Abstract
Pulmonary surfactant protein A (SP-A), a member of the collectin family, plays an important role in innate immune defense of the lung. In this study, we examined the role of SP-A in modulating complement receptor-mediated phagocytosis. Complement receptors (CR), CR3 (CD11b), and CR4 (CD11c) were expressed at reduced levels on the surface of alveolar macrophages from Sp-a–/– compared with Sp-a+/+ mice. Administration of intratracheal SP-A to Sp-a–/– mice induced the translocation of CR3 from alveolar macrophage intracellular pools to the cell surface. Intratracheal challenge with Haemophilus influenza enhanced CR3 expression on the surface of alveolar macrophages from Sp-a–/– and Sp-a+/+ mice, but relative expression remained lower in the Sp-a–/– mice at all time points post-inoculation. The effects of SP-A on macrophage and neutrophil CR3 redistribution between intracellular and cell surface pools were restricted to cells isolated from the lung. SP-A augmented CR3-mediated phagocytosis in a manner that was attenuated by N-glycanase or collagenase treatment of SP-A, implicating the N-linked sugar and collagen-like domains in that function. The binding of CR3 to SP-A was calcium dependent and mediated by the I-domain of CR3 and to a lesser extent by the CR3 lectin domain. Mapping of the domains of SP-A that were required for optimal binding to CR3 revealed that the N-linked sugars were more critical than the collagen-like domain or the extent of oligomeric assembly. We conclude that SP-A modulates the cell surface expression of CR3 on alveolar macrophages, binds to CR3, and enhances CR3-mediated phagocytosis.
The pulmonary collectins, surfactant proteins A and D, are host defense molecules that bind and aggregate bacterial, viral, fungal, and mycobacterial organisms (1, 2). Surfactant protein A (SP-A)2 is one of the most abundant proteins in the epithelial lining fluid, and in humans is expressed throughout the airway from cells comprising the tracheal-bronchial glands, the bronchiolar epithelium, and alveolar type II cells (3). SP-A functions as a broad spectrum opsonin to enhance phagocytosis of a range of pathogenic microbes, including Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Streptococcus pneumoniae, group B streptococci, Haemophilus influenzae, Klebsiella pneumoniae, and Mycoplasma pulmonis (2). SP-A also enhances phagocytosis through direct effects on the phagocytes. Pretreatment with SP-A increases macrophage uptake of E. coli, S. aureus, and P. aeruginosa (4, 5), indicating that SP-A also enhances phagocytosis through a direct effect on the alveolar macrophage. Fc receptor and CR1-mediated phagocytosis of S. aureus are enhanced in the presence of SP-A (6). SP-A promotes mannose receptor activity on alveolar macrophages thereby increasing K. pneumoniae phagocytosis (7). Scavenger receptor expression on alveolar macrophages is enhanced by SP-A resulting in increased uptake of S. pneumoniae (8). Thus, evidence is also accumulating that SP-A alters phagocytosis by augmenting the expression or accessibility of key cell surface receptors.
Activity against diverse microbial species by SP-A is based on pattern recognition by the C-type lectin domain, which binds to carbohydrate, lipid, and glycolipid species, which are abundantly expressed on the surface of pathogens. The scaffolding upon which 18 carbohydrate recognition domains are arrayed is formed by 6 trimers of monomers containing disulfide forming Cys residues in a short N-terminal segment, a triple helix forming collagen-like region, an α-helical coiled-coil forming hydrophobic “neck” region (9).
Complement is an important part of the innate immune system that aids in local inflammatory responses, the recruitment and activation of immune cells, clearance of immune complexes, and direct lysis of some types of bacteria. C3b and iC3b, the covalently bound cleavage fragments of C3, serve as opsonins. Complement receptor 1 (CR1, CD35) binds C3b, whereas CD11b (CR3) and CD11c (CR4) recognize iC3b to enhance phagocytosis of opsonized bacteria.
CR3, also known as Mac-1, CD11b/CD18, is a member of a leukocyte-specific β2-integrin subfamily, which also includes, LFA-1 (CD11a/CD18) and CR4 (CD11c/CD18). The β2-integrins consist of a common β subunit and homologous α subunits (CD11a-c) that bind noncovalently to form an αβ heterodimer (10). CR3 is expressed on mature neutrophils, monocytes, macrophages, and natural killer cells (11) and plays important roles in adhesion, phagocytosis, and oxygen radical production (12–14). All of these functions are impaired in macrophages from CR3-deficient (Cr3–/–) mice (15). iC3b, a complement activation product is an opsonin that binds bacterial pathogens and enhances CR3-mediated phagocytosis. CR3-mediated phagocytosis is important in clearance of group B streptococcus (GBS) (16) and H. influenzae (17), both important pathogens in childhood respiratory disease.
Like most integrins, CR3 is expressed in an inactive state on the cell surface but can switch rapidly and reversibly to the active physiologic ligand-binding state. The CR3 α-chain is involved in ligand recognition and consists of a short C-terminal cytoplasmic tail, a single transmembrane domain, and a long N-terminal extracellular domain. Ligands of CR3, including iC3b, fibrinogen, intracellular adhesion molecule-1 and 2 (ICAM-1 and ICAM-2), and heparin (18), bind to different sites in the integrin extracellular domain, the so-called insertion or I-domain found only in the α-subunit. The lectin site of CR3 is located C-terminal to the I-domain and binds polysaccharides containing mannose, N-acetyl-d-glucosamine, as well as glucose (19).
Phagocytosis and clearance of GBS, H. influenzae (20) and P. aeruginosa are impaired in SP-A gene-targeted mice (21). Although it is clear that reduced phagocytosis in Sp-a–/– mice is due in part to decreased opsonization, other mechanisms by which SP-A regulates innate host defense activities of alveolar macrophages remain unclear. The present study was undertaken to determine the role of CR3 in SP-A-mediated clearance of bacteria from the lung.
EXPERIMENTAL PROCEDURES
Animal Husbandry—The murine SP-A gene locus was targeted by homologous recombination and maintained on a 129 background as previously described. The lungs of Sp-a–/– mice lack detectable SP-A protein (20). Animals were housed and studied under Institutional Animal Care and Use Committee-approved protocols in the animal facility of the Children's Hospital Research Foundation, Cincinnati, OH. Male and female mice of ∼20–25 g (42–56 days old) were used, and all comparisons made were matched for strain and age.
Preparation of Bacteria and Labeling with Fluorescein—A stock culture of GBS and H. influenzae were obtained from clinical isolates. Bacteria for intratracheal injections were prepared as previously described (20). Mice were infected with 1 × 106 cfu of GBS or 1 × 108 cfu of H. influenzae. Bacteria for labeling were grown overnight in tryptic soy broth, pelleted, resuspended in 0.9 ml of phosphate-buffered saline (PBS), and heated to 95 °C for 10 min to kill the bacteria. The heat-killed bacteria were pelleted and resuspended in 1 ml of 0.1 m sodium bicarbonate, pH 9.0. Fluorescein isothiocyanate (FITC, Molecular Probes, Eugene, OR) was added as a 10 mg/ml stock in dimethyl sulfoxide to a final concentration of 0.01 mg/ml, and the suspension was incubated for 1 h in the dark at room temperature with gentle agitation. Labeled bacteria were washed four times with PBS, pH 7.2, to remove unconjugated fluorophore, and finally resuspended in PBS and stored in aliquots of 100 μl at –80 °C.
Complement Receptor Expression—Alveolar macrophages were recovered by bronchoalveolar lavage (BAL). Lungs of uninfected and infected (2 h post-infection) mice were lavaged three times with 1 ml of sterile PBS. H. influenzae (108 cfu) was instilled intratracheally and alveolar macrophages harvested by BAL 2, 6, 12, and 24 h later. Cells from BAL were washed and incubated in FACS buffer (PBS, 0.2% bovine serum albumin fraction V, 0.02% sodium azide) with rat anti-mouse CD16/CD32 antibodies (Fc Block). Separate aliquots were stained with FITC-conjugated mouse CD11b (CR3) or CD11c (CR4) antibodies (BD Pharmingen) for 1 h on ice. Total and external CR3 were determined on cells permeabilized with cytoperm fix solution containing 10 mm Tris-HCl, pH 7.4, 1% Triton X-100, 0.5% Nonidet P-40, and 150 mm NaCl (Cytofix/cytoperm kit, BD Pharmingen). Cell-associated fluorescence was measured on a FACScan flow cytometer, using CELLQuest software (BD Biosciences). For each sample 10,000 events were analyzed and the results were expressed as mean fluorescence intensity. Internal CD11b was determined by subtracting external from total CD11b.
SP-A Preparation—Human SP-A (hSP-A) obtained from patients with alveolar proteinosis was purified by the 1-butanol extraction method of Haagsman et al. (22) and dissolved in HEPES buffer, pH 7.2. Endotoxin contamination was determined with the Limulus Amoebocyte Lysate assay (Sigma) according to the manufacturer's directions, and was less than 60 pg/μl in all hSP-A preparations (<0.06 EU/ml). CR3 expression on alveolar macrophages was determined 24 h after intratracheal injection of 100 μg of SP-A. Collagenase-treated hSP-A was prepared by incubating hSPA (100 μg) in 50 mm Tris-HCl, 1 mm EDTA, 100 mm NaCl, 0.36 mm CaCl2, pH 7.4, buffer containing 187.5 units of collagenase (Sigma) at 37 °C for 72 h. Deglycosylated SPA (100 μg) was prepared by incubating hSP-A in 100 mm NaH2PO4, 10 mm Na2EDTA, pH 6.1, buffer containing 1000 units of peptide N-glycosidase F (New England BioLabs, Ipswich, MA) at 37 °C for 72 h. Samples were then centrifuged in a 30-kDa molecular mass cutoff Centricon column (Millipore, Billerica, MA) and digests were analyzed by SDS-PAGE on a 10–20% Tris glycine gel. The synthesis and purification of the mutant rat recombinant SPA proteins used in this study were as previously described (23). The mutant rSP-A proteins were synthesized in insect cells using baculovirus vectors and purified by affinity chromatography with mannose-Sepharose 6B. SP-A was fluorescently labeled by incubating 1 μg of rSP-A with 0.1 m sodium bicarbonate, pH 9.0, 0.03 μg of Alexa Fluor 488 carboxylic acid succinimidyl ester (Molecular Probes) at room temperature for 3 h followed by dialysis against 5 mm HEPES buffer, pH 7.2, for 72 h at 4 °C to remove unbound label.
CD11b Expression on RAW 264.7 Cells—RAW 264.7 murine macrophage cells were plated at a concentration of 5 × 105 cells per well the day prior to the experiment. The media was removed and cells were washed twice with PBS and 10 or 50 μg/ml of hSP-A added in serum-free media. Following 2 h of incubation at 37 °C, cells were washed twice more with PBS, resuspended in FACS buffer, and stained for CR3 and subjected to flow cytometry for measurement of cell-associated fluorescence.
Peritoneal Macrophages—Peritoneal macrophages were isolated by peritoneal lavage with 5 ml of Dulbecco's modified Eagle's media (Invitrogen) three times, washed with PBS twice, and resuspended in FACS buffer and stained for CR3 as described for BAL macrophages. Cell-associated fluorescence was measured on a FACScan flow cytometer. For each sample 10,000 events were analyzed using CELLQuest software (BD Biosciences) and the results expressed as mean fluorescence intensity.
Neutrophil CR3—Because neutrophils are present in low abundance in the unchallenged lung, mice were infected with a high concentration of H. influenzae (1 × 1010 cfu) to recruit large numbers of neutrophils to the airspace and then lavaged 6 h later (>90% neutrophils in BAL fluid 6 h after infection). Blood was also collected from the inferior vena cava at the same time from challenged mice. Neutrophils from BAL and blood were separated on discontinuous Percoll gradients (Amersham Biosciences) of 1.0865 and 1.1105 g/ml. The gradient was centrifuged at 400 × g for 40 min at 25 °C, and the neutrophil layer was removed and washed twice with Hanks balanced salt solution. CR3 expression on neutrophils was determined as described for alveolar macrophages.
CR3-mediated Phagocytosis in Vitro—A CHO cell line stably expressing human wild-type CR3 called CHO-CR3 cells was kindly provided by Timothy A. Springer, Harvard Medical School (12). The cell line was generated by cotransfecting CHO cells (CHO DG44; containing a deletion of the dihydrofolate reductase gene) with cDNAs for CD11b and CD18 (each in the vector pCDM8) and with the amplifiable vector pCHIP, encoding methotrexate resistance. CHO-K1 cells (CHO-WT) were maintained in α-minimal essential media-supplemented with glutamine, penicillin G, streptomycin, and 10% heat inactivated fetal bovine serum. CHO-CR3 cells were maintained in the same media with the addition of 0.05 μm methotrexate. CHO-WT cells and CHO-CR3 cells were stimulated with PMA (600 ng/ml) to activate CR3 for 30 min followed by incubation with FITC-labeled H. influenzae (1:100, cell:bacteria) for 2 h in the presence of iC3b (3 ng/ml), hSP-A (20 μg/ml), iC3b and hSP-A, collagenase-treated hSP-A (20 μg/ml), or deglycosylated hSPA (20 μg/ml). In addition, phagocytosis experiments were also performed in the presence and absence of 4 mm CaCl2. Cells were incubated in buffer containing 2 mg/ml of trypan blue for 3 min to quench fluorescence of extracellular FITC and eliminate fluorescence associated with bacteria attached to the external surface of the cells. Cell-associated fluorescence was measured on at least 10,000 cells on a FACScan flow cytometer, using CELLQuest software. The results are expressed as fold-increase in mean fluorescence intensity over CHO-WT cells (control).
SP-A and CR3 Co-immunoprecipitation—The Seize® Primary Immunoprecipitation Kit (Pierce) was used for immunoprecipitation of SP-A according to the manufacturer's protocol. Briefly, peritoneal macrophages were collected, lysed in cell lysis buffer (10 mm Tris-HCl, pH 7.5, 15 mm NaCl, 0.5% Nonidet P-40, and 1% Triton X-100 containing protease inhibitor) and incubated overnight at 4 °C with a saturating amount of SP-A. SP-A antibody (Santa Cruz Biotechnology, Santa Cruz, CA), or preimmune serum (control) was added to the Amino-Link® Plus coupling gel suspension and incubated overnight at 4 °C. Following washes, the peritoneal macrophage lysates preincubated with SP-A or control lysates were added to the gel with coupling buffer, pH 7.2, and incubated overnight at 4 °C. Immunoprecipitation buffer was added followed by the Immuno-Pure® IgG elution buffer. Equal amounts of protein were loaded and separated by electrophoresis on a reducing SDS-PAGE gel, transferred to a nitrocellulose membrane, immunoblotted with anti-mouse CD11b antibody (BD Pharmingen), and appropriate secondary antibody conjugated to horseradish peroxidase, and developed by the enhanced chemiluminescence method (Amersham Biosciences).
Binding of SP-A to CR3—SP-A or iC3b were incubated overnight at 4 °C in 100 μl of 0.1 m sodium bicarbonate, pH 8.5, in 96-well plates. The plates were washed twice with Tris-buffered saline (TBS), pH 7.4, and treated with 5% bovine serum albumin and 5% human albumin in TBS for 20 min at room temperature followed by two additional washes with TBS. CHO-WT or CHO-CR3 cells were stimulated with PMA (100 ng/ml) to activate CR3 for 30 min at 37 °C in Dulbecco's modified Eagle's media supplemented with 4.0 mm calcium. Cells were centrifuged at 160 × g for 10 min and washed twice with TBS. The stimulated cells were resuspended in the plating buffer (5 mm HEPES, 0.1% bovine serum albumin in Tyrode's salt solution) and the cell suspension added to 96-well plates previously coated with hSP-A, deglycosylated hSP-A, collagenase-treated hSP-A or iC3b. The plates were then incubated for 1 h at 37 °C with 5% CO2, washed twice with TBS, and the number of bound cells determined by the release of alkaline phosphatase compared with a standard curve generated with a known number of CHO cells. Lysis buffer (100 μl) consisting of 1 m diethanolamine, pH 8, 5 mm MgCl2, 1.5 mm ZnCl2, and 11.5 mm substrate nitrophenyl phosphate disodium salt hexahydrate was added to each well and incubated for 1 h at 37 °C. Color change was read on a Biotech Synergy HT plate reader (Biotek Instruments, Winooski, VT) at an OD of 410 nm. Binding inhibition studies were performed with β-glucan 0 to 1000 μg/1 × 106 cells, or the antibodies M1/70 (Abcam, Cambridge, MA) at 5 μg/1 × 106 cells or CBRM 1/5 (BioLegend, San Diego, CA) at 50 μg/1 × 106 cells. Binding of SP-A to CHO-CR3 cells was also determined by flow cytometry. Cells in Dulbecco's modified Eagle's media with 4 mm Ca2+ were stimulated with 600 ng/ml of PMA for 30 min at 37 °C and incubated with 20 μg/ml of Alexa Fluor 488-labeled untreated hSP-A, collagenase, or N-glycosidase-treated hSP-A, or rSP-A with various mutations for 2.5 h at 37 °C. The cells were then washed five times with PBS resuspended in FACS buffer and analyzed on a FACScan flow cytometer, using CELLQuest software (BD Biosciences). Results were expressed as mean fluorescence intensity of 10,000 events.
Statistical Methods—Receptor expression, phagocytosis, and binding were compared using analysis of variance and the Student's t test. Findings were considered statistically significant at probability levels <0.05.
RESULTS
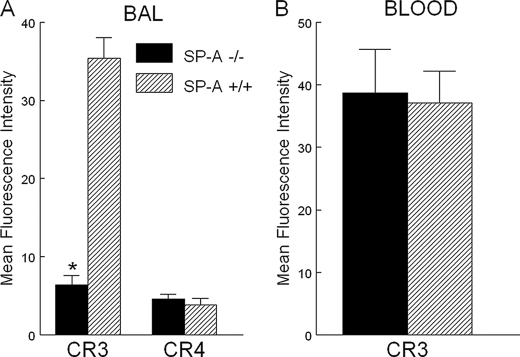
Decreased Complement Receptors on Alveolar Macrophages from Sp-a–/– Mice—Complement receptor expression was determined before and after intratracheal inoculation with H. influenzae. The inoculum sizes of GBS (104 cfu) and H. influenzae (108 cfu) used in the animal experiments were determined based on previous studies (20). Intratracheal administration of bacteria was well tolerated and all animals survived the 24-h study period. BAL cells at baseline consisted of >95% macrophages. The expression of complement receptors CR3 and CR4 was lower on alveolar macrophages isolated from uninfected Sp-a–/– mice than Sp-a+/+ mice, and was further reduced 2 h following GBS or H. influenzae infection (Fig. 1, A and B). Complement receptor expression on Sp-a+/+ alveolar macrophages also fell 2 h post-GBS or H. influenzae infection, but remained greater than Sp-a–/– alveolar macrophages in both cases. The time course of alveolar macrophage CR3 expression after H. influenza inoculation into Sp-a–/– or Sp-a+/+ mice is shown in Fig. 1C. Two hours after H. influenzae infection, CR3 expression was slightly decreased from baseline on alveolar macrophages from both Sp-a–/– and Sp-a+/+ mice. From 2 to 6 h following H. influenzae infection, CR3 expression increased markedly on alveolar macrophages from Sp-a–/– and Sp-a+/+ mice. A further increase in CR3 expression was observed on alveolar macrophages from Sp-a+/+ mice between 12 and 24 h after infection. In contrast, there was no incremental enhancement of CR3 expression on alveolar macrophages between 12 and 24 h following H. influenzae infection in Sp-a–/– mice. Despite the steady increase in CR3 expression from baseline in the H. influenzae-infected mice, alveolar macrophages from Sp-a–/– mice expressed significantly less CR3 at all time points compared with Sp-a+/+ mice (Fig. 1C). Although the cell surface expression of CR3 was lower on alveolar macrophages from Sp-a–/– mice than Sp-a+/+ mice, the internal pools of CR3 were increased in alveolar macrophages from Sp-a–/– mice, (Fig. 1D) and there was no difference in the total cellular pool sizes of CR3 between Sp-a–/– and Sp-a+/+ alveolar macrophages. CR3 protein expression determined by Western blot analysis of alveolar macrophage lysates also revealed that total CR3 was similar between Sp-a–/– and Sp-a+/+ alveolar macrophages (data not shown). These results support a role for SP-A in the regulation of CR3 expression on alveolar macrophages.
FIGURE 1.
SP-A-modulated cell surface expression of complement receptors on alveolar macrophages. Alveolar macrophages were recovered by bronchoalveolar lavage, incubated with the indicated fluorescently labeled anti-complement receptor antibody, and analyzed by flow cytometry. Decreased surface expression of CR3 (panel A) and CR4 (panel B) was observed on alveolar macrophages from Sp-a–/– (solid bar) compared with Sp-a+/+ (hatched bar) mice in uninfected, GBS-infected, and H. influenzae (H FLU)-infected mice. CR3 expression on alveolar macrophages was assessed at intervals for 24 h following H. influenzae infection (104 cfu). CR3 expression on alveolar macrophages fell slightly over the first 2 h post-infection and then increased through 6 h. CR3 expression on AM from Sp-a–/– mice (circle) reached a plateau at 6–24 h, but was significantly less at every time point following H. influenzae infection compared with Sp-a+/+ (triangle) mice (panel C). Total CR3 was similar for Sp-a–/– (solid bar) and Sp-a+/+ (hatched bar) alveolar macrophages and the internal pools of CR3 were increased in alveolar macrophages from Sp-a–/– mice (panel D). Data are mean ± S.E. with n = 8 mice per group, *, p < 0.05 compared with Sp-a+/+ mice for all panels.
Peritoneal Macrophage CR3 Expression Is Similar in Sp-a–/– and Sp-a+/+ Mice—The effect of SP-A on cell surface CR3 expression on macrophages obtained by peritoneal lavage was determined by flow cytometry. CR3 expression was similar for Sp-a–/– and Sp-a+/+ peritoneal macrophages (91.37 ± 1.2 compared with 94.0 ± 0.7 mean fluorescence intensity, respectively, mean ± S.E.). These data suggest that SP-A regulation of CR3 expression is restricted to macrophages isolated from the alveolar compartment.
Decreased CR3 Expression on BAL Neutrophils from Sp-a–/– Mice—To determine whether neutrophil CR3 expression is reduced in the absence of SP-A, neutrophils were isolated from BAL and blood of H. influenzae-infected mice and CR3 expression was determined by flow cytometry. Six h following intratracheal inoculation, neutrophils isolated from the BAL of Sp-a–/– mice expressed much less CR3 compared with alveolar neutrophils isolated from Sp-a+/+ mice (Fig. 2A). CR4 expression, in contrast, was similar on BAL neutrophils isolated from H. influenzae-infected Sp-a–/– and Sp-a+/+ mice. Interestingly, blood neutrophils harvested synchronously with BAL neutrophils from H. influenzae-infected Sp-a–/– and Sp-a+/+ mice, expressed similar levels of CR3 (Fig. 2B). Because the neutrophils in BAL fluid are recruited from the blood, these results support the concept that SP-A regulates CR3 upon entry to the alveolar compartment.
FIGURE 2.
SP-A-modulated cell surface expression of CR3 but not CR4 on alveolar neutrophils. Six hours following H. influenzae infection, neutrophils were isolated from BAL and CR3 and CR4 expression was determined by flow cytometry. Sp-a–/– BAL neutrophils (solid bar) expressed much less CR3 on the cell surface than Sp-a+/+ BAL neutrophils (hatched bar). BAL neutrophil CR4 expression was similar for Sp-a–/– and Sp-a+/+ mice (panel A). In contrast, blood neutrophils obtained synchronously with BAL neutrophils 6 h following H. influenzae infection expressed similar CR3 levels in Sp-a–/– and Sp-a+/+ mice (panel B). Data are mean ± S.E. with n = 6 mice per group, *, p < 0.05 compared with Sp-a+/+ mice for all panels.
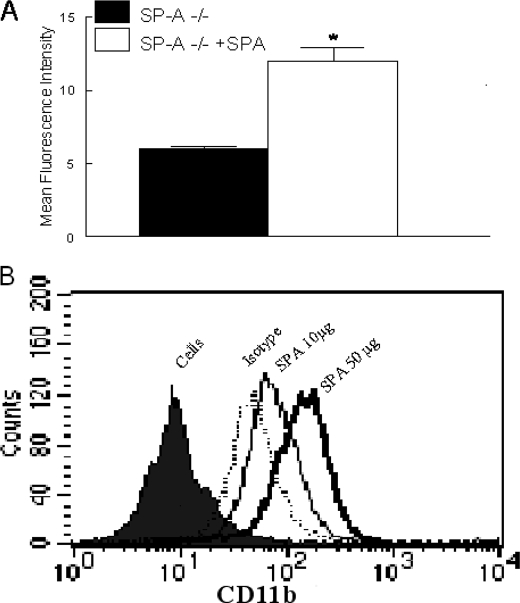
Exogenous SP-A Increases Alveolar Macrophage CR3 in Sp-a–/– Mice—Experiments were performed to determine whether SP-A can acutely modulate CR3 expression. Sp-a–/– mice were treated with intratracheal hSP-A and alveolar macrophages were recovered by BAL 1 day later. CR3 expression was determined by flow cytometry. CR3 expression was significantly enhanced on alveolar macrophages isolated from hSP-A-treated Sp-a–/– mice compared with untreated Sp-a–/– mice (Fig. 3A). In in vitro experiments, RAW 264.7 cells were incubated with various concentrations of hSP-A for 2 h and CR3 expression was determined by flow cytometry. SP-A also enhanced CR3 expression on RAW cells in a dose-dependent manner (Fig. 3B).
FIGURE 3.
Exogenous SP-A enhanced CR3 expression on alveolar macrophages from Sp-a–/– mice. CR3 expression on alveolar macrophages was determined by flow cytometry 24 h after intratracheal injection of 100 μg of SP-A. SP-A treatment significantly enhanced expression of CR3 on alveolar macrophages from Sp-a–/– mice (open bar) compared with untreated Sp-a–/– (solid bar) mice (panel A). Data are mean ± S.E. with n = 8 mice per group, *, p < 0.05 compared with untreated Sp-a–/– mice. RAW cells, a mouse macrophage cell line, were treated with SP-A and CR3 surface expression determined by flow cytometry. SP-A enhanced surface expression of CR3 in a dose-dependent manner (panel B).
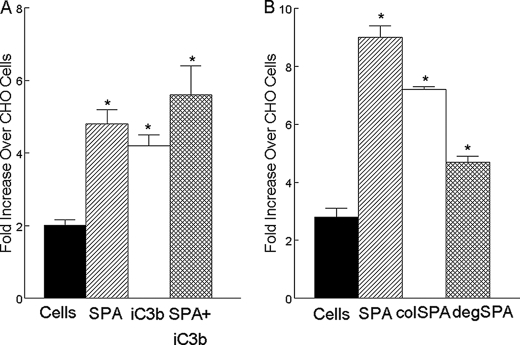
SP-A Enhances CR3-mediated Phagocytosis—The role of SP-A and iC3b in CR3-mediated internalization of bacteria into CHO cells was assessed. Phagocytosis of H. influenzae was greater in CHO-CR3 compared with CHO-WT cells. iC3b, a known opsonin and ligand for CR3, further enhanced CR3-mediated phagocytosis of H. influenzae in the CHO-CR3 cells. SP-A enhanced CR3-mediated phagocytosis by CHO-CR3 cells to an extent that was similar to iC3b. The combination of SP-A and iC3b was not significantly more effective than either component alone in enhancing phagocytosis (Fig. 4A). PMA was required to enhance phagocytosis with iC3b or SP-A by CHO-CR3 cells (data not shown). SP-A enhanced CR3-mediated phagocytosis was similar in the presence and absence of calcium (data not shown). Removal of the asparagine-linked carbohydrate moieties or the collagen-like region from hSP-A using N-glycosidase or collagenase, respectively, diminished the phagocytosis of H. influenzae in CHO-CR-3 cells relative to hSP-A. We conclude that SP-A augments CR3-dependent phagocytosis that is dependent on the active CR3 conformation, and both the collagen like and N-linked sugar domains of SP-A, but not on the presence of calcium.
FIGURE 4.
SP-A enhanced CR3-mediated phagocytosis. The effects of SP-A and iC3b on the phagocytosis of FITC-labeled H. influenzae were determined in CHO-WT cells and CHO-CR3 cells. In the absence of opsonins, the phagocytosis of H. influenzae was greater in CHO-CR3 cells (solid bar) compared with CHO-WT cells. iC3b, a known opsonin and ligand for CR3, enhanced CR3-mediated phagocytosis of H. influenzae in cells that expressed CR3 (open bar, panel A). SP-A enhanced CR3-mediated phagocytosis in CR3 expressing cells (hatched bar, panel A). There was no additive or synergistic effect of SP-A and iC3b on phagocytosis, the combination enhanced CR3-mediated phagocytosis to a similar extent as SP-A or iC3b alone (cross-hatched bar, panel A). SP-A-mediated enhancement of phagocytosis of H. influenzae into CR3 expressing cells was reduced by pre-treatment of SP-A with collagenase (colSPA) or N-glycanase (degSPA), suggesting roles for oligomeric assembly, the collagen like domain and N-linked carbohydrate in this effect (panel B). Data are mean ± S.E. with n = 4 experiments per group, *, p < 0.05 compared with phagocytosis by CHO-CR3 cells.
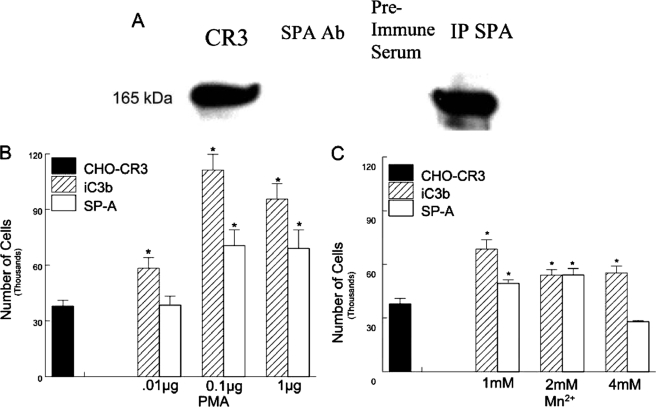
SP-A Binds CR3—To determine whether SP-A interacts with CR3, co-immunoprecipitation experiments were performed. Immunoprecipitation of SP-A using an anti-SP-A antibody from reaction mixtures containing SP-A, peritoneal macrophage lysates followed by immunoblotting with an anti-CR3 antibody, revealed a band consistent with CR3. No CR3 band was apparent from similar immunoprecipitation reactions mixtures in which SP-A was omitted or preimmune serum was substituted for the anti-SP-A antibody (Fig. 5A). To confirm this finding, coated plates with SP-A or iC3b were incubated with CHO-CR3 cells and cell binding determined following lysis of bound cells and determining endogenous alkaline phosphatase release. CHO-CR3 cells bound to iC3b or SP-A were adsorbed to plates, and binding was enhanced in a dose-dependent manner following activation of CR3 with PMA. Mn2+ also enhanced binding of CHO-CR3 cells to plates, but the interaction was not dose dependent (Fig. 5, B and C). The binding of SP-A to CR3 required calcium (data not shown).
FIGURE 5.
Binding of SP-A to CR-3 requires conversion to the active conformation. The interaction of SP-A with CR3 was determined by co-immunoprecipitation. A, in the first lane is an immunoblot of peritoneal cell lysates, the second lane is a control in which SP-A was omitted from incubation with the peritoneal lysate, the third lane shows preimmune serum rather than an anti-SP-A antibody was coupled to the gel suspension that was used for immunoprecipitation, and the fourth lane is an anti-SP-A antibody coupled to a gel suspension shown to co-immunoprecipitate CR3, detected by immunoblot analysis. Binding of SP-A to CR3 was determined in a whole cell assay, in which bound cells are detected by cell lysis and the quantification of associated alkaline phosphatase. Concentrations of PMA from 0.1 to 1 μg were required for sufficient activation of CR3 to bind SP-A (B). Binding of SP-A to CR3 was also enhanced by 1–2 mm Mn2+ (C). iC3b was used as a control. Data are mean ± S.E. of the total number of cells bound with n = 4 experiments per group, *, p < 0.05 compared with binding to non-stimulated CHO-CR3 cells.
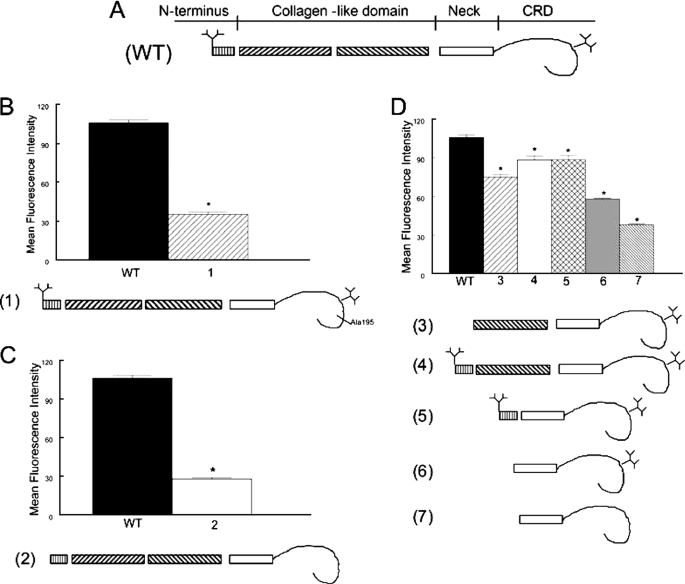
Deletion of the N-Linked Carbohydrates of SP-A Reduces Binding to CR3—The primary structure of rat SP-A is characterized by four sequential domains including a short N-terminal segment, a collagen-like region, a hydrophobic neck domain and a globular C-terminal carbohydrate recognition domain (CRD). Wild-type recombinant SP-A is relatively deficient in hydroxyproline within the collagen-like domain and is modified with less complex, mannose-rich N-linked carbohydrate structures compared with mammalian SP-A, but it is functionally similar to native rat SP-A with respect to interaction with most ligands (24). A panel of mutant recombinant SP-A forms was used to map the domains of SP-A that ligate CR3 (Fig 6). Binding of recombinant rat SP-A (rSP-A) to CR3 was somewhat reduced compared with native human SP-A (106* ± 2 versus 151 ± 15 mean fluorescent intensity, respectively, *, p < 0.05 compared with hSP-A), probably as a result of the incomplete oligomeric assembly and simplified carbohydrate modifications that occur in the invertebrate expression system. Deletion of a portion of the collagen domain or the entire collagen domain of SP-A had minimal effects on binding to CR3 provided that the N-linked carbohydrates of SP-A were present (Fig. 6D, 3 and 5). A mutation in the CRD, which is known to markedly impair the lectin function of SP-A (E195A), greatly reduced binding to CR3 (Fig. 6B). The individual contribution of the N-terminal and C-terminal N-linked carbohydrate domains to SP-A/CR3 binding was tested with mutant SP-As containing single (N1T or N187S, respectively) mutations in the consensus sequence for carbohydrate attachment (not shown). An incremental decrease in binding of SP-A to CR3 was observed with loss of each (N-and C-terminal) N-linked carbohydrate moiety, with the greatest reduction in binding occurring with loss of both N-linked carbohydrates (tandem N1T, N187S mutations) (Fig. 6C). Taken together, the data suggests that both the CRD and the N-linked carbohydrates of SP-A contribute to binding to CR3. The partial CR3 binding, which is retained by the aglyco neck and CRD fragment (Fig. 6D, 7) and after inactivation of the SP-A lectin domain (Fig. 6B) suggest that the neck domain may also play a role in binding. Attempts to introduce truncating mutations into the neck region of SP-A to test this hypothesis were not successful; all mutations resulted in misfolded protein forms that were rapidly degraded. The possibility that SP-A binds to CR3 through a non-lectin interaction with the CRD also cannot be excluded.
FIGURE 6.
Multiple domains of SP-A contribute to binding to CR3. The interaction of fluorescein-labeled SP-A with CR3 was determined by flow cytometry. The structure of the monomeric subunit of wild-type (WT) recombinant SP-A is shown in panel A. Note the branching N-linked carbohydrates attached to the N terminus and the CRD of the protein. In panel B, an E195A mutation that incapacitates the lectin function (1) of the CRD greatly reduced binding of SP-A to CR3. In panel C, tandem N1T and N187S mutations of SP-A, which prevent attachment of N-linked carbohydrates (2) also markedly reduced binding of SP-A to CR3. In panel D, truncated SP-A proteins lacking the N-terminal domain, the N-terminal N-linked carbohydrate attachment site, and first half of the collagen domain but possessing an intact Asn187 carbohydrate attachment (3), lacking the first half of the collagen domain but possessing the N-terminal segment, and both N-linked carbohydrate moieties (4), or lacking the collagen domain but possessing the N-terminal segment, and both carbohydrate moieties (5) retained significant CR3 binding activity. Mutations that resulted in a deletion of the N-terminal segment, the collagen-like domain, and the N-terminal (N1T) carbohydrate moiety (6) reduced binding of SP-A to CR3, and the binding activity that was further diminished by introduction of an additional N187S mutation prevented attachment of the CRD (7). These data implicate the N-linked carbohydrates, the CRD, and perhaps the neck domain in the interaction of SP-A with CR3. Data are mean ± S.E. with n = 4 experiments per group, *, p < 0.05 compared with binding of rat recombinant SP-A.
SP-A Interacts with the I-domain and Lectin Domain of CR3—To map binding domains for SP-A on CR3, competitive inhibition studies were performed with antibodies that recognize the I-domain or sugars that bind the lectin domain of CR3. Antibodies to the I-domain of CR3 (M1/70 and CBRM1/5) reduced binding of SP-A to CR3 by about 85%. Inhibition of the lectin site with β-glucan reduced binding of SP-A to CR3 by about 15% (Fig. 7A). Collagenase treatment of SP-A, which leaves the neck and carbohydrate recognition domain intact, resulted in reduced binding to CR3 compared with native SP-A. Binding of collagenase-treated SP-A to CR3 was further reduced by competitive inhibition of the lectin domain using β-glucan and further still by inhibition of the I-domain of CR3 (Fig. 7B). Deglycosylation of SP-A reduced binding to CR3 compared with native SP-A. Inhibition of the lectin domain using β-glucan did not alter binding of deglycosylated SP-A to CR3, suggesting the N-linked carbohydrates bind the lectin domain of CR3. In contrast, inhibition of the I-domain inhibited binding of deglycosylated SP-A to CR3 (Fig. 7C) indicating that N-linked carbohydrate is not required for binding of SP-A to the I-domain of CR3.
FIGURE 7.
SP-A interacted with the I-domain of CR3. Binding of SP-A to CR3 was determined as outlined in the legend to Fig. 5. β-Glucan (binds the lectin domain of CR3) reduced the binding to SP-A (panel A). Antibodies to the I-domain of CR3 (M1/70 and CBRM1/5) almost completely blocked binding of SP-A to CR3 (panel A). Collagenase treatment of SP-A (colSPA) reduced the binding to CR3 compared with native SP-A. A further reduction in binding of collagenase-treated SP-A to CR3 was observed in the presence of β-glucan and M1/70 antibody (panel B) suggesting a role for the both the I-domain and lectin domain in CR3/SP-A interactions. Deglycosylation of SPA (degSPA) also reduced binding to CR3 compared with native SP-A and this binding was not inhibited in the presence of β-glucan. In contrast, the addition of the M1/70 antibody to the I-domain reduced binding of deglycosylated SP-A to CR3 (panel C), which implicates the collagen domain in the interaction with the I-domain. Data are mean ± S.E. with n = 4 experiments per group, *, p < 0.05 compared with binding of iC3b; #, p < 0.05 compared with human SP-A (panel A). *, p < 0.05 compared with human SPA; #, p < 0.05 compared with collagenase SPA or deglycosylated SPA (panels B and C).
DISCUSSION
Targeted interruption of the SP-A gene resulted in reduced cell surface expression of complement receptors CR3 and CR4 on alveolar macrophages. The total pool size of CR3 receptors was similar for alveolar macrophages from Sp-a–/– and Sp-a+/+ mice, however, CR3 receptors were sequestered in intracellular pools in SP-A–/– alveolar macrophages. The finding that exogenous intratracheal SP-A enhanced surface expression of CR3 on alveolar macrophages isolated from Sp-a–/– mice indicates that SP-A promotes the translocation of CR3 receptor from subcellular compartments to the plasma membrane. CR3 expression was also increased on Sp-a–/– alveolar macrophages following infection with H. influenzae although expression levels in Sp-a–/– mice at all time points post-infection were significantly less than Sp-a+/+ alveolar macrophages. CR3 expression on Sp-a–/– peritoneal macrophages and blood neutrophils was similar to Sp-a+/+ mice suggesting that SP-A regulation of CR3 is restricted to the lung. Binding of SP-A to CR3 was greatest when the N-linked carbohydrates of SP-A were present and this binding was inhibited with antibodies to the I-domain of CR3. SP-A-enhanced CR3-dependent phagocytosis was mediated by both the carbohydrate recognition domain and the collagen-like domain of SP-A. These data support a novel mechanism by which SP-A influences CR3-mediated phagocytosis, both by direct binding to CR3 and by regulating CR3 expression on alveolar macrophages.
Complement is present in the uninfected lung although at reduced levels compared with the serum. Complement levels, expression, and function are altered in acute lung injury. For example, alveolar lining fluid isolated by BAL from rats exposed to intratracheal lipopolysaccharide (LPS) have a greater ability to deposit C3b onto bacteria through complement activation than alveolar lining fluid isolated by BAL from control rats (25). CR3 and CR4 are involved in phagocytosis of complement-opsonized particles. Previous studies have provided evidence that collectin family members interact with the complement system. C1q, the recognition component of the classical complement pathway, inhibited mannose binding lectin-enhanced phagocytosis and SP-A-enhanced phagocytosis suggesting similar mechanisms of receptor-mediated recognition for phagocytosis (9). Mannose-binding lectin but not SP-A or SP-D, can initiate the complement cascade. Additionally, SP-A inhibits C1q from associating with C1r and C1s to form the intact C1 complex required to activate complement suggesting an anti-inflammatory role of SP-A (26). Last, alveolar macrophages from SP-D-deficient mice express reduced CR3 similar to alveolar macrophages from the Sp-a–/– mice (27). Sp-a–/– mice do not have reduced levels of SP-D in the lung (1) suggesting both collectins are necessary for maintaining CR3 in alveolar macrophages. These studies provide evidence for interaction of collectins with the complement pathway.
Pulmonary bacterial infection enhanced CR3 expression on alveolar macrophages. However, significantly less CR3 was expressed on alveolar macrophages from Sp-a–/– mice at all time points after infection. Collectins enhance phagocytosis, chemotaxis, and oxygen radical production through a variety of receptors. SP-A opsonizes S. aureus and facilitates monocyte phagocytosis by binding the C1q receptor (28). SP-A interacts directly with CD14, a LPS receptor and modulates the LPS-induced cellular responses (29). In addition, the carbohydrate recognition domain of SP-A binds to Toll-like receptor (TLR)-4 and MD-2, critical signaling receptors for LPS preventing LPS induced signaling (30). Pre-treatment of macrophages with SP-A prior to incubation with K. pneumoniae increases uptake in a mannan-reversible manner, suggesting that SP-A increases activity of the mannose receptor. SP-A is also reported to enhance adherence of M. tuberculosis to macrophages by enhancing activity of the mannose receptor (31). SP-A augments scavenger receptor-A expression and phagocytosis of S. pneumoniae by alveolar macrophages independent of its binding to the bacteria (8). In the current study, alveolar macrophages from Sp-a–/– mice expressed less CR3 on the cell surface compared with alveolar macrophages from wild-type mice. Subcellular pools of CR3 were greater in the Sp-a–/– mice. In vivo treatment of Sp-a–/– mice with SP-A enhanced CR3 expression on alveolar macrophages and in vitro treatment of RAW cells, a macrophage cell line, with SP-A-enhanced CR3 expression suggesting that SP-A is important for recruitment of CR3 to the cell surface. These in vitro and in vivo studies support a role of SP-A in the regulation of alveolar macrophage CR3 cellular pools.
CR3 was reduced on alveolar macrophages and neutrophils but not peritoneal macrophages or blood neutrophils from Sp-a–/– mice. Restriction of SP-A effects to macrophages derived from the lung has been previously demonstrated. In vitro, SP-A induces oxygen radical production by rat alveolar macrophages but not peritoneal macrophages, neutrophils, or monocytes (32). Similarly, SP-A enhances production of colony-stimulating factors from alveolar macrophages but not peritoneal macrophages or alveolar fibroblasts (33). Scavenger receptor A expression on alveolar macrophages is up-regulated by SP-A, whereas no effect is observed on peritoneal macrophages (8). It is not surprising that SP-A regulation of complement receptors is specific for cells found in the alveolar space where SP-A is predominately found.
SP-A enhanced CR3-mediated phagocytosis. Chinese hamster ovary cells transfected with CR3 were used to determine the role of SP-A in CR3-mediated phagocytosis. CR3 binds the complement activation product, iC3b, fixed on bacterial pathogens and triggers phagocytosis via receptor priming induced by co-ligation of the CR3 lectin site by microbial surface polysaccharides (34, 35). Bone marrow, peritoneal, and alveolar macrophages (36, 37) from CR3–/– mice exhibit impaired phagocytosis of Mycobacterium tuberculosis (36, 37) and CR3–/– mice have increased mortality with overwhelming sepsis following intraperitoneal infection with S. pneumonia (38). In the current study, opsonization of H. influenzae with SP-A enhanced CR3-mediated phagocytosis to a similar extent as that observed with iC3b opsonization and increased SP-A-mediated phagocytosis was independent of calcium. SP-A augmented CR3-mediated phagocytosis in a manner that was attenuated by N-glycosidase or collagenase treatment of SP-A, implicating the N-linked sugar and collagen-like domains in that function. Enhanced phagocytosis by SP-A may be mediated through opsonization, direct receptor-mediated alveolar macrophage activation, and/or agglutination of bacteria resulting in presentation of multiple bacteria for engulfment.
SP-A binds CR3 and this interaction occurs only when CR3 is in the active conformation. The increase in CR3 affinity can be accomplished by a variety of different stimuli including: 1) cellular stimulation with chemotactic factors, cytokines, or phorbol esters; 2) the divalent cation, Mn2+; and 3) cross-linking of functionally related cell-surface receptors (18). In the present study, increasing concentrations of Mn2+ enhanced binding of SP-A to CR3; however, the highest concentration (4 mm) did not enhance binding. Similarly, low concentrations of PMA were not sufficient to enhance binding of SP-A to CR3 but concentrations above 0.1 μg enhanced binding. SP-A is a calcium-dependent lectin and binding to macrophages, LPS, immunoglobulin, and Toll-like receptor 4 have also been shown to depend on calcium. Binding of SP-A to CD14 is independent of calcium and this binding has been shown to occur through the neck region of SP-A. In the current study, in the absence of calcium, SP-A did not bind CR3 supporting a role for the lectin domain in the interaction.
Mapping experiments with mutant recombinant SP-A forms revealed the importance of the CRD and the N-linked carbohydrates of SP-A for CR3. Mutations of SP-A preventing attachment of N-linked carbohydrates to SP-A (resulting in an aglyco form of the protein) resulted in the greatest reduction in binding to CR3. Inactivation of the lectin function within the CRD of SP-A, through introduction of an E195A substitution, which is known to block calcium dependent binding to many carbohydrate and lipid ligands (23), also markedly reduced binding of SP-A to CR3. These results, combined with the observation of retained CR3 binding of the mutant form of SP-A containing only the neck and CRD domains (and lacking N-linked carbohydrate), also implicate the neck region or a non-lectin region of the CRD in the interaction between SP-A and CR3. Collectively, these data indicate that the CRD, the N-linked carbohydrate, and possibly the neck domains of SP-A all contribute to the binding of SP-A to CR3.
Reciprocal experiments were performed to determine the domains of CR3 that are required for binding to SP-A. Binding of SP-A to CR3 was inhibited by blocking antibodies to the I-domain of CR3. The I-domain of CR3 is found only in the α subunit of the β2-integrin and is known to directly mediate binding to a number of ligands including iC3b, ICAM-1, and the coagulation proteins fibrinogen and factor X (18). Partial blockade of the binding of SP-A to CR3 was also observed with β-glucan. The β-glucan binding site (lectin site) of CR3 has previously been mapped to a region of CD11b located C-terminal to the I-domain. Small soluble β-glucans were shown to saturate cell surface CR3 molecules. The lectin site of CR3 has broad specificity for certain polysaccharides containing mannose and N-acetylglucosamine as well as glucose (35). Binding of deglycosylated SP-A to CR3 was not inhibited by β-glucan suggesting it is the N-linked carbohydrates of SP-A that bind the lectin domain of CR3 rather than the lectin domain of CR3 binding to the carbohydrates attached to SP-A.
In summary, SP-A increased the surface expression of CR3 on alveolar macrophages. Up-regulation of CR3 by SP-A provides a mechanism for enhanced CR3-dependent phagocytosis of inhaled bacteria. The reduced phagocytic clearance and fatal bacterial infections that occur in humans and animal models of leukocyte adhesion deficiency resulting from reduced leukocyte CD11/CD18 expression attests to the importance of CR3 to innate immune defense (18). We speculate that reduction of SP-A in the lungs of patients with inflammatory lung diseases such as idiopathic pulmonary fibrosis and cystic fibrosis (9) may down-regulate alveolar macrophage CR3 expression and enhance susceptibility to pneumonia and sepsis.
Acknowledgments
We thank Lizhen Wu for assistance with animal husbandry and Huixing Wu for preparation of the mutant surfactant proteins.
This work was supported, in whole or in part, by National Institutes of Health Grant HL071522 (to A. M. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SP-A, surfactant protein A; CR3, complement receptor-3; SP-D, surfactant protein D; ICAM, intracellular adhesion molecule; GBS, group B streptococcus; BAL, bronchoalveolar lavage; CHO, Chinese hamster ovary; PMA, phorbol 12-myristate 13-acetate; LPS, lipopolysaccharide; TBS, Tris-buffered saline; WT, wild type; CRD, carbohydrate recognition domain.
References
- 1.LeVine, A. M., and Whitsett, J. A. (2001) Microbes Infect. 3 161–166 [DOI] [PubMed] [Google Scholar]
- 2.Korfhagen, T. R. (2001) Am. J. Respir. Cell Mol. Biol. 25 668–672 [DOI] [PubMed] [Google Scholar]
- 3.Khoor, A., Gray, M. E., Hull, W. M., Whitsett, J. A., and Stahlman, M. T. (1993) J. Histochem. Cytochem. 41 1311–1319 [DOI] [PubMed] [Google Scholar]
- 4.Manz-Keinke, H., Plattner, H., and Schlepper-Schafer, J. (1992) Eur. J. Cell Biol. 57 95–100 [PubMed] [Google Scholar]
- 5.Ohmer-Schrock, D., Schlatterer, C., Plattner, H., and Schlepper-Schafer, J. (1995) J. Cell Sci. 108, 3695–3702 [DOI] [PubMed] [Google Scholar]
- 6.Tenner, A. J., Robinson, S. L., Borchelt, J., and Wright, J. R. (1989) J. Biol. Chem. 264 13923–13928 [PubMed] [Google Scholar]
- 7.Kabha, K., Schmegner, J., Keisari, Y., Parolis, H., Schlepper-Schaeffer, J., and Ofek, I. (1997) Am. J. Physiol. 272 L344–L352 [DOI] [PubMed] [Google Scholar]
- 8.Kuronuma, K., Sano, H., Kato, K., Kudo, K., Hyakushima, N., Yokota, S., Takahashi, H., Fujii, N., Suzuki, H., Kodama, T., Abe, S., and Kuroki, Y. (2004) J. Biol. Chem. 279 21421–21430 [DOI] [PubMed] [Google Scholar]
- 9.Wright, J. R. (1997) Physiol. Rev. 77 931–962 [DOI] [PubMed] [Google Scholar]
- 10.Larson, R. S., and Springer, T. A. (1990) Immunol. Rev. 114 181–217 [DOI] [PubMed] [Google Scholar]
- 11.Ross, G. D., and Vetvicka, V. (1993) Clin. Exp. Immunol. 92 181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meerschaert, J., and Furie, M. B. (1995) J. Immunol. 154 4099–4112 [PubMed] [Google Scholar]
- 13.Fallman, M., Andersson, R., and Andersson, T. (1993) J. Immunol. 151 330–338 [PubMed] [Google Scholar]
- 14.von Asmuth, E. J., van der Linden, C. J., Leeuwenberg, J. F., and Buurman, W. A. (1991) J. Immunol. 147 3869–3875 [PubMed] [Google Scholar]
- 15.Lu, H., Smith, C. W., Perrard, J., Bullard, D., Tang, L., Shappell, S. B., Entman, M. L., Beaudet, A. L., and Ballantyne, C. M. (1997) J. Clin. Investig. 99 1340–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell, J. R., Baker, C. J., and Edwards, M. S. (1991) Infect. Immun. 59 1978–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noel, G. J., Katz, S., and Edelson, P. J. (1988) J. Infect. Dis. 157 85–90 [DOI] [PubMed] [Google Scholar]
- 18.Ehlers, M. R. (2000) Microbes Infect. 2 289–294 [DOI] [PubMed] [Google Scholar]
- 19.Xia, Y., and Ross, G. D. (1999) J. Immunol. 162 7285–7293 [PubMed] [Google Scholar]
- 20.LeVine, A. M., Whitsett, J. A., Gwozdz, J. A., Richardson, T. R., Fisher, J. H., Burhans, M. S., and Korfhagen, T. R. (2000) J. Immunol. 165 3934–3940 [DOI] [PubMed] [Google Scholar]
- 21.LeVine, A. M., Kurak, K. E., Bruno, M. D., Stark, J. M., Whitsett, J. A., and Korfhagen, T. R. (1998) Am. J. Respir. Cell Mol. Biol. 19 700–708 [DOI] [PubMed] [Google Scholar]
- 22.Haagsman, H. P., Hawgood, S., Sargeant, T., Buckley, D., White, R. T., Drickamer, K., and Benson, B. J. (1987) J. Biol. Chem. 262 13877–13880 [PubMed] [Google Scholar]
- 23.McCormack, F. X., Stewart, J., Voelker, D. R., and Damodarasamy, M. (1997) Biochemistry 36 13963–13971 [DOI] [PubMed] [Google Scholar]
- 24.McCormack, F. X., Kuroki, Y., Stewart, J. J., Mason, R. J., and Voelker, D. R. (1994) J. Biol. Chem. 269 29801–29807 [PubMed] [Google Scholar]
- 25.Bolger, M. S., Ross, D. S., Jiang, H., Frank, M. M., Ghio, A. J., Schwartz, D. A., and Wright, J. R. (2007) Am. J. Physiol. 292 L748–L759 [DOI] [PubMed] [Google Scholar]
- 26.Watford, W. T., Wright, J. R., Hester, C. G., Jiang, H., and Frank, M. M. (2001) J. Immunol. 167 6593–6600 [DOI] [PubMed] [Google Scholar]
- 27.Senft, A. P., Korfhagen, T. R., Whitsett, J. A., and LeVine, A. M. (2007) Am. J. Physiol. 292 L469–L475 [DOI] [PubMed] [Google Scholar]
- 28.Geertsma, M. F., Nibbering, P. H., Haagsman, H. P., Daha, M. R., and van Furth, R. (1994) Am. J. Physiol. 267 L578–L584 [DOI] [PubMed] [Google Scholar]
- 29.Sano, H., Sohma, H., Muta, T., Nomura, S., Voelker, D. R., and Kuroki, Y. (1999) J. Immunol. 163 387–395 [PubMed] [Google Scholar]
- 30.Yamada, C., Sano, H., Shimizu, T., Mitsuzawa, H., Nishitani, C., Himi, T., and Kuroki, Y. (2006) J. Biol. Chem. 281 21771–21780 [DOI] [PubMed] [Google Scholar]
- 31.Gaynor, C. D., McCormack, F. X., Voelker, D. R., McGowan, S. E., and Schlesinger, L. S. (1995) J. Immunol. 155 5343–5351 [PubMed] [Google Scholar]
- 32.van Iwaarden, F., Welmers, B., Verhoef, J., Haagsman, H. P., and van Golde, L. M. (1990) Am. J. Respir. Cell Mol. Biol. 2 91–98 [DOI] [PubMed] [Google Scholar]
- 33.Blau, H., Riklis, S., Kravtsov, V., and Kalina, M. (1994) Am. J. Physiol. 266 L148–L155 [DOI] [PubMed] [Google Scholar]
- 34.Ross, G. D., Cain, J. A., and Lachmann, P. J. (1985) J. Immunol. 134 3307–3315 [PubMed] [Google Scholar]
- 35.Thornton, B. P., Vetvicka, V., Pitman, M., Goldman, R. C., and Ross, G. D. (1996) J. Immunol. 156 1235–1246 [PubMed] [Google Scholar]
- 36.Melo, M. D., Catchpole, I. R., Haggar, G., and Stokes, R. W. (2000) Cell. Immunol. 205 13–23 [DOI] [PubMed] [Google Scholar]
- 37.Hu, C., Mayadas-Norton, T., Tanaka, K., Chan, J., and Salgame, P. (2000) J. Immunol. 165 2596–2602 [DOI] [PubMed] [Google Scholar]
- 38.Prince, J. E., Brayton, C. F., Fossett, M. C., Durand, J. A., Kaplan, S. L., Smith, C. W., and Ballantyne, C. M. (2001) J. Immunol. 166 7362–7369 [DOI] [PubMed] [Google Scholar]