Abstract
The dominant gain-of-function polyglutamine repeat diseases, in which the initiating mutation is known, allow development of models that recapitulate many aspects of human disease. To the extent that pathology is a consequence of disrupted fundamental cellular activities, one can effectively study strategies to ameliorate or protect against these cellular insults. Model organisms allow one to identify pathways that affect disease onset and progression, to test and screen for pharmacological agents that affect pathogenic processes, and to validate potential targets genetically as well as pharmacologically. Here, we describe polyglutamine repeat diseases that have been modeled in a variety of organisms, including worms, flies, mice, and non-human primates, and discuss examples of how they have broadened the therapeutic landscape.
Features of Polyglutamine Diseases
Polyglutamine repeat diseases are caused by the expansion of an unstable CAG repeat in the coding region of the respective disease gene (1), leading to an abnormally long polyQ3 tract, which causes dominant pathogenesis. These protein conformation diseases are particularly insidious because they typically become manifest later in life after having children. Nine such disorders have been described (1, 2), including HD, SBMA, DRPLA, and several ataxias (SCA1–3) (6–7, 17). Although the genes affected have divergent functions, polyQ diseases share several common features, including neuronal dysfunction and loss in the central nervous system. Clinical features include ataxia and other movement disorders, loss of cognitive ability, and psychiatric disabilities.
Brief Summary of PolyQ Proteins
Pathology associated with polyQ repeat disease impacts a range of cellular mechanisms, with both distinct and common themes. For instance, pathology is often accompanied by accumulation of the full-length protein or of proteolytically processed fragments containing the extended polyQ repeats (Fig. 1). Mechanistic insights have emerged from disease models that have informed mechanisms for another polyQ disease or that have proven consistent across diseases. A role for normal disease protein function has emerged: for ATXN1 (ataxin-1), the presence of the mutation within the protein modifies the cellular levels of at least two distinct protein complexes that are involved in mediating disease pathology (3), and normal activities of huntingtin (Htt) may be compromised (1, 4). Furthermore, mutant protein accumulation is often accompanied by mislocalization in the nucleus (e.g. expanded repeat Htt). A key role for post-translational modifications in pathogenesis has been revealed by studies in both SCA1 and HD (reviewed in Refs. 1, 5, and 6). Brains of SCA2 patients contain a small N-terminal polyQ-containing fragment that is associated with pathology (7), although SCA2 is the only polyQ disease in which the symptoms are not tightly associated with the accumulation of nuclear or cytoplasmic aggregates. In SCA3 or Machado-Joseph disease, the most common dominantly inherited ataxia worldwide, mutant ataxin-3 interacts with the ubiquitin protease cascade (reviewed in Refs. 1 and 8). SCA6 is caused by an expansion in the α1A calcium channel gene (CACNA1), which functions as a voltage-dependent Ca2+ channel protein that normally resides in the plasma membrane (1). ATX7, the gene affected in SCA7, is a subunit of the GCN5 histone acetyltransferase complex that may alter a matrix-associated nuclear structure and/or disrupt nucleolar function as well (1). SCA17, caused by mutant TATA-binding protein (1), highlights a role for transcription in the polyQ diseases. SBMA, caused by a glutamine-expanded androgen receptor gene (1), is one of the few polyQ repeat diseases (with SCA17) in which the normal function of the affected protein is well established. Disease is androgen-dependent and appears to involve translocation of the activated receptor to the nucleus, with profound therapeutic implications for reduction of androgen production or interference with androgen function (9).
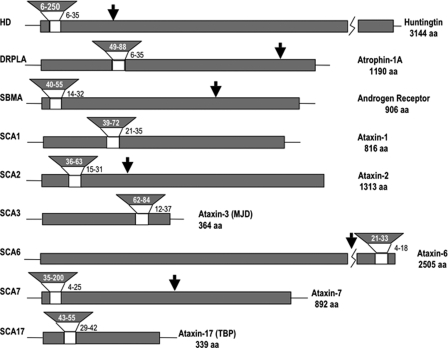
FIGURE 1.
Structures of polyQ disease proteins. The polyQ expansion is indicated by inverted wedges. The polyQ repeat size in normal individuals is adjacent to the unshaded portion, with disease range expansion in the shaded portion. Arrows above the diagram indicate the smallest claimed pathogenic fragments (10, 12, 19). aa, amino acids; MJD, Machado-Joseph disease; TBP, TATA-binding protein.
Why PolyQ Diseases Are Particularly Amenable to Development of Model Systems
PolyQ diseases are particularly amenable to modeling in a variety of animals because the triggering event, e.g. repeat expansion, is so clearly defined. Because they share a common feature of expanded polyQ, it has been tempting to speculate that a common pathophysiology underlies the polyQ disease processes; however, there is no direct proof for that assumption. Formally, disease could stem from either a common pathophysiology resulting from unique properties of expanded polyQ peptides or from altered function of each of the affected proteins caused by the incorporation of expanded polyQs. The observation that expanded polyQ peptides alone can cause neurodegenerative disease and pathology suggests that at least some of the pathophysiology is a consequence of the special properties of expanded polyQ (10). In particular, the amyloid structures formed by expanded polyQ proteins share a number of structurally distinct conformations consistent with other amyloidogenic proteins, and antibodies directed against different conformers can block cellular toxicity in cell culture systems (5, 6, 11). These observations suggest common structural contributions to pathophysiology.
Despite recent progress, questions remain as to the pathogenic agent, i.e. is it a particular conformational form, loss of normal function, a particular proteolytic fragment, or a particular modified form of the protein? Questions also remain as to the primary events that trigger pathogenesis versus compensatory or incidental cellular responses. Modeling them in a variety of animal systems has proven effective in elucidating the molecular and cellular consequences of these expansions that lead to disease.
Overview of Animal Models
Worms—Worms have been used to model HD, SCA3, and pure polyQ disease (Table 1) (reviewed in Ref. 12). Parker et al. (27) found impaired neuronal function in Caenorhabditis elegans caused by the overexpression of Htt exon 1 with 88 or 128 glutamine repeats. The neuronal dysfunction was polyQ length-dependent and was not accompanied by cell death. Faber et al. (26) also observed that dysfunction of C. elegans sensory neurons as a consequence of overexpression of Htt exon 1-Q150 was not associated with cell death; however, coexpression of a subthreshold dose of a toxic form of oncostatin M resulted in CED-3 caspase-dependent apoptotic cell death of the affected neurons. The Htt exon 1-Q150 model was also used to check the therapeutic effectiveness of potential drugs for treating HD (13).
TABLE 1.
Historical time line of polyQ repeat diseases mimicked in model organisms
To limit the number of references, each of the models listed is contained within the following reviews: Refs. 1, 8, 10, 12, 19, and 20. GFP, green fluorescent protein; YAC, yeast artificial chromosome; BAC, bacterial artificial chromosome.
| Year | C. elegans | Drosophila | Mouse | Primates |
|---|---|---|---|---|
| 1995 | Transgenic SCA1 (Burright et al.), transgenic SBMA (Bingham et al.) | |||
| 1996 | First HD model (R6/2 mouse), exon 1 expressed (Mangiarini et al.) | |||
| 1998 | SCA3 model (Warrick et al.), N171 Htt (first HD model) (Jackson et al.) | Full-length Htt cDNA (HD) (Reddy et al.), N171 Htt (HD) (Schilling et al.) | ||
| 1999 | Full-length Htt cDNA with 150Q (Faber et al.) | YAC full-length Htt (Hayden mouse) (Hodgson et al.), truncated DRPLA (Schilling et al.), full-length transgenic DRPLA (Sato et al.), HD knock-in (Levine et al.) | ||
| 2000 | PolyQ-GFP model (Satyal et al.) | SCA1 model (Fernandez-Funez et al.), polyQ model (Marsh et al.), polyQ model (Kazemi-Estarjani et al.) | Inducible exon 1 model (HD) (Yamamoto et al.), HD knock-in (Wheeler et al.) | |
| 2001 | PolyQ model (Parker et al.) | 1st third of Htt (Laforet et al.), transgenic SCA7 (La Spada et al.), truncated transgenic SBMA (Abel et al.), polyQ model (Adachi et al.), HD knock-in (Lin et al.) | ||
| 2002 | SMBA model (Takeyama et al.) | Knock-in SCA1 model (v et al.), YAC SCA3 model (Cemal et al.) | ||
| 2003 | Knock-in SCA1 model (Yoo et al.) | |||
| 2004 | Transgenic SCA3 (Goti et al.), YAC SBMA model (Sopher et al.) | |||
| 2005 | N117 short stop (Slow et al.) | |||
| 2006 | Transgenic SCA2 (Aguiar et al.) | |||
| 2007 | Transgenic SCA7 (LaTouche et al.) | |||
| 2008 | Full-length Htt (HD) (Romero et al.) | BAC HD model (Gray et al.) | HD exon 1 model (Yang et al.) |
In an SCA3 model, the expression of either truncated or full-length SCA3/Machado-Joseph disease protein throughout the C. elegans nervous system led to impaired synaptic transmission and a disrupted UPS, resulting in accumulation of polyQ aggregates and morphological abnormalities (14). Morimoto and co-workers (12, 15) developed a C. elegans model of the common features of polyQ diseases by expressing different lengths of polyQ stretches with no additional flanking sequences. They used this model to carry out an RNA interference modifier screen for interacting proteins and concluded that the polyQ diseases share a common property of protein aggregation/misfolding with several chaperones involved.
Flies—The first transgenic Drosophila model of a human neurodegenerative disease was SCA3, followed closely by a transgenic model of HD and of pure polyQ tracts (Table 1) (10). Fly models have significantly contributed to our growing understanding of the molecular bases of these diseases (e.g. HD, polyQ repeat, SCA1, SCA3, SCA7, and SBMA) (for reviews, see Refs. 16 and 17).
Dominant diseases are modeled by “humanizing” a fly so that transgenic animals express the expanded polyQ form of a human disease gene or its processed fragments. Transgenic flies containing a tissue-specific promoter fused to the yeast Gal4 transcription factor are crossed with flies containing the gene of interest fused to the yeast upstream activator sequence such that offspring will express the human disease gene only in selected tissues in a controlled manner (10). Typically, expression is driven in neurons (e.g. the elav driver) or in all cells of the eye (e.g. the gmr driver), although other drivers are also used, including glial-specific and neuron-specific drivers. Several readily quantifiable assays of neurotoxicity have been used, including measuring the loss of visible photoreceptor neurons in the eye, monitoring developmental lethality of the organism, measuring longevity (life span), and assaying a number of behavioral phenotypes such as motor function (for reviews, see Refs. 10, 16, and 18).
Mice—Mouse models for each of the polyQ diseases have been generated, including HD, SCA1–3, SCA7, SBMA, DRPLA, and polyQ (Table 1) (1, 19, 20), and recapitulate disease pathology with characteristic behavioral phenotypes that can be quantitatively assessed. For Htt, mice with different length Htt fragments as well as knock-in and knock-out models have been made (19), with historically the most widely used being the first transgenic model (R6/2) expressing a pathogenic exon 1 fragment with ∼150 repeats. Homozygous knock-out of the endogenous Htt gene in mice proved to be embryonic lethal and could be rescued by the mutant human gene (5, 6), indicating that expanded polyQ Htt still retains significant normal activity. Several full-length knock-in models of HD have been made (for review, see Ref. 19), although symptoms in these mice are milder. Yeast and bacterial artificial chromosome models expressing the full-length expanded human Htt gene have been generated and recapitulate aspects of disease (19, 21). Lentiviral injection of truncated mutant Htt into rat brain causes neurological phenotypes, thus allowing analysis in an animal with a larger brain mass (reviewed in Ref. 21).
A DRPLA model expressing full-length Atropin-1 (19) shows characteristic symptoms (e.g. ataxia, tremors, abnormal movements, seizures, and premature death) and accumulates an N-terminal fragment of the mutant protein in cell nuclei, indicating a possible role of proteolysis in the pathogenesis. Continuing the theme of protein accumulation in disease states, all of the proteins translated from various SCA transgenes were found to accumulate in the nucleus of cerebellar neurons with the sole exception of SCA2, which remained visibly localized in the cytoplasm in mouse brains (7).
Both truncated and full-length (19, 22, 23) forms of the expanded androgen receptor have been expressed in mice, displaying many of the phenotypes that are characteristic of SBMA, including its gender specificity (i.e. males are affected more severely than females), the presence of nuclear aggregates in neurons, and dependence on activation of the androgen receptor, raising the possibility for therapeutic intervention by modulating androgen activity. For example, leuprorelin (a luteinizing hormone-releasing hormone agonist that reduces testosterone release) was found to be highly beneficial in mice overexpressing the mutant protein (19).
Non-human Primates—HD has recently been modeled in a rhesus macaque (24) using a lentiviral vector to deliver green fluorescent protein-tagged Htt exon 1 with 84 CAG repeats into unfertilized monkey egg cells (Table 1). Interestingly, these initial proof-of-principle experiments indicate that the Htt transgenes used are extremely toxic, with some animals exhibiting movement dysfunction at birth or within a few days. This was quite a surprise given the more leisurely course of pathology observed with similar constructs in flies and mice. Although more work is needed, future generations of such animals may be extremely valuable in testing therapeutic strategies and addressing issues such as whether primates are uniquely sensitive to expanded polyQ insult.
Target Identification and Validation
Model organisms remain an essential tool in the search for therapeutic strategies for neurodegenerative diseases. They reliably recapitulate the various disease processes being studied, provide a more complete cellular context, and allow for quantitative analysis of behavior and neuropathological features of disease. The primary value of model organisms therefore lies in their ability to provide a cost- and time-effective means to perform two main functions: 1) target identification and 2) target validation by testing the value of specific therapeutic strategies and drugs in relieving disease severity.
Model Organisms in the Identification of Modifiers
Genes whose activities can be manipulated to affect pathogenesis represent potential therapeutic targets. The use of model organisms to identify potential disease modifiers is limited only by the imagination of the investigator, and model organisms have led to the identification of new therapeutic targets in several cases. For example, early studies in Drosophila revealed histone deacetylase inhibitors as useful in the treatment of HD (10). Subsequent studies demonstrated that similar strategies were effective in mouse models, and ongoing clinical trials are promising (1, 10, 19). Model organisms also allow the use of genetic screens to identify potential modifiers in an unbiased manner. For example, screens of Drosophila models of SCA1 and SCA3 identified loci that alter disease progression (10, 25); the impact of altered Apaf1 in affecting polyQ disease progression was demonstrated in flies (10); the role of the PQE protein was noted using worms (12, 26); and the attractiveness of HIP1 (huntingtin-interacting protein 1) in therapeutic strategies was noted using both worms and mice (27). The presence and repeat length of other endogenous polyQ repeat proteins are found to influence pathogenesis in Drosophila (10, 28). These examples illustrate the utility of multiple model organisms in facilitating the speed of preclinical testing.
Model organisms also offer a number of tools to identify and validate post-translational modifications such as phosphorylation, SUMOylation, proteolytic processing, and the pathways that regulate these processes. The importance of phosphorylation of SCA1 (reviewed in (1) and the relevance of Htt phosphorylation and kinase pathways were illustrated in flies and mice (29–31). The importance of lysine modifications has been demonstrated in flies for Htt (10). These examples suggest that these modification pathways may be productively targeted for therapeutic intervention.
Identifying Pathways as Targets
On a broader scale, model organisms allow the dissection of cellular pathways and the testing of their relevance to pathology. A number of studies in worms, flies, and mice have illustrated the role of chaperones in impacting polyQ diseases (1, 10, 32, 33), thus identifying several new therapeutic avenues. Expanding on this, Gidalevitz et al. (34) not only discovered new therapeutic strategies but importantly provided valuable new insight into protein homeostasis that has relevance for many diseases and for our understanding of basic cell biology. Their studies demonstrated that the presence of expanded polyQ proteins facilitates overall protein misfolding of cellular proteins. This link was recently confirmed in Drosophila through studies of HDAC6, suggesting that it may serve as a molecular link between the two pathways (35). Recent studies with a fly model of SCA3 (10) and mouse models of HD (36) point to the importance of global changes in the UPS in animals challenged with expanded mutant proteins and reveal new mechanisms of pathogenesis that relate to the previous global chaperone studies in worms (37). The use of model organisms to identify modifiers of polyQ repeat aggregation (2, 10, 37, 38) has led to a convergence of observations with different toxic challenges in different organisms (fly, worm, and mouse) and all pointing to global disruption of the UPS and lysosomal pathways in contributing to pathogenesis. These studies of polyQ pathology have led to a better understanding of the trafficking and interactions in cells between various degradative processes such as autophagy and the proteasome (1, 39, 40), and by identifying pathways critical to pathology, they invite further exploration of this avenue of pathogenic modulation.
A particularly exciting recent study used Drosophila to demonstrate that microRNA pathways modulate polyQ disease pathology, which raises new therapeutic avenues (41). Other recent studies using a Drosophila model of SCA3 also demonstrated a contribution of RNA to toxicity that is independent of translation (42) and demonstrated that toxicity decreases with imperfect repeats (e.g. CAG versus CAACAG). Using the power of genetics, this study was able to identify genetic modifiers of the RNA toxicity and demonstrate that both the fly muscleblind gene and the human counterpart can modulate translation-independent RNA toxicity in Drosophila.
Using the same Drosophila model of SCA3, Jung and Bonini (43) discovered that CAG repeat instability is noticeably enhanced by transcription and that it is modulated by nuclear excision repair. Most importantly, they found that altered levels of CBP (cAMP-responsive element-binding protein-binding protein), a histone acetyltransferase whose decreased activity is implicated in polyQ disease, also lead to altered DNA repair activity. Pharmacological treatment to normalize acetylation suppresses instability. Thus, toxic consequences of pathogenic polyQ protein may include enhancing repeat instability. Such experiments would have been extremely costly and time-consuming to perform in mammals and illustrate powerfully how the tools available in different model organisms can be productively used to open a whole new landscape of potential therapeutic intervention.
Target Validation
Model organisms are an essential tool for validating the results of high-throughput screening for potential therapeutic targets. Although extremely attractive for their high volume and direct testing of compounds, high-throughput biochemical screens are all based on assumptions that affecting a particular biochemical process will have therapeutic value. It is absolutely essential that these assumptions be tested in vivo. For example, yeast two hybrid/co-immunoprecipitation screens for interacting proteins identified a number of putative targets that tested positively for therapeutic efficacy in Drosophila, which provided powerful validation for that screening approach (44). Other screens for modifying or interacting loci have proven valuable, e.g. Branco et al. for SCA1 and HD (50) and Lam et al. for SCA1 (51). Finally, high-throughput screening in cells with subsequent validation of targets in Drosophila has identified the profilin-actin assembly pathway as an attractive therapeutic target for several polyQ diseases (45, 46).
Disease models are also an essential component for validating potential drugs for further testing during preclinical studies. For example, a drug that exhibits its effect even when the presumed protein target has been removed genetically points to alternative drug targets that may be responsible for the in vivo effect. For example, genetic manipulations and small molecules in worms (47) and flies (48) have been used in combination to reveal the complex relationship between resveratrol and sirtuins and between longevity and neuroprotection. Worms and flies have also been widely used to screen for disease-suppressing drugs (Refs. 13 and 49 and reviewed in Ref. 10), and flies have been used to test for effective drug combinations in HD (10). An example of the effectiveness of this type of approach can be found with Dimebon (Medivation, Inc.), the safety and tolerability of which were originally well proven in humans for other indications. Following testing that demonstrated effectiveness in suppressing HD-like pathology in a Drosophila model,4 phase II clinical trials have produced promising results in terms of efficacy in improving cognitive function in HD patients. Such examples stand as powerful validation for the utility of animal models in validating therapeutic strategies. Overall, these and many other studies beyond the scope of this review demonstrate the power of model organisms to identify and validate targets and further demonstrate that such studies can be a very effective way to rapidly discover critical cellular mechanisms and to improve the likelihood of success in human trials of polyQ disease therapies.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants NS045283 (to J. L. M.) and NS52789 (to L. M. T.). This work was also supported by Hereditary Disease Foundation Grant HDF-24085, the Huntington's Disease Society of America, and the High Q Foundation/CHDI, Inc. This is the fifth of five articles in the Unstable Nucleotide Repeat Minireview Series. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: polyQ, polyglutamine; HD, Huntington disease; SBMA, spinal and bulbar muscular atrophy (Kennedy disease); DRPLA, dentatorubral-pallidoluysian atrophy; SCA, spinocerebellar ataxia; Htt, huntingtin; UPS, ubiquitin-proteasome system.
J. L. Marsh and L. M. Thompson, unpublished data.
References
- 1.Orr, H. T., and Zoghbi, H. Y. (2007) Annu. Rev. Neurosci. 30 575–621 [DOI] [PubMed] [Google Scholar]
- 2.Katsuno, M., Banno, H., Suzuki, K., Takeuchi, Y., Kawashima, M., Tanaka, F., Adachi, H., and Sobue, G. (2008) Curr. Mol. Med. 8 221–234 [DOI] [PubMed] [Google Scholar]
- 3.Lim, J., Crespo-Barreto, J., Jafar-Nejad, P., Bowman, A. B., Richman, R., Hill, D. E., Orr, H. T., and Zoghbi, H. Y. (2008) Nature 452 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattaneo, E., Zuccato, C., and Tartari, M. (2005) Nat. Rev. Neurosci. 6 919–930 [DOI] [PubMed] [Google Scholar]
- 5.Dorval, V., and Fraser, P. E. (2007) Biochim. Biophys. Acta 1773 694–706 [DOI] [PubMed] [Google Scholar]
- 6.Shao, J., and Diamond, M. I. (2007) Hum. Mol. Genet. 16 R115–R123 [DOI] [PubMed] [Google Scholar]
- 7.Huynh, D. P., Figueroa, K., Hoang, N., and Pulst, S. M. (2000) Nat. Genet. 26 44–50 [DOI] [PubMed] [Google Scholar]
- 8.Riess, O., Rub, U., Pastore, A., Bauer, P., and Schols, L. (2008) Cerebellum 7 125–137 [DOI] [PubMed] [Google Scholar]
- 9.Adachi, H., Waza, M., Katsuno, M., Tanaka, F., Doyu, M., and Sobue, G. (2007) Neuropathol. Appl. Neurobiol. 33 135–151 [DOI] [PubMed] [Google Scholar]
- 10.Marsh, J. L., and Thompson, L. M. (2006) Neuron 52 169–178 [DOI] [PubMed] [Google Scholar]
- 11.Kayed, R., and Glabe, C. G. (2006) Methods Enzymol. 413 326–344 [DOI] [PubMed] [Google Scholar]
- 12.Brignull, H. R., Morley, J. F., Garcia, S. M., and Morimoto, R. I. (2006) Methods Enzymol. 412 256–282 [DOI] [PubMed] [Google Scholar]
- 13.Voisine, C., Varma, H., Walker, N., Bates, E. A., Stockwell, B. R., and Hart, A. C. (2007) PLoS ONE 2 e504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan, L. A., Bauer, P. O., Miyazaki, H., Lindenberg, K. S., Landwehrmeyer, B. G., and Nukina, N. (2006) J. Neurochem. 98 576–587 [DOI] [PubMed] [Google Scholar]
- 15.Brignull, H. R., Moore, F. E., Tang, S. J., and Morimoto, R. I. (2006) J. Neurosci. 26 7597–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bilen, J., and Bonini, N. M. (2005) Annu. Rev. Genet. 39 153–171 [DOI] [PubMed] [Google Scholar]
- 17.Sang, T. K., Li, C., Liu, W., Rodriguez, A., Abrams, J. M., Zipursky, S. L., and Jackson, G. R. (2005) Hum. Mol. Genet. 14 357–372 [DOI] [PubMed] [Google Scholar]
- 18.Sang, T. K., and Jackson, G. R. (2005) NeuroRx 2 438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bates, G. P., and Gonitel, R. (2006) Mol. Biotechnol. 32 147–158 [DOI] [PubMed] [Google Scholar]
- 20.Ramaswamy, S., McBride, J. L., and Kordower, J. H. (2007) ILAR J. 48 356–373 [DOI] [PubMed] [Google Scholar]
- 21.Heng, M. Y., Detloff, P. J., and Albin, R. L. (2008) Neurobiol. Dis. 32 1–9 [DOI] [PubMed] [Google Scholar]
- 22.Katsuno, M., Adachi, H., Tanaka, F., and Sobue, G. (2004) J. Mol. Med. 82 298–307 [DOI] [PubMed] [Google Scholar]
- 23.Bingham, P. M., Scott, M. O., Wang, S., McPhaul, M. J., Wilson, E. M., Garbern, J. Y., Merry, D. E., and Fischbeck, K. H. (1995) Nat. Genet. 9 191–196 [DOI] [PubMed] [Google Scholar]
- 24.Yang, S. H., Cheng, P. H., Banta, H., Piotrowska-Nitsche, K., Yang, J. J., Cheng, E. C., Snyder, B., Larkin, K., Liu, J., Orkin, J., Fang, Z. H., Smith, Y., Bachevalier, J., Zola, S. M., Li, S. H., Li, X. J., and Chan, A. W. (2008) Nature 453 921–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bilen, J., and Bonini, N. M. (2007) PLoS Genet. 3 1950–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faber, P. W., Voisine, C., King, D. C., Bates, E. A., and Hart, A. C. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 17131–17136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker, J. A., Metzler, M., Georgiou, J., Mage, M., Roder, J. C., Rose, A. M., Hayden, M. R., and Neri, C. (2007) J. Neurosci. 27 11056–11064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Ramahi, I., Perez, A. M., Lim, J., Zhang, M., Sorensen, R., de Haro, M., Branco, J., Pulst, S. M., Zoghbi, H. Y., and Botas, J. (2007) PLoS Genet. 3 e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lievens, J. C., Iche, M., Laval, M., Faivre-Sarrailh, C., and Birman, S. (2008) Hum. Mol. Genet. 17 882–894 [DOI] [PubMed] [Google Scholar]
- 30.Pardo, R., Colin, E., Regulier, E., Aebischer, P., Deglon, N., Humbert, S., and Saudou, F. (2006) J. Neurosci. 26 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warby, S. C., Chan, E. Y., Metzler, M., Gan, L., Singaraja, R. R., Crocker, S. F., Robertson, H. A., and Hayden, M. R. (2005) Hum. Mol. Genet. 14 1569–1577 [DOI] [PubMed] [Google Scholar]
- 32.Al-Ramahi, I., Lam, Y. C., Chen, H. K., de Gouyon, B., Zhang, M., Perez, A. M., Branco, J., de Haro, M., Patterson, C., Zoghbi, H. Y., and Botas, J. (2006) J. Biol. Chem. 281 26714–26724 [DOI] [PubMed] [Google Scholar]
- 33.Landles, C., and Bates, G. P. (2004) EMBO Rep. 5 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gidalevitz, T., Ben-Zvi, A., Ho, K. H., Brignull, H. R., and Morimoto, R. I. (2006) Science 311 1471–1474 [DOI] [PubMed] [Google Scholar]
- 35.Pandey, U. B., Nie, Z., Batlevi, Y., McCray, B. A., Ritson, G. P., Nedelsky, N. B., Schwartz, S. L., DiProspero, N. A., Knight, M. A., Schuldiner, O., Padmanabhan, R., Hild, M., Berry, D. L., Garza, D., Hubbert, C. C., Yao, T. P., Baehrecke, E. H., and Taylor, J. P. (2007) Nature 447 859–863 [DOI] [PubMed] [Google Scholar]
- 36.Bennett, E. J., Shaler, T. A., Woodman, B., Ryu, K. Y., Zaitseva, T. S., Becker, C. H., Bates, G. P., Schulman, H., and Kopito, R. R. (2007) Nature 448 704–708 [DOI] [PubMed] [Google Scholar]
- 37.Brignull, H. R., Morley, J. F., and Morimoto, R. I. (2007) Adv. Exp. Med. Biol. 594 167–189 [DOI] [PubMed] [Google Scholar]
- 38.Boeddrich, A., Gaumer, S., Haacke, A., Tzvetkov, N., Albrecht, M., Evert, B. O., Muller, E. C., Lurz, R., Breuer, P., Schugardt, N., Plassmann, S., Xu, K., Warrick, J. M., Suopanki, J., Wullner, U., Frank, R., Hartl, U. F., Bonini, N. M., and Wanker, E. E. (2006) EMBO J. 25 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia, K., Hart, A. C., and Levine, B. (2007) Autophagy 3 21–25 [DOI] [PubMed] [Google Scholar]
- 40.Winslow, A. R., and Rubinsztein, D. C. (2008) Biochim. Biophys. Acta 1782 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bilen, J., Liu, N., Burnett, B. G., Pittman, R. N., and Bonini, N. M. (2006) Mol. Cell 24 157–163 [DOI] [PubMed] [Google Scholar]
- 42.Li, L. B., Yu, Z., Teng, X., and Bonini, N. M. (2008) Nature 453 1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung, J., and Bonini, N. (2007) Science 315 1857–1859 [DOI] [PubMed] [Google Scholar]
- 44.Kaltenbach, L. S., Romero, E., Becklin, R. R., Chettier, R., Bell, R., Phansalkar, A., Strand, A., Torcassi, C., Savage, J., Hurlburt, A., Cha, G. H., Ukani, L., Chepanoske, C. L., Zhen, Y., Sahasrabudhe, S., Olson, J., Kurschner, C., Ellerby, L. M., Peltier, J. M., Botas, J., and Hughes, R. E. (2007) PLoS Genet. 3 e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burnett, B. G., Andrews, J., Ranganathan, S., Fischbeck, K. H., and DiProspero, N. A. (2008) Neurobiol. Dis. 30 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao, J., Welch, W. J., DiProspero, N. A., and Diamond, M. I. (2008) Mol. Cell. Biol. 28 5196–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker, J. A., Arango, M., Abderrahmane, S., Lambert, E., Tourette, C., Catoire, H., and Neri, C. (2005) Nat. Genet. 37 349–350 [DOI] [PubMed] [Google Scholar]
- 48.Pallos, J., Bodai, L., Lukacsovich, T., Purcell, J. M., Steffan, J. S., Thompson, L. M., and Marsh, J. L. (2008) Hum. Mol. Genet. 17 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varma, H., Cheng, R., Voisine, C., Hart, A. C., and Stockwell, B. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14525–14530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Branco, J., Al-Ramahi, I., Ukani, L., Perez, A. M., Fernandez-Funez, P., Rincon-Limas, D., and Botas, J. (2008) Hum. Mol. Genet. 17 376–390 [DOI] [PubMed] [Google Scholar]
- 51.Lim, J., Crespo-Barreto, J., Jafar-Nejad, P., Bowman, A. B., Richman, R., Hill, D. E., Orr, H. T., and Zoghbi, H. Y. (2008) Nature 452 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.