Abstract
The fragile X syndrome results from expansions as well as deletions of the repeating CGG·CCG DNA sequence in the 5′-untranslated region of the FMR1 gene on the X chromosome. The relative frequency of disease cases promoted by these two types of mutations cannot be ascertained at present because the routine clinical assay monitors only expansions. At least 30 articles have been reviewed that document the involvement of deletions of part or all of the CGG·CCG repeats along with varying extents of DNA flanking regions as well as very small mutations including single base pair changes. Studies of deletion mutants of CGG·CCG tracts in Escherichia coli plasmids revealed a similar spectrum of mutagenic products. The triplet repeat tract in a non-B conformation is the mutagen, not the sequence per se in the right-handed B helix. Hence, molecular investigations in a simple model organism may generate useful initial information toward therapeutic strategies for this disease.
General Overview: Genetic Instability and Hereditary Neurological Diseases
Substantial progress has been made in the past 20 years in our understanding of the pathophysiology, genetics, and biochemistry of approximately 20 neurological diseases associated with simple sequence amplification (1, 2). These data serve as the overarching subject of this minireview series. Dynamic mutations involving the role of DNA hairpin loops or slipped strand conformations with differing relative stabilities of repeating tri-, tetra-, or pentanucleotide tracts are involved in these expansions and deletions (reviewed in Refs. 1 and 3–6). The diseases, including fragile X syndrome, myotonic dystrophy, Huntington disease, and Friedreich ataxia, are reviewed elsewhere (1), along with their inheritance patterns, chromosomal localizations, protein products, and loci of the repeat sequences. In type 2 diseases, for example, the repeat expansions are massive (thousands of repeats), whereas in type 1 diseases, the TRS2 are in coding regions and elicit a modest expansion of a polyamino acid tract (usually glutamine, but alanine in some diseases) (1, 2). The clinical observation of anticipation, the decrease in age of onset, and the increase in severity with progression through a family pedigree are observed with most, but not all, of these diseases. Usually, a more severe neurological syndrome is observed in patients with longer repeat tracts. Substantial work over the past decade has demonstrated that the expansions and deletions are mediated by DNA replication, repair, and recombination, probably acting in concert (reviewed in Refs. 1 and 3–7). The slippage of the repeating DNA complementary strands to form non-B DNA structures, such as hairpin loops or slipped strand conformations, with differing relative stabilities is an important component in the mechanism (1, 3–7).
In general, the genetic instabilities in the simple repeating sequences are found within the TRS, not in the flanking regions, for the majority of these neurological disorders (1, 2). However, a large number of articles (>30) have described a variety of classical mutations, such as deletions, found in the DNA of fragile X syndrome patients in the vicinity of the CGG·CCG repeats. This behavior seems to be more frequent for the fragile X syndrome than for other hereditary neurological diseases (8). To focus on the molecular mechanisms of the mutagenic spectra found in deletions related to fragile X syndrome, I shall not consider other folate-sensitive fragile sites (1, 2, 8) that also have CGG·CCG expansions.
Complex Family of Types of Mutations, Both Deletions and Expansions, Causes the Fragile X Syndrome
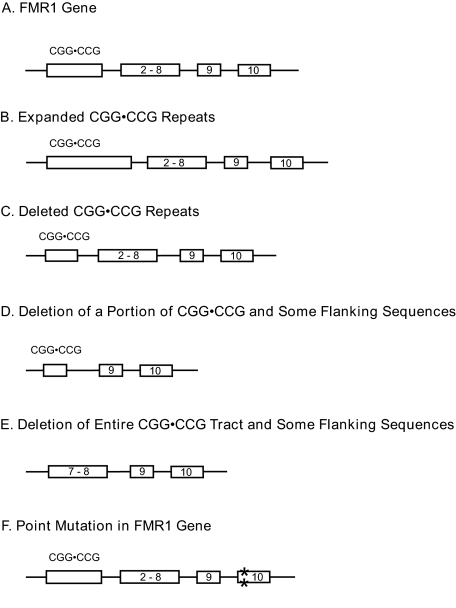
Deletions as well as expansions are important mutagenic processes as related to the fragile X syndrome (Fig. 1). The principal mutation responsible for the fragile X syndrome is generally considered to be the expansion of an untranslated CGG·CCG repeat in the 5′-untranslated region of the FMR1 gene on the X chromosome (1, 8). This mutation is associated with the hypermethylation of the proximal CpG island and the triplet repeat region that gives rise to the down-regulation of the FMR1 gene. An absence or a reduction in the amount of the corresponding protein (FMRP) is responsible for the etiology of the disease. Hence, any type of mutation in the FMR1 gene, including the CGG·CCG expansion, might lead to the disease. However, the extent to which the notion that expansions are the predominant mechanism remains to be proven. At least 30 articles on patient DNAs describe point mutations, 2-bp changes, deletions of varying sizes (including the entire gene), and genomic rearrangements that affect part of or the entire gene. Therefore, base pair changes ranging from 1 unit to the entire gene may disrupt the function of FMRP or prevent the protein from being formed, thus giving rise to the fragile X syndrome. In general, the prevailing thought within the fragile X molecular biology community is that the CGG·CCG repeat expansions account for at least 95% of the mutations (1, 8). However, no quantitative data are currently available3 because the routine clinical testing for the suspected fragile X syndrome focuses only on the CGG·CCG repeat expansion. Accordingly, a large number of other types of mutations may be responsible for the disease phenotype but are currently unrecognized because of the screening methods that are typically employed.
FIGURE 1.
Mutagenic spectra found in fragile X clinical phenotypes. This schematic diagram is not drawn to scale.
Interestingly, a Google search revealed 290,000 web sites for expansions along with 158,000 sites for fragile X deletions; a PubMed search showed about the same ratio (160 articles on expansions and 101 articles on deletions). I was surprised to learn that the bias was only modestly skewed toward expansions. Thus, on a relative basis, we know a large amount about deletions as the causative mutation.
Expansions and Deletions of CGG·CCG Repeats—As stated above, studies on patient materials show that expansions as well as deletions of the CGG·CCG repeats are the mutations responsible for the fragile X syndrome (Fig. 1B) (1). However, the ascertainment bias inherent in the clinical assays may skew our understanding toward the expansion mechanisms. It may be noted that extensive mosaicism is observed and that most of our knowledge is based on Southern blot analyses only. Regardless, the massive expansions of this TRS are an important mutagenic mechanism that elicits the disease etiology (reviewed in Refs. 1, 4, and 6–8). However, at least one case is known of a patient with a deleted region only in the CGG·CCG repeat sequence, leaving 15 pure repeats in the DNA (Fig. 1C) (9). Estivill and co-workers (9) state that the methylation status in this patient is normal but that the level of FMRP may not be enough to prevent the clinical abnormalities of the fragile X syndrome. Deletions of the CGG·CCG repeats are common, especially compared with expansions, in molecular biological investigations in model organisms such as E. coli and Saccharomyces cerevisiae (10–15). In these studies, the triplet repeat contractions were confined to the repeat sequence and did not extend into flanking sequences.
Deletions of a Portion of CGG·CCG Repeats and Some Flanking Sequences—Several authors have described deletions containing a portion of the CGG·CCG repeats and various lengths of flanking sequences ranging from 30 bp to 1.6 kb (Fig. 1D) (16–23). In addition, some authors believe that the mutations in the CGG·CCG repeats that are less than a full mutation may be due to a deletion event following an expansion of the TRS. The longest deletion (1.6 kb) was reported in the initial discovery (17, 18) of this behavior. At least 35 other patients' phenotypes have been characterized within this general category (19). Thus, the general behavior of mutations extending from the TRS into flanking sequences is a common event; therefore, one might surmise that the TRS plays some role in initiating the deletion processes, which is clearly demonstrated below in model systems. The concept of the CGG·CCG repeats undergoing substantial expansions followed by deletions of a portion of the repeat sequence along with a flanking tract (17, 20) is an intriguing idea but is difficult to document in human systems, where only the end product of the genetic event can be studied. Clearly, further work in model systems will be needed to evaluate this concept.
Deletion of All CGG·CCG Repeats and Some Flanking Sequences—At least 16 articles (24–39) describe large deletions of the entire CGG·CCG repeat tracts along with varying extents of DNA flanking sequences (Fig. 1E). These deletions range in size from several base pairs to as much as ∼13 megabases of DNA along with all of the FMR1 gene and some flanking DNA sequences (36). Several other articles describe the deletion of the entire FMR1 gene and smaller extents of the flanking DNA tracts. The DNA of at least 24 patients was studied. Hence, the phenomenon of gross deletions of the FMR1 gene causing the fragile X syndrome is not an isolated event.
Point Mutations—To this point, the previously described types of mutations are believed to be promoted by the presence of long CGG·CCG repeats. However, this subsection on very small mutations is included for the sake of inclusiveness of the types of mutations that are involved in fragile X syndrome. However, no evidence exists to support the role of long CGG·CCG repeats in the formation of the mutations described in this subsection.
Several cases of point mutations or 2-bp changes have been observed (Fig. 1F). Willems and co-workers (40) found a patient with the fragile X phenotype and without cytogenetic expression of FMRP with a single point mutation but with a CGG·CCG repeat of normal length and an unmethylated CpG island. Also, two different patients were found with intragenic loss-of-function mutations, a single de novo nucleotide deletion in one and a 2-bp change in the other; both patients displayed the classical features of fragile X syndrome (41). Furthermore, three unrelated fragile X patients were found with a C-to-T point mutation at the 14th nucleotide in intron 10 with normal length CGG·CCG repeats (Fig. 1F) (42). Clearly, these point mutations are sufficient for the development of the fragile X syndrome. Alternatively, three unrelated patients were identified with silent mutations in exon 1 (16, 17); it is not unexpected that silent mutations would be found in FMR1 exons.
Studies in Model Systems
Conformation(s) of Non-B DNA Fragile X Syndrome Triplet Repeats as Mutagenic Agents—A recent investigation has revealed that fragile X repeats (CGG·CCG) are potent inducers of complex multiple-site rearrangements and/or gross deletions in flanking DNA sequences in E. coli plasmids (43). DNA sequence analyses of mutant clones revealed the influence of the length (24, 44, or 73 repeats) and the orientation of the repeat region relative to the unidirectional origin of replication and its transcription status.
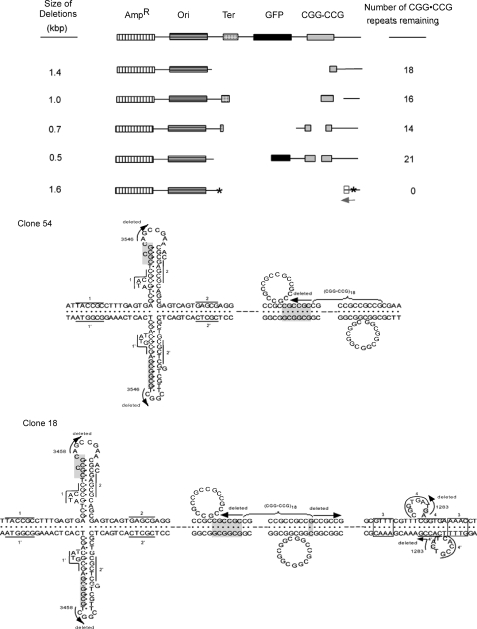
Complex rearrangements occurred in the mutant clones because some products contained deletions, inversions, and insertions, and some products had only gross deletions. Fig. 2 (upper panel) shows the types of multiple-site deletions and rearrangements that were found, ranging in size from 0.5 to 1.6 kb. Furthermore, the CGG·CCG repeats repeatedly induced, up to 22 times, the formation of identical (to the base pair) mutagenic products, indicating the powerful nature of the complex processes involved (43). Also, the mutations were bidirectional from the TRS. The healed junctions had CG-rich microhomologies of 1–6 bp, CG-rich regions, and putative cruciforms and slipped structures (Fig. 2, lower panel). Thus, essentially the entire mutagenic spectrum observed in patients with fragile X syndrome has been found in this E. coli model system. Accordingly, I submit that this bacterial system has numerous advantages for investigating the molecular aspects of these processes compared with human systems. Indeed, all biochemical processes (replication, repair, and recombination) responsible for the genetic instability of repeating tri-, tetra-, and pentanucleotides were first demonstrated in E. coli or yeast (1, 3, 4, 7) and then later studied and co-opted in eucaryotic systems. Obviously, investigations on neurological or developmental issues must be addressed in eucaryotic systems, but molecular questions on the instability processes may be broached in these simpler systems, which are genetically tractable. Also, earlier investigations found deletions in DNA flanking sequences to the CGG·CCG repeats in another E. coli and a COS-1 cell system (44, 45).
FIGURE 2.
Upper panel, restriction maps of the mutant clones induced by long tracts of CGG·CCG repeats in plasmids in E. coli. AmpR, ampicillin resistance gene; Ori, pUC19 origin of replication; Ter, transcription terminator cassette; GFP, LacZ-green fluorescent protein fusion gene; CGG·CCG, (CGG·CCG)n tract where n = 0 (control), 24, 44, and 73 repeats. The CGG·CCG tracts were cloned into the pGFPT vector. Microinsertions are designated with asterisks. For further details, see Fig. 2 in Ref. 43. kbp, kilobase pairs. Lower panel, non-B DNA structures predicted from the sequences flanking the healed junctions of clones with single (clone 54) or double (clone 18) deletions. The sequences read from the 5′- to the 3′-ends of the top strands. The arrows show the directions of the sequences that were deleted, and the numbers at these arrows designate their healed junction positions. Nucleotides in shaded boxes indicate homology at breaks identified by the DNA sequencing data. The dashed lines between the nucleotides present the continuous intervening sequences. However, the slipped structures and the deletions may be anywhere inside the CGG·CCG repeat tracts. The numbers beside the lines indicate the base pairs with direct repeat homology shown for both DNA strands, and the numbers above the boxes present base pairs with inverted repeat homology. Clone 54 shows a cruciform and a slipped strand structure, whereas clone 18 derived from pRW5501 presents a cruciform and two slipped structures. These conformations are representative of the types of non-B structures found. Other types of non-B structures are found at other breakpoint junctions (46, 47, 50). For further details, see supplemental Fig. 1 in Ref. 43.
Models were constructed to explain the mechanisms involved in the formation of the complex multiple-site DNA rearrangements induced by the CGG·CCG repeat tract (43). The four critical sequences and/or DNA conformational features (see Fig. 4 in Ref. 43) are apparently operative in a defined set of sequential steps to generate these complex rearrangements. The non-B DNA structures (cruciforms and slipped structures) flanking the healed regions, which may also have an influence on the induction of these mutagenic events, were described (43). However, the breaks occurred inside the CGG·CCG tracts in all clones with single and double deletions. Thus, this sequence and/or conformation is a significant trigger for the complex DNA rearrangements. This model requires the presence of homologous sequences that serve as a substrate for double-strand break repair followed by recombination repair, which leads to deletion of the intervening sequences; healed junctions in the mutant progeny were observed at non-B DNA structures (43). The types of enzymatic systems involved in these rearrangements have been reviewed (1, 4).
Relatively few studies have been reported on the molecular aspects of the genetic instabilities in CGG·CCG repeats compared with myotonic dystrophy type 1 and 2 and Friedreich ataxia repeats because of the extreme difficulty in working with these highly unstable sequences (11). Furthermore, DNA sequence analyses are difficult because of the repetitive GC-rich arrays, which cause extensive slippage during both template preparation and sequencing reactions. Thus, the CTG·CAG, CCTG·CAGG, and GAA·TTC repeat genetic instabilities have been more extensively investigated than the CGG·CCG repeats.
Non-B DNA Conformation(s) of Other Repeat Sequences as Mutagenic Agents—Not unexpectedly, other types of long repeat tracts of CTG·CAG, CCTG·CAGG, and GAA·TTC also induce gross deletions and inversions in model systems (13, 46, 47). These repeat sequences are integral to the etiology of myotonic dystrophy types 1 and 2 and Friedreich ataxia, respectively. Behaviors similar to those described above for CGG·CCG repeats were observed except that the fragile X sequence was the most potent, on a base pair basis, of any of the other repeats in causing the mutagenesis (43). Furthermore, the fragile X TRS induced a larger mutation spectrum (Fig. 2) than the other repeat sequences. The long repeats of CTG·CAG, CCTG·CAGG, and GAA·TTC caused deletions of most or all of the repeats and the flanking DNA sequences. Deletions of 0.6–1.8 kb were found as well as inversions in E. coli and two types of mammalian fibroblast-like cells. Under certain conditions, 30–50% of the products of episome replication/transcription in COS-7 cells contained gross deletions. The breakpoint junctions revealed the presence of direct or inverted repeat homologies in all cases. Also, the presence of non-B folded conformations (i.e. slipped structures, cruciforms, or triplexes) at or near the breakpoints was predicted in all cases. Increased negative superhelical density on the plasmids in vivo enhanced the genetic instability of the TRS (13) as expected because it stabilized the formation of these non-B conformations (1, 3, 7).
Gross deletions and other genomic rearrangements have been documented in patient materials for other hereditary neurological diseases, especially Friedreich ataxia (48). However, many more complex mutations have been reported for fragile X syndrome than for the other diseases (1, 2, 8).
A long-standing but critical question for many years has been the extent to which the DNA sequences involved trigger these mutagenic reactions versus the non-B DNA conformations that may be adopted in vivo. Wojciechowska et al. (47) definitively demonstrated by three experimental strategies that the non-B DNA conformations are critical for these mutagenic mechanisms, not the sequences per se. Hence, future work should be aimed at evaluating the role of the non-B DNA conformational features and their thermodynamically most stable state in the mutagenic processes. Also, another prior investigation revealed that long CTG·CAG tracts induced deletions and rearrangements of flanking sequences during recombination in Chinese hamster ovary cells (49).
Future Challenges
The genetic basis for the fragile X syndrome is a variety of types of mutations, including point mutations, double mutations, and deletions of several types of varying lengths up to ∼13 megabases that either include or do not include the CGG·CCG repeats. However, the disease is generally attributed to the massive expansion of the TRS, but the extent of the bias between the different types of mutations is uncertain at present because the clinical assays monitor only expansions, not deletions. Mechanistic studies in E. coli and mammalian cells provide suitable models for evaluating the mechanisms and enzymatic systems responsible for these instabilities. Further investigations in these systems may be useful for initial steps toward therapeutic intervention for this disease.
An important goal of many investigations related to fragile X syndrome is to understand the broad aspects of the pathophysiology of the disease as modulated by TRS lengths and the consequences of these expansion processes. Understanding the enzymatic systems that involve the DNA transactions causing the expansions (4) is critical. The routine clinical testing for this disease focuses on the length of the TRS, thereby not specifically monitoring some of the other types of mutations such as described in this review. Broader based molecular biological studies on patient samples will be required to better understand the extent of the involvement of various types of mutations within the spectra described herein. An understanding of the molecular mechanisms requires appropriate studies in a wide range of systems, including E. coli, yeast, human cells, mice, etc.; obviously, our goal is to understand the mechanisms in humans. Dramatic advances have been made in our understanding of the mechanisms of human hereditary neurological diseases (1–8), underscoring the importance of a wide range of strategies for molecular investigations.
The recognition of the roles of non-B DNA structures in human disease offers new strategies for controlling the mutagenic processes. Because the non-B DNA structures, not the DNA sequence per se in the orthodox right-handed B conformations, are the mutagenic agents, methodologies that will reduce the propensity of these tracts to adopt the unorthodox structures may be beneficial. This relationship between non-B DNA structures and genomic disorders has been a multidecade goal of the DNA structural field (1, 3, 7, 47, 50). An extremely large number of questions concerning the role of non-B DNA structures in mutagenesis are outstanding (see box 3 in Ref. 50). Hence, this topic is fertile for future investigations.
Supplementary Material
Acknowledgments
I thank Drs. B. Kosmider, M. Wojciechowska, M. Napierala, D. L. Nelson, and A. Bacolla for helpful advice and comments on the manuscript and Dr. Kosmider for assistance with the figures. I express appreciation to J. E. Larson for 40 years of research on non-B DNA structures. All research materials from this laboratory have been transferred to S. M. Mirkin (Sergei.Mirkin@Tufts.edu).
This work was supported, in whole or in part, by National Institutes of Health Grant ES11347. This work was also supported by the Friedreich's Ataxia Research Alliance, the Seek-a-Miracle Foundation, and the Robert A. Welch Foundation. This is the first of five articles in the Unstable Nucleotide Repeat Minireview Series. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: TRS, triplet repeat sequence(s); FMRP, fragile X mental retardation protein.
D. L. Nelson, personal communication.
References
- 1.Wells, R. D., and Ashizawa, T. (eds) (2006) Genetic Instabilities and Neurological Diseases, 2nd Ed., Academic Press, San Diego, CA
- 2.Orr, H. T., and Zoghbi, H. Y. (2007) Annu. Rev. Neurosci. 30 575–621 [DOI] [PubMed] [Google Scholar]
- 3.Mirkin, S. M. (2006) Curr. Opin. Struct. Biol. 16 351–358 [DOI] [PubMed] [Google Scholar]
- 4.Wang, G., and Vasquez, K. M. (2006) Mutat. Res./Fundam. Mol. Mech. Mutagen. 598 103–119 [Google Scholar]
- 5.Wang, Y. H. (2007) Front. Biosci. 12 4731–4741 [DOI] [PubMed] [Google Scholar]
- 6.Kovtun, I. V., and McMurray, C. T. (2008) Cell Res. 18 198–213 [DOI] [PubMed] [Google Scholar]
- 7.Wells, R. D., Dere, R., Hebert, M., Napierala, M., and Son, L. S. (2005) Nucleic Acids Res. 33 3785–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penagarikano, O., Mulle, J. G., and Warren, S. T. (2007) Annu. Rev. Genomics Hum. Genet. 8 109–129 [DOI] [PubMed] [Google Scholar]
- 9.Mila, M., Castellvi-Bel, S., Sanchez, A., Lazaro, C., Villa, M., and Estivill, X. (1996) J. Med. Genet. 33 338–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang, S., Ohshima, K., Shimizu, M., Amirhaeri, S., and Wells, R. D. (1995) J. Biol. Chem. 270 27014–27021 [DOI] [PubMed] [Google Scholar]
- 11.Shimizu, M., Gellibolian, R., Oostra, B. A., and Wells, R. D. (1996) J. Mol. Biol. 258 614–626 [DOI] [PubMed] [Google Scholar]
- 12.Bacolla, A., Gellibolian, R., Shimizu, M., Amirhaeri, S., Kang, S., Ohshima, K., Larson, J. E., Harvey, S. C., Stollar, B. D., and Wells, R. D. (1997) J. Biol. Chem. 272 16783–16792 [DOI] [PubMed] [Google Scholar]
- 13.Napierala, M., Bacolla, A., and Wells, R. D. (2005) J. Biol. Chem. 280 37366–37376 [DOI] [PubMed] [Google Scholar]
- 14.Balakumaran, B. D., Freudenreich, C. H., and Zakian, V. A. (2000) Hum. Mol. Genet. 9 93–100 [DOI] [PubMed] [Google Scholar]
- 15.Pelletier, R., Krasilnikova, M. M., Samadashwily, G. M., Lahue, R., and Mirkin, S. M. (2003) Mol. Cell. Biol. 23 1349–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gronskov, K., Hallberg, A., and Brondum-Nielsen, K. (1998) Hum. Genet. 102 440–445 [DOI] [PubMed] [Google Scholar]
- 17.Meijer, H., de Graaff, E., Merckx, D. M., Jongbloed, R. J., de Die-Smulders, C. E., Engelen, J. J., Fryns, J.-P., Curfs, P. M., and Oostra, B. A. (1994) Hum. Mol. Genet. 3 615–620 [DOI] [PubMed] [Google Scholar]
- 18.Wiegers, A. M., Curfs, L. M., Meijer, H., Oostra, B., and Fryns, J.-P. (1994) Genet. Couns. 5 377–380 [PubMed] [Google Scholar]
- 19.de Graaff, E., Rouillard, P., Willems, P. J., Smits, A. P., Rousseau, F., and Oostra, B. A. (1995) Hum. Mol. Genet. 4 45–49 [DOI] [PubMed] [Google Scholar]
- 20.Mannermaa, A., Pulkkinen, L., Kajanoja, E., Ryynanen, M., and Saarikoski, S. (1996) Am. J. Med. Genet. 64 293–295 [DOI] [PubMed] [Google Scholar]
- 21.de Graaff, E., de Vries, B. B., Willemsen, R., van Hemel, J. O., Mohkamsing, S., Oostra, B. A., and van den Ouweland, A. M. (1996) Am. J. Med. Genet. 64 302–308 [DOI] [PubMed] [Google Scholar]
- 22.Hammond, L. S., Macias, M. M., Tarleton, J. C., and Pai, G. S. (1997) Am. J. Med. Genet. 72 430–434 [PubMed] [Google Scholar]
- 23.Petek, E., Kroisel, P. M., Schuster, M., Zierler, H., and Wagner, K. (1999) Am. J. Med. Genet. 84 229–232 [PubMed] [Google Scholar]
- 24.Grasso, M., Faravelli, F., Lo Nigro, C., Chiurazzi, P., Sperandeo, M. P., Argusti, A., Pomponi, M. G., Lecora, M., Sebastio, G. F., Perroni, L., Andria, G., Neri, G., and Bricarelli, F. D. (1999) Am. J. Med. Genet. 85 311–316 [DOI] [PubMed] [Google Scholar]
- 25.Wohrle, D., Kotzot, D., Hirst, M. C., Manca, A., Korn, B., Schmidt, A., Barbi, G., Rott, H.-D., Poustka, A., Davies, K. E., and Steinbach, P. (1992) Am. J. Hum. Genet. 51 299–306 [PMC free article] [PubMed] [Google Scholar]
- 26.Gedeon, A. K., Baker, E., Robinson, H., Partington, M. W., Gross, B., Manca, A., Korn, B., Poustka, A., Yu, S., Sutherland, G. R., and Mulley, J. C. (1992) Nat. Genet. 1 341–344 [DOI] [PubMed] [Google Scholar]
- 27.Tarleton, J. Richie, R., Schwartz, C., Rao, K., Alysworth, A. S., and Lachiewicz, A. (1993) Hum. Mol. Genet. 2 1973–1974 [DOI] [PubMed] [Google Scholar]
- 28.Albright, S. G., Lachiewicz, A. M., Tarleton, J. C., Rao, K. W., Schwartz, C. E., Richie, R., Tennison, M. B., and Aylsworth, A. S. (1994) Am. J. Med. Genet. 51 294–297 [DOI] [PubMed] [Google Scholar]
- 29.Trottier, Y., Imbert, G., Poustka, A., Fryns, J.-P., and Mandel, J. L. (1994) Am. J. Med. Genet. 51 454–457 [DOI] [PubMed] [Google Scholar]
- 30.Gu, Y., Lugenbeel, K. A., Vockley, J. G., Grody, W. W., and Nelson, D. L. (1994) Hum. Mol. Genet. 3 1705–1706 [DOI] [PubMed] [Google Scholar]
- 31.Schmidt, M., Robertson, A., and Crawford, M. (1994) Am. J. Med. Genet. 51 451. [DOI] [PubMed] [Google Scholar]
- 32.Quan, F., Zonana, J., Gunter, K., Peterson, K. L., Magenis, R. E., and Popovich, B. W. (1995) Am. J. Hum. Genet. 56 1042–1051 [PMC free article] [PubMed] [Google Scholar]
- 33.Hirst, M., Grewal, P., Flannery, A., Slatter, R., Maher, E., Barton, D., Fryns, J.-P., and Davies, K. (1995) Am. J. Hum. Genet. 56 67–74 [PMC free article] [PubMed] [Google Scholar]
- 34.Quan, F., Grompe, M. Jakobs, P., and Popovich, B. W. (1995) Hum. Mol. Genet. 4 1681–1684 [DOI] [PubMed] [Google Scholar]
- 35.Schmucker, B., Ballhausen, W. G., and Pfeiffer, R. A. (1996) Hum. Genet. 98 409–414 [DOI] [PubMed] [Google Scholar]
- 36.Wolff, D. J., Gustashaw, K. M., Zurcher, V., Ko, L., White, W., Weiss, L., Van Dyke, D. L., Schwartz, S., and Willard, H. F. (1997) Hum. Genet. 100 256–262 [DOI] [PubMed] [Google Scholar]
- 37.Gronskov, K., Hjalgrim, H., Bjerager, M. O., and Brondum-Nielsen, K. (1997) Am. J. Hum. Genet. 61 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia, A. D., de Diego, Y., Oostra, B. A., Willemsen, R., and Mirta Rodriguez, M. (2000) Hum. Genet. 106 366–369 [DOI] [PubMed] [Google Scholar]
- 39.Fan, H., Booker, J. K., McCandless, S. E., Shashi, V., Fleming, A., and Farber, R. A. (2005) Am. J. Med. Genet. 136 214–217 [DOI] [PubMed] [Google Scholar]
- 40.De Boulle, K., Verkerk, A. J. M. H., Reyniers, E., Vits, L., Hendrickx, J., Van Roy, B., Van Den Bos, F., de Graaff, E., Oostra, B. A., and Willems, P. J. (1993) Nat. Genet. 3 31–35 [DOI] [PubMed] [Google Scholar]
- 41.Lugenbeel, K. A., Peier, A. M., Carson, N. L., Chudley, A. E., and Nelson, D. L. (1995) Nat. Genet. 10 483–485 [DOI] [PubMed] [Google Scholar]
- 42.Wang, Y.-C., Lin, M.-L., Lin, S. J., Li, Y.-C., and Li, S.-Y. (1997) Hum. Mutat. 10 393–399 [DOI] [PubMed] [Google Scholar]
- 43.Kosmider, B., and Wells, R. D. (2007) DNA Repair 6 1850–1863 [DOI] [PubMed] [Google Scholar]
- 44.Hirst, M. C., and White, P. J. (1998) Nucleic Acids Res. 26 2353–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edamura, K. N., and Pearson, C. E. (2005) Hum. Genet. 118 301–304 [DOI] [PubMed] [Google Scholar]
- 46.Wojciechowska, M., Bacolla, A., Larson, J. E., and Wells, R. D. (2005) J. Biol. Chem. 280 941–952 [DOI] [PubMed] [Google Scholar]
- 47.Wojciechowska, M., Napierala, M., Larson, J. E., and Wells, R. D. (2006) J. Biol. Chem. 281 24531–24543 [DOI] [PubMed] [Google Scholar]
- 48.Zuhlke, C. H., Dalski, A., Habeck, M., Straube, K., Hedrich, K., Hoeltzenbein, M., Konstanzer, A., Hellenbroich, Y., and Schwinger, E. (2004) Eur. J. Hum. Genet. 12 979–982 [DOI] [PubMed] [Google Scholar]
- 49.Meservy, J. M., Sargent, R. G., Iyer, R. R., Chan, F., McKenzie, G. J., Wells, R. D., and Wilson, J. H. (2003) Mol. Cell. Biol. 23 3152–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wells, R. D. (2007) Trends Biochem. Sci. 32 271–278 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.