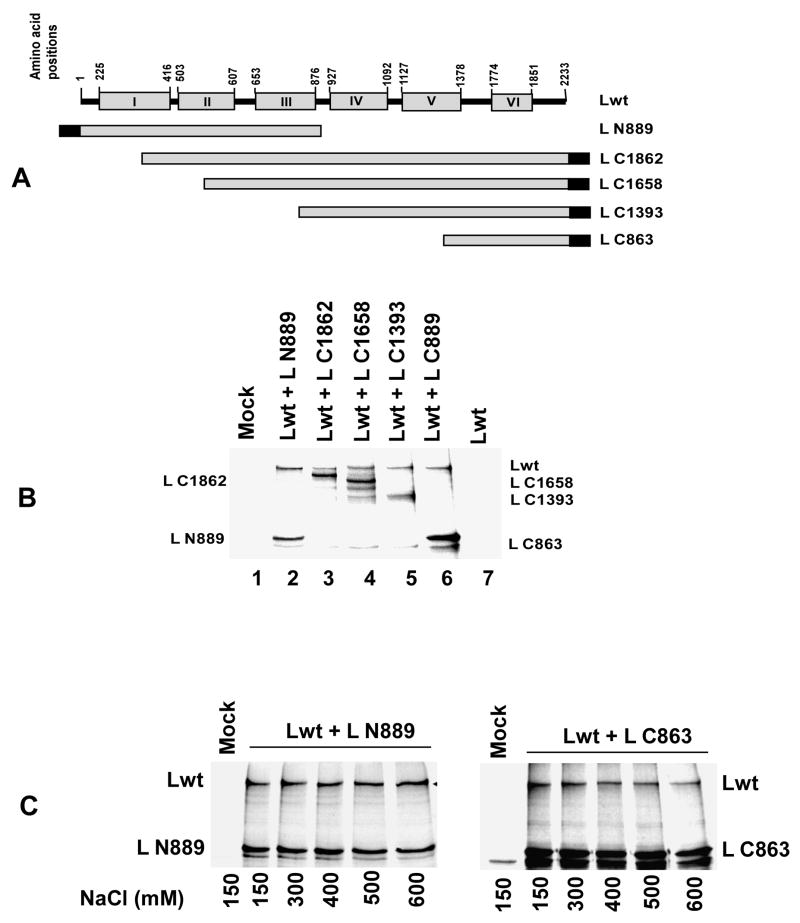
Earlier reports suggested that L proteins of paramyxovirus may form oligomers and that the oligomerization domain resides within the N-terminal region (Cevik et al., 2003; Cevik et al., 2004; Cevik et al., 2007; Smallwood and Moyer, 2004); for HPIV3, the domain resides within the N-terminal 889 amino acids (Smallwood and Moyer, 2004). These studies were carried out primarily by co-expression of Lwt with only the C-terminally truncated L proteins followed by immunoprecipitation. To determine the precise boundary of the oligomerization domain within the L protein, we co-expressed, this time, a series of N-terminally deleted L proteins (Fig. 1A), tagged with FLAG epitope at their C-termini, with non-tagged Lwt followed by immunoprecipitation with FLAG-antibody as described in Materials and Methods. As a positive control, we used (C-terminally deleted) N-terminally FLAG-tagged L N889 as previously used by Smallwood and Moyer (2004). Surprisingly, all of the N-terminally truncated L proteins interacted with Lwt (Fig 1B). These results were unexpected since L C863 and L N889 (the former does not overlap with the latter) interacted with almost equal efficiency with Lwt, suggesting that there may not be a precise oligomerization domain within the L protein. Both L- fragments (L N889 and L C863) formed highly stable complexes with Lwt since immunoprecipitates could be washed with NaCl concentration as high as 600 mM (Fig. 1C). These results strongly suggested that the L-L self-association is mediated possibly via the entire length by hydrophobic interactions.
Fig. 1.
(A) Schematic representation (not to the scale) of full length HPIV3 L protein (2233 amino acids) and deletion mutants. The boxes indicate the conserved regions I-VI. The conserved regions were identified by aligning HPIV3 and SeV L proteins and by comparison with other L proteins (Poch et al., 1990). A series of N-terminally deleted L proteins, which are FLAG-tagged at their C-terminus, were co-expressed with non-tagged full length L (Lwt) in HeLa cells as described in Materials and Methods. L C1862, L C1658, L C 1393 and L C863 carry C-terminal 1862, 1658, 1393 and 863 amino acids respectively. N-terminally FLAG epitope tagged L N889 was also co-expressed with Lwt as positive control (Smallwood et al., 2004). (B) Analysis of L-L interaction by co-immunoprecipitation assay. Proteins expressed in HeLa cells were labeled with 35S and were immunoprecipitated with anti-FLAG antibody and analyzed by SDS-PAGE followed by autoradiography as described in text. The migration positions of full length L and L deletion mutant proteins are indicated. All truncated L proteins (including L C863, which lacks 1370 amino acids from the N-terminus) interacted with Lwt. Lwt, without any FLAG tagged truncated L mutant was used as negative control (lane 7) (C) Analysis of L-L interaction by co-immunoprecipitation assay using different salt concentrations. Lwt and L N889 or L C863 were co-expressed in HeLa cells, labeled with 35S-methionine and immunoprecipitated using anti-FLAG antibody. Co-precipitated proteins were washed with graded NaCl concentrations. Both L N889 and L C863 interacted with Lwt up to 600 mM NaCl.
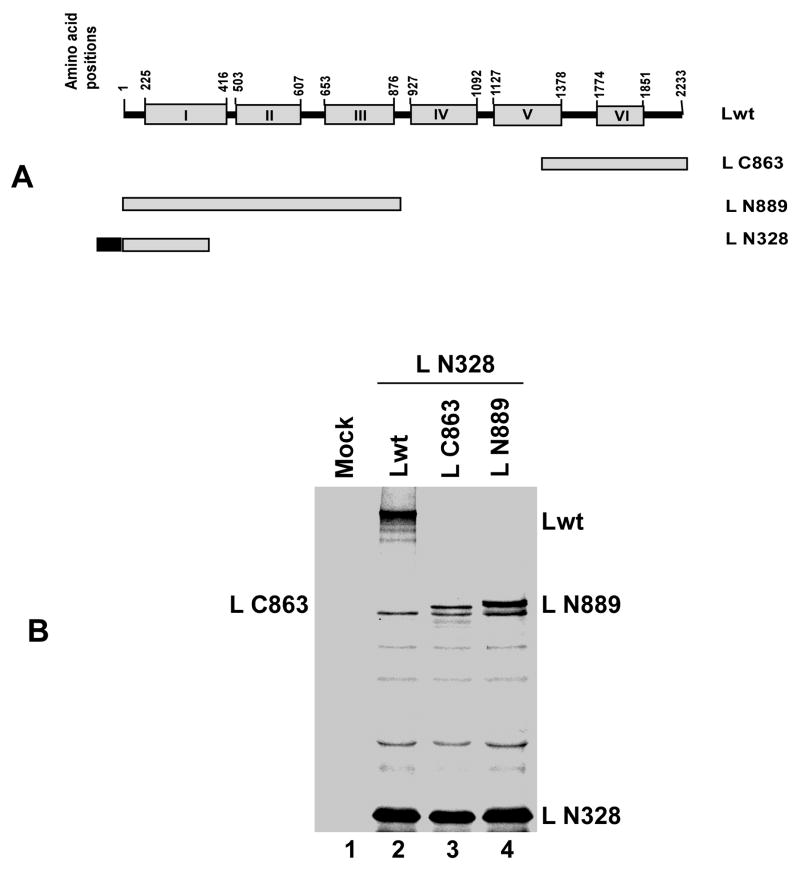
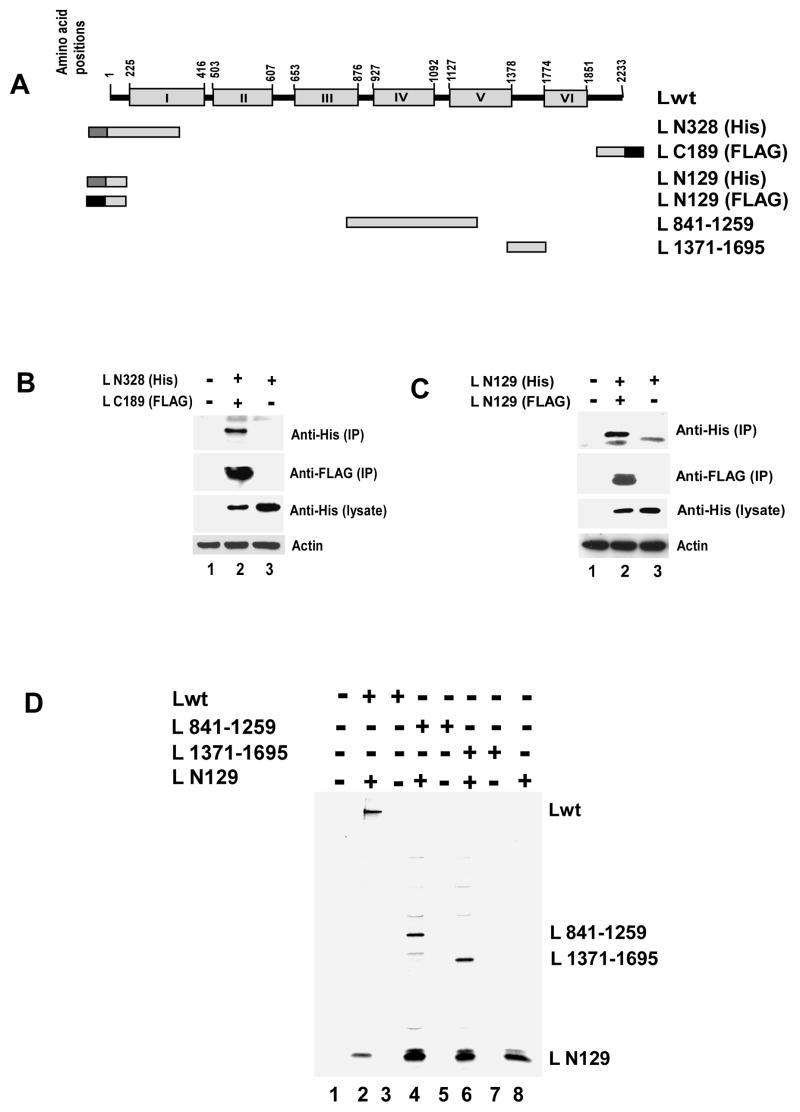
Next, we shortened the N-terminal portion to 328 amino acids (L N328). FLAG tagged L N328 was co-expressed with Lwt or L N889 or L C863 (Fig 2A) and immunoprecipitated with FLAG antibody. As shown in Fig 2B, Lwt, L N889 and L C863 were efficiently co-immunoprecipated with L N328 indicating that the latter shorter N-terminal fragment can also effectively interact with the Lwt. Furthermore, even a shorter fragment from the C-terminal side, L C189, could also effectively interact with L N328, His tagged at the N-terminus (Fig. 3A) as shown by immunoprecipitation with FLAG antibody followed by Western blot analysis by His antibody (Fig. 3B). Additionally, as shown in Fig. 3C, with similar experiments involving co-expression and immunoprecipitation of the differentially tagged N-terminal 129 amino acids of the L protein (L N129), both fragments interacted with each other. Finally, when L N129 and L fragments expressing 419 amino acids (L 841-1259) or 325 amino acids (L 1371-1695) in the mid region of the protein (Fig. 3A) were co expressed, L N129 interacted strongly with L 841-1259 and L 1371-1695 (Fig. 3D), suggesting that the observed L-L associations are mediated not via a defined oligomerization domain but via a non-specific interaction leading possibly to aggregation.
Fig. 2.
(A) Schematic representation (not to the scale) of Lwt (2233 amino acids) and deletion mutants. N-terrminally FLAG epitope tagged N-terminal 328 amino acids (L N328) were expressed together with Lwt or L C863 or L N889. (B) Analysis of L-L interaction by co-immunoprecipitation assay. 35S-labeled proteins were immunoprecipitated with anti-FLAG antibody and analyzed by SDS-PAGE followed by autoradiography as described in text. The migration positions of full length L and L deletion mutant proteins are indicated. L N328 interacted with Lwt, L C863 and L N889.
Fig 3.
(A) Schematic representation (not to the scale) of Lwt and deletion mutants. (B) Analysis of L-L interaction by co-immunoprecipitation assay. C-terrminally FLAG-tagged C-terminal 189 amino acids (L C189) were expressed together with N-terminally His tagged N-terminal 328 amino acids (L N328). Cells were lysed and immunoprecipitated using anti-FLAG antibody coated beads. Western blot analyses were carried out using anti-His or anti-FLAG monoclonal antibodies for immunoprecipitated (IP) as well as total proteins (lysate). L N328 was co-immunoprecipitated with L C189 (lane 2 in upper and second panel from top). Similar level of expression of L N328 was confirmed in lysates (lane 2 and 3 in third panel from top) and anti-β actin antibody was used to verify the loading control (lower panel) (C) N- terminal 129 amino acids (L N129) were differentially tagged (His or FLAG) and co-expressed in HeLa cells. Lysates were prepared and immunoprecipitation was carried out using anti-FLAG antibody coated beads. Western blot analysis with anti-His and anti-FLAG antibody showed that L N129 self interacts (lane 2, top panel and second panel from top). His tagged L N129 was expressed at similar level in lysates (lane 2 and 3 in third panel from top) and synthesis of actin was used as loading control (lower panel) (D) FLAG-tagged L N129 was co-expressed with L 841-1259 or L 1371-1695. Proteins were labeled with 35S as described in text. Analysis of L-L interaction by co-immunoprecipitation assay showed L N129 efficiently interacts with both the L fragments.
L-L interaction leads to aggregation
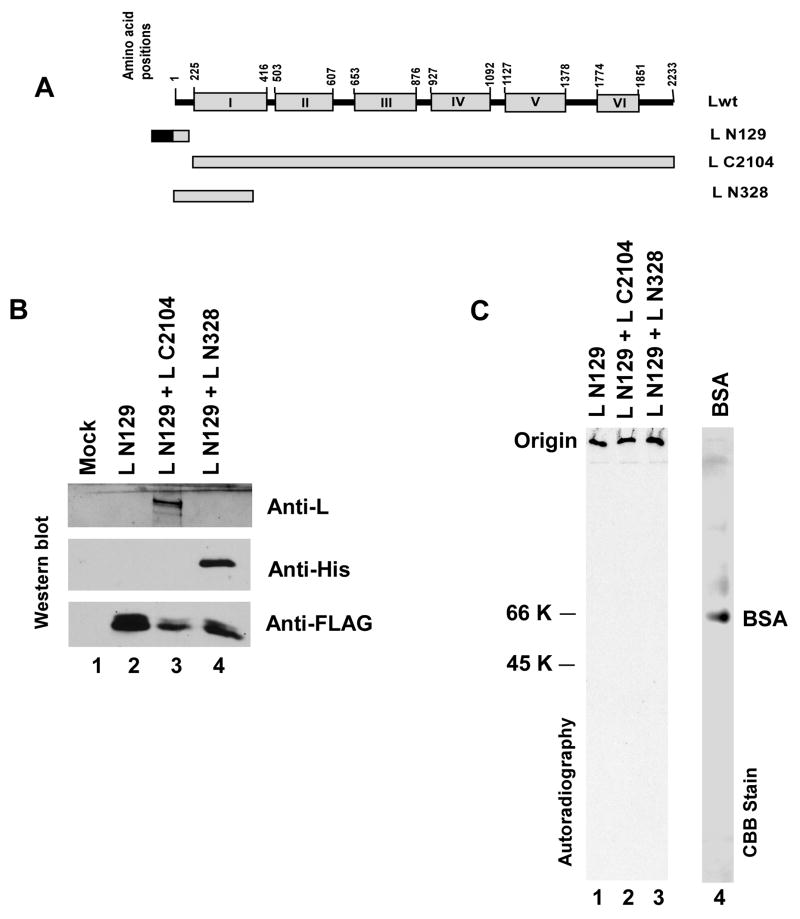
To gain a better insight into the mode of L-L interaction, FLAG-tagged L N129, was expressed either singly or together with non-tagged L C2104 (130–2233 amino acids) or L N328 (Fig. 4A). The expressed proteins were labeled with 35S and purified by affinity chromatography as described in Materials and Methods. The presence of L N129 in the eluent and co-purification of L C2104 or L N328 with L N129 were confirmed by SDS-PAGE followed by Western blot analysis against anti-L (for the detection of L C2104), anti-His (for the detection of L N328) and anti-FLAG (for the detection of L N129) antibodies (Fig 4B). Moreover, when membrane contains immobilized protein samples was stained with ponceau S no antibody bands were detected (data not shown). Therefore, the protein samples were free from antibody coated beads. An aliquot of the protein samples were then run in PAGE (10 % resolving and 4.5% stacking) under native condition. Serum albumin was used as control. As shown in Fig 4C none of the 35S labeled protein samples entered the gel but remained at the origin instead. On the other hand, albumin entered the gel and migrated according to its molecular weight (66 kDa). Similar results were also observed when both 15% and 6 % native PAGE were used (data not shown). These results strongly support the contention that L N129 interacted with itself (when singly expressed) as well as with L C2104 or L N328 in a non-specific manner and formed an apparently high molecular weight mass by aggregation, which failed to enter the gel, thus, could not be resolved in native PAGE.
Fig 4.
(A) Schematic representation (not to the scale) of Lwt and deletion mutants. N-terminally FLAG-tagged L N129 was expressed together with L deletion mutants, L C2104 (N-terrminally 130 amino acid deleted) or His-tagged L N328 (contains 1–328 amino acids). (B) Eluted proteins were run on SDS-PAGE and Western blot analyses were performed using anti-L, anti-His and anti-FLAG antibodies for the detection of L C2104, L N328 and L N129, respectively. (C) Purified 35S labeled protein samples were run on 10 % native PAGE. No 35S labeled protein samples (lane 1–3) has entered in gel and remained at the origin. BSA (lane 4), as detected by CBB stain, migrated according to its expected size (66 kDa).
Role of P protein in L-L self-association
It is important to underscore the point that the above studies were carried out by the expression of Lwt and its mutants in the absence of P protein. Since the L protein does not remain free in the infected cells under physiological condition, rather complexed with the P protein, it was important to study the fate of the observed L-L self-association in the presence of P protein. Before we initiated this study, first we needed to precisely locate the P protein binding site within the L protein to compare Lwt and L fragment association in the presence of P protein that binds or does not bind to the specific L fragments.
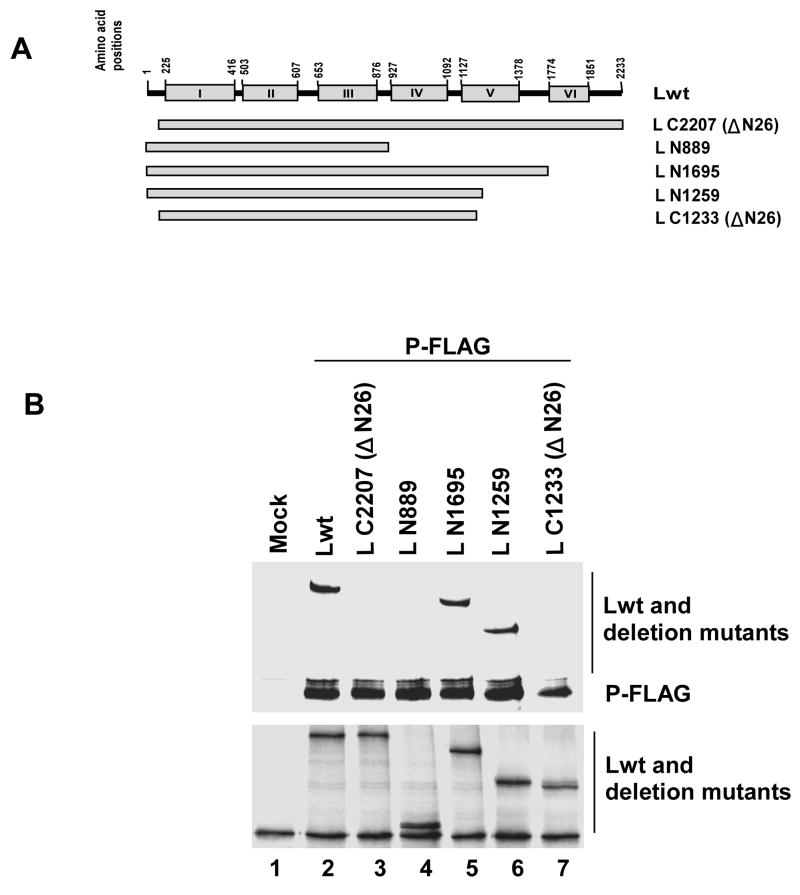
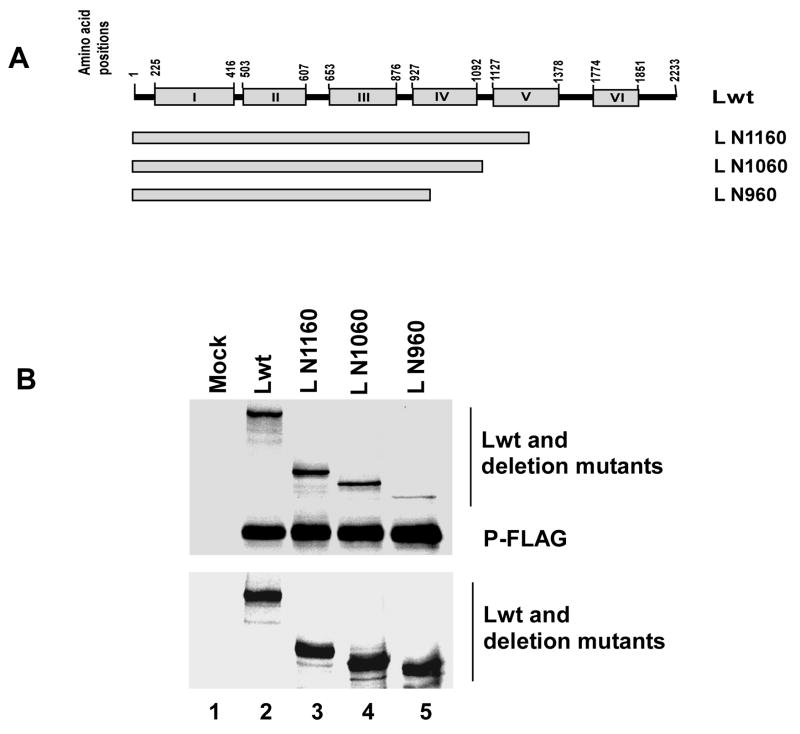
We previously mapped the P protein binding domain to within the highly conserved N-terminal region (26 amino acids) of HPIV3 L protein (Malur et al., 2002b) and by mutational analysis we demonstrated that this region is essential for transcription. A recent report, however, described that a C-terminally truncated L protein, L N889, containing the N-terminal 26 amino acids (Smallwood and Moyer, 2004) failed to interact with the P protein, suggesting that for binding to P a larger fragment of N-terminal L may be necessary. To identify the minimum length of L protein required for L-P interaction we made a series of N- and C- terminally truncated L proteins— L C2207 (NΔ26), L N889, L N1695, L N1259 and L N1233 (ΔN26) as shown in Fig. 5A. The Lwt or the mutant L proteins were co-expressed with C-terminally FLAG-tagged P protein. Cells were labeled with 35S and immunoprecipitated with anti-FLAG antibody as described in Materials and Methods. Both N- and C-terminally truncated L proteins, L C2207 (NΔ26) and L N889 (Fig 5B, lane 3 and 4, upper panel), as expected, failed to co-precipitate with P-FLAG protein, confirming the previous results (Malur et al., 2002b; Smallwood and Moyer, 2004). However, the two C-terminally truncated L proteins (L N1259 and L N1695), which increased the length of L N889 by 370 and 806 amino acids, respectively, were effectively co-precipitated with the P protein (lane 6 and 5, upper panel). Also, in control lane, Lwt (lane 2, upper panel), was co-precipitated with the P protein. When the N-terminal 26 amino acids were subsequently deleted from L N1259, L N1233 (ΔN26), the mutant again failed to bind to the P protein (lane 7, upper panel). These results strongly suggested that the entire N-terminal 1259 amino acids are required for P protein binding presumably to fold the L protein properly. The expression of all truncated L proteins were at similar levels (Fig. 5B, lower panel) since they interacted efficiently with a FLAG-tagged L N129. To determine the precise length of L protein required to bind to P protein, we constructed three additional C-terminally truncated non-tagged L proteins designated L N1160, L N1060, L N960 (Fig 6A). As shown in Fig 6B, all truncated L proteins bound to P-FLAG but the L N960 (lane 5, upper panel) bound less efficiently (compare lane 2, 3 and 4 upper panel). Quantitative analysis using a phosphoimager showed that both L N1160 and L N1060 bound similar to Lwt, whereas, L N960 bound only to 26.8% of Lwt. Moreover, any small internal deletion (40 amino acids) within the N-terminal 1060 amino acids led to loss of L-P interaction (data not shown). Based on these results we conclude that a minimum length of N-terminal 1060 amino acids of L protein is presumably required for efficient binding to the P protein; removal of N-terminal 26 amino acids disrupts the structural integrity of the L protein.
Fig. 5.
(A) Schematic representation (not to the scale) of Lwt and deletion mutants. The mutant L protein, L C2207 (ΔN26), lacks 26 amino acids from the N-terminus and carries the entire C-terminal 2207 amino acids and L N889, carries intact N-terminal 889 amino acids but has lost C-terminal 1344 amino acids. L N1695 and L N1259 carry N-terminal 1695 and 1259 amino acids respectively. The mutant L N1233 (ΔN26) was constructed by deleting N-terminal 26 amino acids from L N1259. (B) Analysis of L-P interaction by co-immunoprecipitation assay. 35S-labeled cell extracts prepared after transfection were immunoprecipitated with anti-FLAG antibody and analyzed by SDS-PAGE followed by autoradiography as described in text (top). Levels of expression of full length L and the L mutant proteins were detected by their interaction with L N129 (bottom).
Fig 6.
(A) Schematic representation (not to the scale) of Lwt and deletion mutants. The C-terminally truncated non-tagged L proteins (L N1160, L N1060 and L N960) express N-terminal 1160, 1060 and 960 amino acids respectively. (B) Analysis of L-P interaction by co-immunoprecipitation assay. 35S-labeled cell extracts prepared after transfection were immunoprecipitated with anti-FLAG antibody and analyzed by SDS-PAGE followed by autoradiography as described in text (top). Levels of expression of full length L and the L mutant proteins were detected their interaction with L N129 (bottom). The migration positions of L and P proteins are indicated.
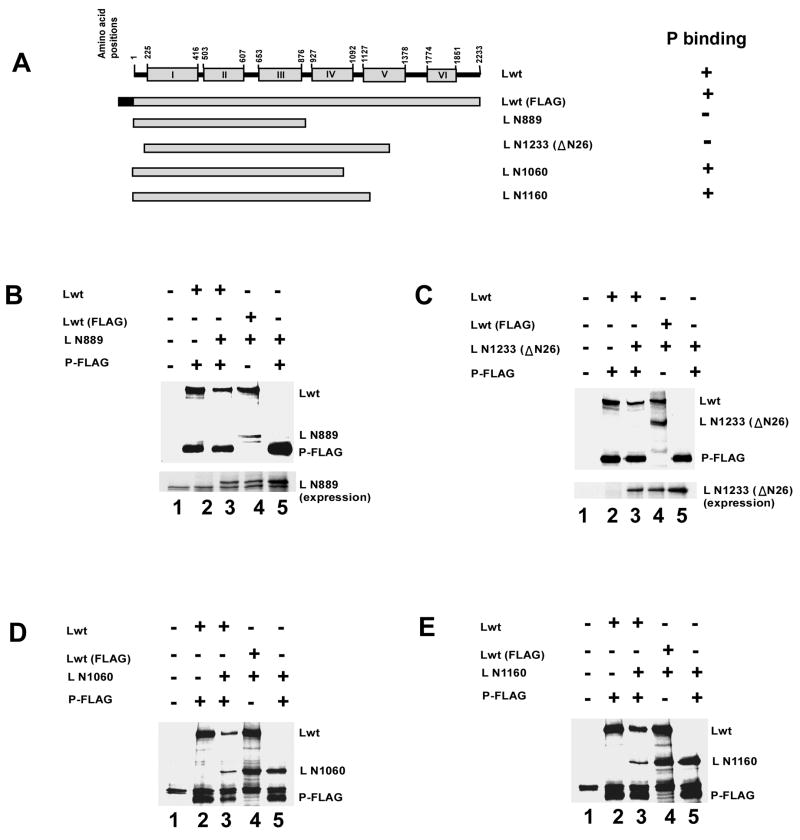
Having established the P protein binding domain in L protein, we then proceeded to study the effect of co-expression of P protein in the presence of L fragments that either bind to P protein or unable to interact. Accordingly, we co-expressed L N889 and L N1233(Δ26) (which bind with Lwt but not with P) and Lwt with or without P-FLAG (Fig 7A). Immunoprecipitation of lysates with anti-FLAG antibody showed that Lwt when bound to P-FLAG failed to pull-down L N889 and L N1233(Δ26) (lane 3 in Fig. 7B and 7C). On the other hand, as expected, N-terminally FLAG tagged Lwt co-immunoprecipitated efficiently both L N889 and L N1233(Δ26) in the absence of P protein (lane 4 in Fig. 7B and 7C). Efficient expression level of both L N889 and L N1233(Δ26) were confirmed (Figure 7B and 7C, lower panel). In a separate experiment, Lwt was co-expressed with L N1060 or L N1160 (which bind to P protein) and with FLAG-tagged P protein (Fig. 7A). After immunoprecipitation with FLAG antibody, as expected, both Lwt and L N1060 or L N1160 were pulled down by P-FLAG demonstrating efficient interaction of P protein with both the mutants (lane 3 in Fig. 7D and 7E). These results strongly suggest that L protein, in the absence of P protein, forms an aggregated complex with L N889 or L N1233(Δ26), but co-expression of P protein efficiently prevents such complex formation resulting in the formation of RdRp.
Fig. 7.
(A) Schematic representation (not to the scale) of full length HPIV3 L protein (Lwt, 2233 amino acids) and deletion mutants. Mutant L proteins, L N889 and L N1233(ΔN26), do not bind with P protein but interact with Lwt. These truncated L proteins were co-expressed with Lwt in absence as well as in the presence of P-FLAG and immunoprecipitated. (B) Analysis of L-L interaction by co-immunoprecipitation assay. 35S-labeled cell extracts prepared after transfection were immunoprecipitated with anti-FLAG antibody and analyzed by SDS-PAGE followed by autoradiography as described in text. The migration positions of Lwt, L N889 and P protein are indicated. Lwt interacts efficiently with L N889 in absence of P protein (lane 4, upper panel). However, P protein bound Lwt failed to interact with L N889 (lane 3, upper panel). Similar level of expression of L N889 was detected in lysates (lower panel). (C) Similarly, P protein bound Lwt did not interact with L N1233(ΔN26) (lane 3, upper panel) although, they interacted efficiently in the absence of P protein (lane 4, upper panel). (D) In control experiment, L N1060, which interacts with P protein was co-expressed with Lwt either in presence or in absence of the P protein. Co-immunoprecipitation experiment showed that L N1060 and Lwt both can interact with P-FLAG (lane 3). (E) Similarly, L N1160 (that interacts with P protein) when co-expressed with Lwt and P-FLAG, both Lwt and L N1160 interacted with P protein.
Distribution of L protein inside the cell in the presence or absence of P protein
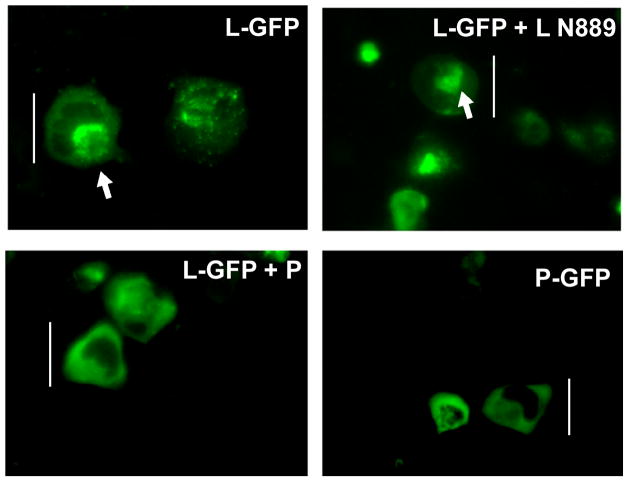
To visualize directly the fate of the L protein inside the cell in the presence and absence of the P protein, L protein was fused with GFP (L-GFP) as described in Materials and Methods and expressed in HeLa cells either singly or with L N889 or P protein. After 20 hr post-transfection live cells were observed under a fluorescent microscope. As shown in Fig 8A, when the L-GFP was expressed alone, in ~80% of the GFP expressing cells, the fluorescence within the cells were found localized as aggregated bodies within the cytoplasm. Similar fluorescence pattern was observed when L was co-expressed with L N889. In direct contrast, in cells where L-GFP was co-expressed with P protein a uniformly diffused fluorescence pattern covering the entire cytoplasm was observed. Similar fluorescence pattern was observed when P-GFP was expressed alone. Expression of P and P-GFP were detected by Western blot analysis using anti-RNP antibody (data not shown). These results strongly suggest that P protein upon binding to the L protein prevented L-L self-aggregation leading to redistribution of L-P complex inside the cell.
Fig 8.
(A) L-GFP was expressed in HeLa cells as described in Materials and Methods. L-GFP was also co-expressed either with L N889 or P protein. P-GFP was expressed as control. L-GFP when expressed singly or co-expressed with L N889 formed localized aggregated bodies in cytoplasm. When P protein was co-expressed with L-GFP, fluorescence was observed throughout the cytoplasm indicating P protein prevented aggregation of L-GFP. Similar diffused pattern of fluorescence was observed when P-GFP was expressed alone. A white line in each figure indicates the length of the cell and a white arrow indicates the point of aggregation inside the cell.
Polymerase activity of Lwt in the presence of L fragments and P proteins
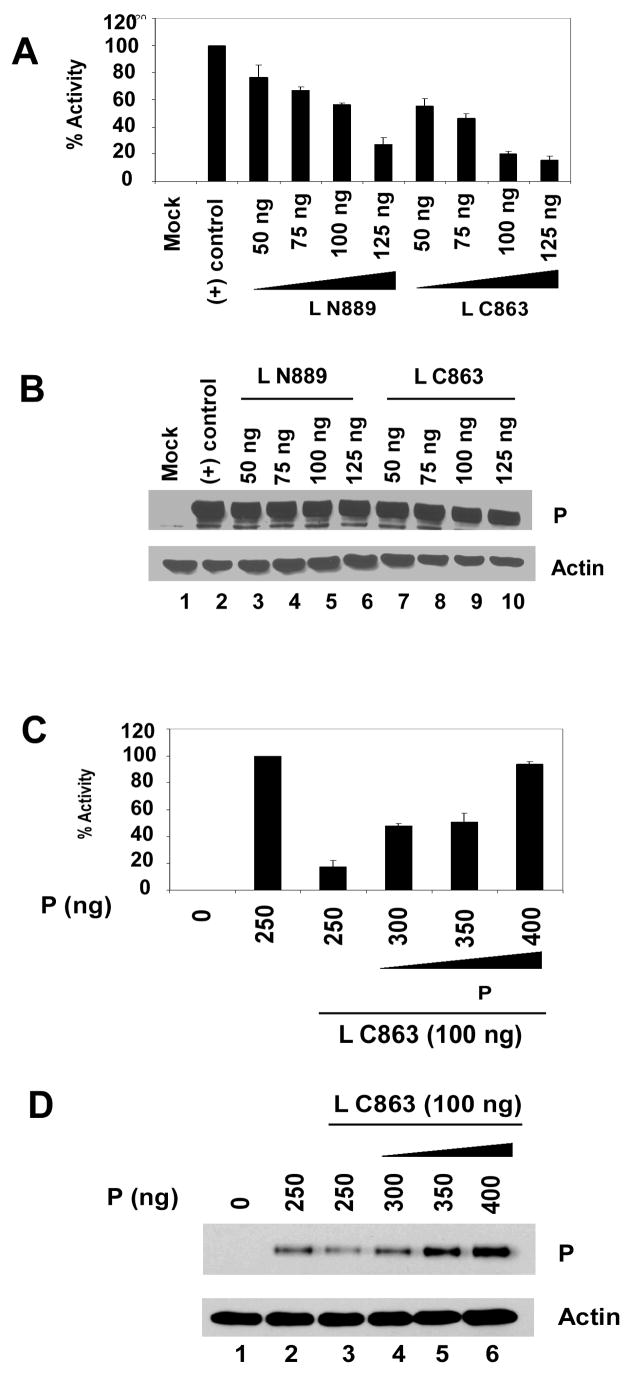
To assess the effect of L-L self-aggregation and its prevention by P protein on transcription, we used a HPIV3 minireplicon system previously used in our laboratory (Hoffman and Banerjee, 2000). Following infection of HeLa cells with recombinant vaccinia virus, HPIV3 MG (−) minireplicon expressing luciferase reporter gene (200 ng) and with support plasmids encoding N (500 ng), P (250 ng) and Lwt (100 ng) carrying a T7 promoter, were transfected. In the first round of transcription of the negative sense minireplicon RNA, encapsidated with the N protein is formed which then is used as template by the (L-P) heterocomplex. The RdRp activity was assayed by measuring luciferase activity of the cell lysate. When the assay was carried out with limiting amount of P protein (250 ng plasmid), the expression of L N889 or L C863 inhibited minigenome transcription in a dose dependent manner, suggesting that the fragments interacted with Lwt and rendered it functionally inactive (Fig 9A). Similar level of P protein expression was confirmed in lysates by Western blot analysis using anti-RNP antibody (Fig. 9B, upper panel) and synthesis of actin was used as loading control (Fig. 9B, lower panel).
Fig. 9.
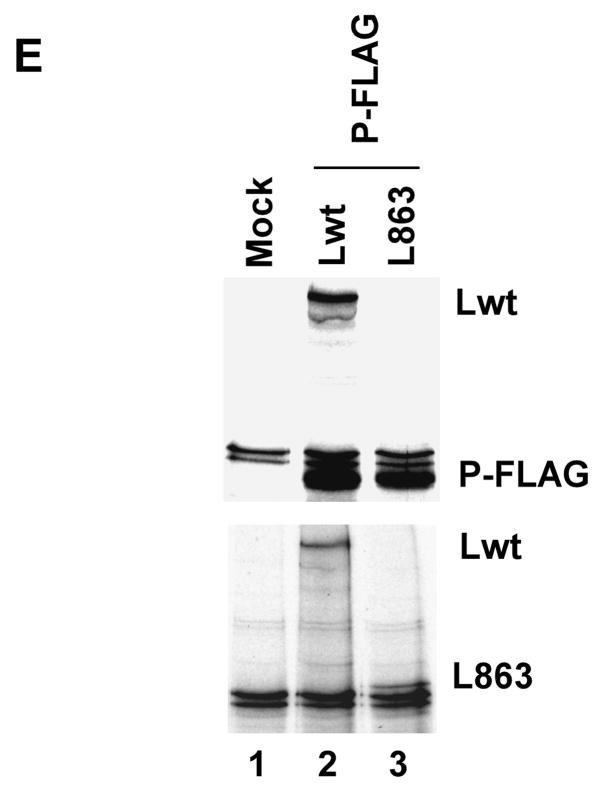
(A) Activity of mutant L proteins in minigenome transcription in vivo. HeLa cells were infected with modified vaccinia virus expressing T7 polymerase and transfected with pHPIV3-MG(−) and the support plasmids encoding N, P, and L proteins. At 24 h postinfection, cell extracts were prepared and luciferase activity was determined. Activity of L proteins were suppressed when 100 ng L N889 or L C863 were co-transfected. (B) Similar level of P protein expression was detected by Western blot analysis with anti-RNP antibody (upper panel) and anti-β actin antibody was used as loading control. (C) 100 ng L C863 was co-transfected with increased amount of P plasmid (250–400 ng). The negative effect on transcription by L C863 was recovered with increase P protein synthesis in a dose dependent manner. Transcriptional activity was restored up to 93.6% by addition of excess P protein (400 ng). (D) Western blot analysis of the lysates obtained from the cells expressing the above mentioned proteins. Anti-RNP antibody was used for the detection of P protein (upper panel) and anti-β actin antibody was used to estimate the amount of lysates loaded on gel (lower panel). (E) Lwt and L C863 were co-expressed with P-FLAG and labeled with 35S and immunoprecipitated with anti-FLAG beads. Lwt interacted with P-FLAG (lane 2, upper panel), while L C863 failed to interact (lane 3, lower panel). Both the proteins were expressed well (lane 2 and 3, lower panel).
Next, the same minigenome assay was carried out in the presence of L C863 (100 ng plasmid) and increased synthesis of P protein (300–400 ng plasmid). As shown in Fig 9C, the transcription was restored in a dose dependent manner with increasing amount of P plasmid transfection; in the presence of same amount of L C863 and increased P plasmid concentration (400 ng) the transcription was restored up to 93.6 % of the control. Increased level of P protein synthesis was verified by Western blot analysis using anti-RNP antibody (Fig. 9D, upper panel). Synthesis of actin was used as loading control (Fig. 9D, lower panel). As expected, L C863 did not interact with P protein (Fig 9E) although the mutant L protein interacts strongly with Lwt (Fig 1B). These results strongly suggest that the P protein effectively competes with L C863 to bind to the Lwt, resulting in increased availability of Lwt for RNA synthesis. Identical result was obtained when we used another L mutant (L N129) to test the ability of P protein to prevent negative effect on transcription exerted by the L mutant (data not shown).
Discussion
The paramyxoviral L proteins form a stable complex with their cognate P protein in the infected cells to carry out RNA synthesis (Lamb and Parks, 2007). Although the oligomeric nature of the P proteins has been well established (Tarbouriech et al., 2000), the precise subunit composition of the RdRp vis-à-vis the L protein in the complex remains unknown. Recently, it was reported that several paramyxovirus L proteins form oligomers when expressed in cells which is mediated via the N-terminal domain of the L protein (Cevik et al., 2003, Cevik et al. 2004, Cevik et al. 2007; Smallwood and Moyer, 2004). These conclusions were based primarily from co-immunoprecipitation studies exclusively using C-terminally truncated L proteins. To better understand the L protein structure, we wanted to first locate the boundaries within the L protein in which the presumptive oligomerization domain lies. In the present study, we made a series of both N- and C-terminally truncated L proteins of HPIV3 and tested their ability to interact with Lwt by co-immunoprecipitation. Not only did the C-terminally deleted L mutants pulled down Lwt as reported earlier, but all N-terminally deleted L mutants did so as well (Fig. 1). Interestingly, the mutant L protein, L C863, which did not contain the N-terminal 1370 amino acids, hence did not contain the putative oligomerization domain, also efficiently interacted with Lwt (Fig. 1). Moreover, these L-L interactions were resistant to high NaCl concentration (600 mM) suggesting that the proteins may not interact via ionic interactions, rather by some hydrophobic interactions. In subsequent deletion studies, we found that a small C-terminal L protein (189 amino acids) could also interact with the N-terminal 328 amino acids (Fig. 3). Surprisingly, L fragments originated from the mid-region of L (L 841-1259 and L 1371-1695, expressing 419 amino acids and 325 amino acids respectively) also interacted with L N129 (Fig. 3). Thus, we were unable to locate any specific oligomerization domain within the L protein. Rather, that the L protein, due to its large size, containing multiple domain structure and exposed hydrophobic regions, self-interacted leading to “aggregate” formation instead of ordered oligomerization via a specific domain. This contention is further supported by the fact that no truncated L protein or protein complexes could be resolved in native PAGE (Fig. 4). As an additional confirmation of the above results, we attempted to analyze the sedimentation profile of the L protein when expressed singly or co-expressed with P protein in a glycerol gradient. However, our attempts to carry out such experiments produced inconsistent results possibly due to the low level of expression of L without P protein (Horikami et al, Virology, 1992), which made the detection of L protein difficult after ultracentrifugation. However, co-expression of P proteins indeed increased the amount soluble L protein suggesting a possible role of P protein in the prevention of L-L self association (data not shown).
To further investigate the role of P protein in L-L self association P protein was co-expressed with Lwt in the presence or in the absence of truncated L mutants. When two deletion mutants (L N889 and L N1233ΔN26, which individually do not bind with P protein though they interacted with Lwt) were co-expressed with Lwt in the presence of P protein, no L-L interaction was seen (Fig. 7). However, as observed earlier (Fig. 1) in the absence of P protein two L deletion mutants interacted efficiently with Lwt (Fig. 7). These results clearly indicate that P protein for its strong affinity interacts with L protein to form the L-P complex, thus, preventing self-association with the L mutants. The observed L-P complex formation was further confirmed when we expressed L-GFP in the presence or absence of P protein in HeLa cells. The fluorescence emanated from GFP was readily detected as aggregated bodies within the cytoplasm of the most cells (~80%) in the absence of P. However, in the presence of P protein a diffused fluorescence pattern, similar to the fluorescence pattern of P-GFP expression alone, was observed indicating P protein interacted with L protein that caused the diffused pattern and, thus, prevented self-aggregation of L protein.
Finally, we used the minireplicon transcription system to test whether P protein restores the function of Lwt in the presence of such L fragments. The L C863 mutant strongly inhibited transcription in a dose-dependent manner, when expressed in the presence of limiting amount of P protein (Fig. 9). However, with increased P protein expression, the negative effect on RNA synthesis by the L N863 was overcome, also in a dose-dependent manner (Fig. 9). Similar result was obtained when another mutant L protein (L N129) was used instead of L C863 (data not shown). These results strongly suggest that P protein prevented non-functional self-interaction between Lwt and truncated L proteins, thus, increasing the effective concentration of (L-P) complex formation for increased RNA synthesis.
It is noteworthy that previous result (Smallwood et al., 2002a) demonstrated intragenic complementation of two inactive L mutants to occur in an in vitro reconstitution reaction, albeit, at an extremely low level. We tested this possibility in our in vivo minireplicon system using LΔ1215-1255 and LΔ1396-1436 along with the P protein. We were unable to obtain any restoration of transcription activity (data not shown). The apparent discrepancy between the two results may reside in using two different systems, i.e. in vitro and in vivo reconstitution reactions.
It is well documented that the native fold of a protein is determined by its amino acid sequences and spontaneous refolding in vitro is usually efficient for small, single-domain proteins (Anfinsen, 1973; Dobson and Karplus, 1999). However, under physiological condition, proteins, particularly those of multiple domains and large molecular weights, are often tend to misfold due to inappropriate interactions among regions of the folding peptide chains leading to aggregation (Hartl and Hayer-Hartl, 2002). Various chaperone factors inside the cells interact with newly synthesized peptide chains in order to prevent self-association into disordered, non-native molecular complexes (Hartl and Hayer-Hartl, 2002). The studies presented in this report support the contention that the P protein, by virtue of its strong affinity for the L protein gains access to the newly synthesized N-terminal region of the L protein, effectively folds before the rest of the growing L polypeptide chain could interact with the L protein, a function typically manifested by chaperone proteins in cells. We, thus, propose that a tetrameric P (analogy to SeV P) must interact with a monomeric L to provide this chaperone function. Further detail structural studies would be needed to precisely establish the subunit composition of the RdRp.
Materials and methods
Cells and viruses
HeLa cells were used for all experiments in this study. Recombinant vaccinia virus vTF7-3 expressing bacteriophage T7 RNA polymerase was propagated in HeLa cells.
Plasmid constructs
pGEM4 L (Galinski et al., 1988), containing L cDNA of HPIV3 was used for experiments involving expression of full length, non-tagged HPIV3 L protein (2233 amino acids) in HeLa cells. The same HPIV3 L cDNA clone was used as template for all the genetic manipulations using standard molecular biology techniques to construct deletion mutants as well as introducing FLAG or His epitopes or fusing GFP to the proteins. Detailed cloning strategies are available upon request. All constructs were verified by sequencing.
Coexpression and immunoprecipitation of proteins
HeLa cells in a 6-well plate were infected with vTF7-3 at a multiplicity of infection (MOI) of 1. At 1 h post infection, the cells were transfected with the plasmids encoding full length or mutant L (2.5 μg) or P (0.5 μg) proteins of HPIV3 using Lipofectin (Invitrigen). At 12 h post infection, the medium was replaced with 2 ml of methionine and cesteine free DMEM, and the incubation was continued at 37°C. At 14 h post-infection, the cells were labeled with 50 μCi of [35S]-methionine and cysteine (Parkin-Elmer) in 2 ml of methionine and cysteine free DMEM for 6 h. At 20 h of post infection, cells were washed with cold PBS, lysed with buffer (Roche or a buffer containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, 0.5% NP40, 1 μg/ml aprotinin) and immunoprecipitated with anti-FLAG antibody (Sigma, St. Louis, Mo.) conjugated to agarose beads by following the manufacturer’s protocol. The immunoprecipitated complexes were washed with buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, unless otherwise mentioned. The co-immunoprecipitated proteins were resolved in SDS-PAGE and visualized by autoradiography or Western blot analysis.
Purification of FLAG tagged proteins and native PAGE
HeLa cells were transfected with plasmids encoding FLAG tagged N-terminal 129 amino acids of L protein (L N129). HeLa cells were also co-transfected with FLAG LN129 and a plasmid expressing non-tagged C-terminal 2104 amino acids (L C2104) or a plasmid expressing N-terminally His-tagged N-terminal 328 amino acids (L N328). Expressed proteins were labeled with 35S as described above, purified by affinity chromatography using anti-FLAG agarose beads and 1X FLAG peptide following manufacturer’s protocol (Sigma). Eluted proteins were mixed with native sample buffer containing no SDS or reducing agent (62.5 mM Tris-HCL, pH 6.8, 40% glycerol, 0.01% Bromophenol Blue) and run on native PAGE (4.5% staking and 10% or 15% or 6 % resolving). Samples were not heated prior to loading on gel and electrophoreses were carried out at low current (10 mA). Bovine serum albumin (BSA) was run as control. Gels were stained with Coomassie brilliant blue (CBB) for the detection of BSA and 35S-labelled proteins were detected by autoradiography. A fraction of the eluted protein samples were separated by 10% SDS-PAGE and Western blotted using anti-L, anti-His and anti-FLAG antibodies to identify the proteins.
Fluorescence microscopy of GFP fused proteins
HeLa cells grown in 12 well plates were transfected with 0.5 μg of plasmid expressing a C-terminally GFP fused L protein of HPIV3 (L-GFP). L-GFP was also co-expressed in the same way with N-terminal 889 amino acids (L N889) or P protein. A C-terminally GFP fused P protein and empty vector (pGEM4) was also expressed as control. After 20 h post transfection live cells were observed under a fluorescent microscope. After taking representative images cells were lysed and western blotted to confirm expression of P protein.
In vivo minigenome assay
The in vivo HPIV3 minigenome assay was performed as it was described earlier (Hoffman and Banerjee, 2000) with slight modifications. In brief, HeLa cell monolayers in 12 well plates, grown to 90% confluency, were infected with recombinant vaccinia virus vTF7-3, at an MOI of 3. After 1 h of infection cells were washed with PBS and transfected with HPIV3 minigenome plasmid HPIV3-MG(−) (200 ng) carrying luciferase reporter gene together with supporting plasmids carrying N (500 ng), P (250 ng) and L (100 ng) using Lipofectin (Invitrogen) according to manufacturer’s protocol. After 5 h, transfection medium was removed, cells were washed with PBS and incubated with Dulbecco’s modified Eagle’s medium (DMEM) for 24 hr. Cells were lysed in 150 μl of lysis buffer, from which 1.5 μl aliquots were used to determine luciferase activity in a luminometer (Victor) according to the manufacturer’s instructions (Luciferase Assay Kit; Roche).
Western Blot analysis
Protein samples were run on SDS-PAGE and transferred to nitrocellulose membrane as per standard procedure. Membranes were incubated with appropriate primary and secondary antibodies and proteins were visualized by chemiluminescence (GE Healthcare). For the detection of P protein of HPIV3 a polyclonal anti-RNP antibody (Malur et al., 2002b) was used. Anti-FLAG M2 monoclonal (Sigma) and anti-His monoclonal (Abcam Inc., Cambridge, MA) antibodies were used respectively for the detection of FLAG and His tagged truncated L proteins of HPIV3. For the detection of L C2104 a polyclonal anti-L antibody (raised in rabbit against C-terminal region of HPIV3 L protein expressed in E. coli) was used. Nitrocellulose membranes were also incubated with anti-β actin antibody (Sigma) to estimate the amount of proteins loaded on gel.
Acknowledgments
This work was supported by a grant from United States Public Health Services (AI-32027 to A.K.B.). We are thankful to Sourav Misra for critically reviewing the manuscript.
Footnotes
We have corrected the N-terminal 915 amino acids mentioned by Smallwood and Moyer (2004), as N-terminal 889 amino acids based on L protein of 2233 amino acids expressed from ORF of L cDNA sequence of Galinski et al. (1988), whereas the previous report used an incorrect start codon that predicted L protein of 2258 amino acids (Durbin et al., 1999 and P. Collins personal communication).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Banerjee AK. Transcription and replication of rhabdoviruses. Microbiol Rev. 1987;51:66–87. doi: 10.1128/mr.51.1.66-87.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Barik S, De BP. Gene expression of nonsegmented negative strand RNA viruses. Pharmacol Ther. 1991;51:47–70. doi: 10.1016/0163-7258(91)90041-j. [DOI] [PubMed] [Google Scholar]
- Cevik B, Smallwood S, Moyer SA. Two N-terminal regions of the Sendai virus L RNA polymerase protein participate in oligomerization. Virology. 2007;363:189–197. doi: 10.1016/j.virol.2007.01.032. [DOI] [PubMed] [Google Scholar]
- Cevik B, Holmes DE, Vrotsos E, Feller JA, Smallwood S, Moyer SA. The phosphoprotein (P) and L binding sites reside in the N-terminus of the L subunit of the measles virus RNA polymerase. Virology. 2004;327:297–306. doi: 10.1016/j.virol.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Cevik B, Smallwood S, Moyer SA. The L-L oligomerization domain resides at the very N-terminus of the sendai virus L RNA polymerase protein. Virology. 2003;313:525–536. doi: 10.1016/s0042-6822(03)00342-8. [DOI] [PubMed] [Google Scholar]
- Chandrika R, Horikami SM, Smallwood S, Moyer SA. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology. 1995;213:352–363. doi: 10.1006/viro.1995.0008. [DOI] [PubMed] [Google Scholar]
- Chen M, Ogino T, Banerjee AK. Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J Virol. 2007;81:13478–13485. doi: 10.1128/JVI.01244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ogino T, Banerjee AK. Mapping and functional role of the self-association domain of vesicular stomatitis virus phosphoprotein. J Virol. 2006;80:9511–9518. doi: 10.1128/JVI.01035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary SK, Malur AG, Huo Y, De BP, Banerjee AK. Characterization of the oligomerization domain of the phosphoprotein of human parainfluenza virus type 3. Virology. 2002;302:373–382. doi: 10.1006/viro.2002.1668. [DOI] [PubMed] [Google Scholar]
- Davis NL, Arnheiter H, Wertz GW. Vesicular stomatitis virus N and NS proteins form multiple complexes. J Virol. 1986;59:751–754. doi: 10.1128/jvi.59.3.751-754.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Green TJ, Lu S, Luo M. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J Virol. 2006;80:2808–2814. doi: 10.1128/JVI.80.6.2808-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM, Karplus M. The fundamentals of protein folding: bringing together theory and experiment. Curr Opin Struct Biol. 1999;9:92–101. doi: 10.1016/s0959-440x(99)80012-8. [DOI] [PubMed] [Google Scholar]
- Durbin AP, McAuliffe JM, Collins PL, Murphy BR. Mutations in the C, D, and V open reading frames of human parainfluenza virus type 3 attenuate replication in rodents and primates. Virology. 1999;261:319–330. doi: 10.1006/viro.1999.9878. [DOI] [PubMed] [Google Scholar]
- Feller JA, Smallwood S, Horikami SM, Moyer SA. Mutations in conserved domains IV and VI of the large (L) subunit of the sendai virus RNA polymerase give a spectrum of defective RNA synthesis phenotypes. Virology. 2000;269:426–439. doi: 10.1006/viro.2000.0234. [DOI] [PubMed] [Google Scholar]
- Galinski MS, Mink MA, Pons MW. Molecular cloning and sequence analysis of the human parainfluenza 3 virus gene encoding the L protein. Virology. 1988;165:499–510. doi: 10.1016/0042-6822(88)90594-6. [DOI] [PubMed] [Google Scholar]
- Grdzelishvili VZ, Smallwood S, Tower DR, Hall L, Hunt DM, Moyer SA. A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J Virol. 2005;79:7327–7337. doi: 10.1128/JVI.79.12.7327-7337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hoffman MA, Banerjee AK. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology. 2000;269:201–211. doi: 10.1006/viro.2000.0223. [DOI] [PubMed] [Google Scholar]
- Horikami SM, Curran J, Kolakofsky D, Moyer SA. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J Virol. 1992;66:4901–4908. doi: 10.1128/jvi.66.8.4901-4908.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karron RA, Collins PL. Parainfluenza viruses. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins, a Wolters Kluwer Business; Philadelphia, Pa: 2007. pp. 1497–1526. [Google Scholar]
- Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott Williams & Wilkins, a Wolters Kluwer Business; Philadelphia, Pa: 2007. pp. 1449–1496. [Google Scholar]
- Li J, Rahmeh A, Morelli M, Whelan SP. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol. 2008;82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Fontaine-Rodriguez EC, Whelan SP. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol. 2005;79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang JT, Whelan SP. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci U S A. 2006;103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A, Bhattacharya R, Basak S, Shaila MS, Chattopadhyay D, Roy S. P-protein of Chandipura virus is an N-protein-specific chaperone that acts at the nucleation stage. Biochemistry. 2004;43:2863–2870. doi: 10.1021/bi035793r. [DOI] [PubMed] [Google Scholar]
- Majumder A, Basak S, Raha T, Chowdhury SP, Chattopadhyay D, Roy S. Effect of osmolytes and chaperone-like action of P-protein on folding of nucleocapsid protein of Chandipura virus. J Biol Chem. 2001;276:30948–30955. doi: 10.1074/jbc.M011705200. [DOI] [PubMed] [Google Scholar]
- Malur AG, Gupta NK, De BP, Banerjee AK. Analysis of the mutations in the active site of the RNA dependent RNA polymerase of human parainfluenza virus type 3 (HPIV 3) Gene Expr. 2002a;10:93–100. [PMC free article] [PubMed] [Google Scholar]
- Malur AG, Choudhary SK, De BP, Banerjee AK. Role of a highly conserved NH(2)-terminal domain of the human parainfluenza virus type 3 RNA polymerase. J Virol. 2002b;76:8101–8109. doi: 10.1128/JVI.76.16.8101-8109.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PS, Banerjee AK. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J Virol. 1988a;62:2658–2664. doi: 10.1128/jvi.62.8.2658-2664.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PS, Banerjee AK. Resolution of multiple complexes of phosphoprotein NS with nucleocapsid protein N of vesicular stomatitis virus. J Virol. 1988b;62:2651–2657. doi: 10.1128/jvi.62.8.2651-2657.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Poch O, Delarue M, Bishop DHL, Bouloy M. Rift valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol. 1994;75:1345–1352. doi: 10.1099/0022-1317-75-6-1345. [DOI] [PubMed] [Google Scholar]
- Ogino T, Kobayashi M, Iwama M, Mizumoto K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J Biol Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol. 1990;71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- Sánchez AB, de la Torre JC. Genetic and biochemical evidence for an oligomeric structure of the functional L polymerase of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2005;79:7262–7268. doi: 10.1128/JVI.79.11.7262-7268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu MS, Menonna JP, Cook SD, Dowling PC, Udem SA. Canine distemper virus L gene: sequence and comparison with related viruses. Virology. 1993;193:50–65. doi: 10.1006/viro.1993.1102. [DOI] [PubMed] [Google Scholar]
- Sleat DE, Banerjee AK. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol. 1993;67:1334–1339. doi: 10.1128/jvi.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood S, Cevik B, Moyer SA. Intragenic complementation and oligomerization of the L subunit of the sendai virus RNA polymerase. Virology. 2002a;304:235–245. doi: 10.1006/viro.2002.1720. [DOI] [PubMed] [Google Scholar]
- Smallwood S, Ryan KW, Moyer SA. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology. 1994;202:154–163. doi: 10.1006/viro.1994.1331. [DOI] [PubMed] [Google Scholar]
- Smallwood S, Moyer SA. The L polymerase protein of parainfluenza virus 3 forms an oligomer and can interact with the heterologous Sendai virus L, P and C proteins. Virology. 2004;318:439–450. doi: 10.1016/j.virol.2003.09.045. [DOI] [PubMed] [Google Scholar]
- Smallwood S, Easson CD, Feller JA, Horikami SM, Moyer SA. Mutations in conserved domain II of the large (L) subunit of the Sendai virus RNA polymerase abolish RNA synthesis. Virology. 1999;262:375–383. doi: 10.1006/viro.1999.9933. [DOI] [PubMed] [Google Scholar]
- Smallwood S, Hovel T, Neubert WJ, Moyer SA. Different substitutions at conserved amino acids in domains II and III in the Sendai L RNA polymerase protein inactivate viral RNA synthesis. Virology. 2002b;304:135–145. doi: 10.1006/viro.2002.1644. [DOI] [PubMed] [Google Scholar]
- Tarbouriech N, Curran J, Ruigrok RW, Burmeister WP. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat Struct Biol. 2000;7:777–781. doi: 10.1038/79013. [DOI] [PubMed] [Google Scholar]