Abstract
The regulation of lipid homeostasis by insulin is mediated in part by the enhanced transcription of the gene encoding SREBP-1c (sterol regulatory element-binding protein-1c). Nascent SREBP-1c is synthesized and embedded in the endoplasmic reticulum (ER) and must be transported to the Golgi in coatomer protein II (COPII) vesicles where two sequential cleavages generate the transcriptionally active NH2-terminal fragment, nSREBP-1c. There is limited indirect evidence to suggest that insulin may also regulate the posttranslational processing of the nascent SREBP-1c protein. Therefore, we designed experiments to directly assess the action of insulin on the post-translational processing of epitope-tagged full-length SREBP-1c and SREBP-2 proteins expressed in cultured hepatocytes. We demonstrate that insulin treatment led to enhanced post-translational processing of SREBP-1c, which was associated with phosphorylation of ER-bound nascent SREBP-1c protein that increased affinity of the SREBP-1c cleavage-activating protein (SCAP)-SREBP-1c complex for the Sec23/24 proteins of the COPII vesicles. Furthermore, chemical and molecular inhibitors of the phosphoinositide 3-kinase pathway and its downstream kinase protein kinase B (PKB)/Akt prevented both insulin-mediated phosphorylation of nascent SREBP-1c protein and its posttranslational processing. Insulin had no effect on the proteolysis of nascent SREBP-2 under identical conditions. We also show that in vitro incubation of an active PKB/Akt enzyme with recombinant full-length SREBP-1c led to its phosphorylation. Thus, insulin selectively stimulates the processing of SREBP-1c in rat hepatocytes by enhancing the association between the SCAP-SREBP-1c complex and COPII proteins and subsequent ER to Golgi transport and proteolytic cleavage. This effect of insulin is tightly linked to phosphoinositide 3-kinase and PKB/Akt-dependent serine phosphorylation of the precursor SREBP-1c protein.
Sterol regulatory element-binding proteins (SREBPs)3 are transcription factors that regulate expression of genes controlling cholesterol homeostasis and de novo fatty acid synthesis (1–7). SREBP-1a and SREBP-1c, which differ only in their first exon, are derived from a single gene through the use of alternative promoters, whereas SREBP-2 is encoded by a separate gene (8). Although there is clearly some functional overlap among the three SREBP isoforms (5), these proteins regulate different metabolic pathways. SREBP-1c preferentially affects transcription of genes that regulate de novo lipid synthesis, whereas SREBP-2 regulates genes involved in cholesterol biosynthesis and metabolism. The SREBP-1a isoform transactivates both lipogenic and cholesterogenic genes (9). In addition, the three SREBP isoforms exhibit differential tissue-specific expression. In replicating tumor cell lines, SREBP-1a constitutes greater than 90% of the SREBP-1 pool; conversely, SREBP-1c is the predominant isoform in liver and adipose tissue (9). Increased hepatic levels of nuclear SREBP-1c are thought to mediate the development of hyperlipidemia in type II diabetes and hyperinsulinemia (10–12). Nutritional and hormonal factors have been shown to regulate expression of SREBP-1c and its downstream regulatory targets (10, 13–15). Insulin induces the expression of SREBP-1c mRNA and nascent precursor protein (10, 16, 17). Glucagon opposes this effect of insulin via its second messenger cAMP (18). Newly synthesized SREBPs contain two transmembrane domains that are embedded in the endoplasmic reticulum (ER) with the NH2- and COOH-terminal sequences exposed to the cytoplasm. Following transport from ER to Golgi, the transcriptionally active NH2-terminal segments of SREBPs are liberated by two successive cleavages; the first cleavage in the loop extending into the vesicular lumen is carried out by site 1 protease (S1P), and the second cleavage is executed within the NH2-proximal transmembrane domain by site 2 protease (S2P).
Regulation of post-translational proteolysis has been studied most extensively in the case of SREBP-2 and SREBP-1a, both of which are regulated primarily by sterols. Within the ER, the COOH-terminal domains of SREBPs are associated with the integral membrane protein SREBP cleavage-activating protein (SCAP) (1, 4). SCAP contains a sterol-sensing domain similar to that in hydroxymethylglutaryl-CoA reductase. In cholesterol-replete cells, SCAP is associated within the ER with the ER retention proteins Insig (insulin-induced gene)-1 or Insig-2 (19). In the absence of cholesterol, SCAP undergoes a conformational change, dissociates from Insig (20), and becomes associated with the Sec23 and Sec24 components of a newly budding COPII-coated vesicle (21). The SCAP and SREBP are incorporated into COPII-coated vesicles and transported to the Golgi where SREBP is cleaved to release the transcriptionally active NH2-terminal fragment (5, 22).
Insulin is a potent inducer of de novo lipogenesis, and it is well known that insulin elevates the expression of SREBP-1c mRNA and protein. However, it is not known whether, in addition to transcriptional up-regulation, insulin also increases the proteolytic processing of SREBP-1c to specifically increase the rate of generation of the mature, transcriptionally active nSREBP-1c. The observation of rapid induction of nSREBP-1c by insulin in hepatocytes treated with the liver X receptor activator TO901317 in the studies of Hegarty et al. (23) was interpreted as reflecting enhanced proteolysis of the precursor SREBP-1c. Because rigorous measurements of the rates of translation, stability, and proteasomal degradation of the precursor SREBP-1c and nSREBP-1c were not made in these studies, these data may be subject to additional interpretation. To directly determine the effect of insulin to promote proteolytic processing of SREBP-1c independent of effects on production of endogenous nascent SREBP-1c, we examined the rate of generation of the transcriptionally active NH2-terminal fragment of SREBP-1c (nSREBP-1c) from an epitope-tagged nascent SREBP-1c whose transcription was driven by a cytomegalovirus (CMV) promoter. These studies were conducted in the presence of the proteasomal inhibitor ALLN to assess generation of the active NH2-terminal fragment of SREBP-1c independent of changes in rates of degradation. We demonstrate that insulin selectively stimulated processing of constitutively expressed SREBP-1c but not SREBP-2. We show further that insulin promoted the association of the SCAP-SREBP1c complex with the COPII vesicle proteins Sec23/Sec24 in rat hepatocytes and that this process is tightly linked to phosphoinositide 3-kinase (PI3K) and Akt/PKB-dependent phosphorylation of nascent SREBP-1c.
MATERIALS AND METHODS
Reagents—Insulin, aprotinin, acetylleucyl-leucyl-norleucinal (ALLN), dithiothreitol, leupeptin, sodium orthovanadate, sodium deoxycholate, phenylmethylsulfonyl fluoride, compactin, hydroxypropyl-β-cyclodextrin, and Hepes were purchased from Sigma. LY294002 and wortmannin were obtained from Calbiochem. Akt inhibitor II (SH-5) and Akt inhibitor III (SH-6) were procured from Invitrogen. [32P]Orthophosphoric acid (8500 Ci/mm) and [γ-32P]ATP (6000 Ci/mm) were purchased from PerkinElmer Life Sciences. Dominant negative (pdelta p85), constitutively active (p110) subunits of PI3K, His-GSK3β/pET29B and His-GSK3β (S9A)/pET29B were purchased from Addgene Inc. (Cambridge, MA).
Preparation of Recombinant Adenovirus Expressing SREBPs—Ad-HisSREBP-1cFLAG encodes rat full-length SREBP-1c, containing six tandem copies of His epitope tag at the NH2 terminus and 3× FLAG tag at the COOH terminus under the control of CMV promoter. To generate this adenovirus, the full-length rat hepatocyte SREBP-1c cDNA was amplified by PCR (forward primer, 5′-cgtgtcgacatggattgcacatttgaag containing an engineered SalI restriction site, and reverse primer, 5′-gcaaagcttctatgactgttggcgccg containing an engineered HindIII restriction site). The PCR product was digested with SalI and HindIII and ligated into pShuttle-CMV vector (Stratagene; La Jolla, CA) between the SalI and HindIII sites. Then a fragment encoding a BglII site, six tandem copies of the His epitope tag, and a SalI site were introduced between the BglII and SalI sites of the created plasmid. Then a fragment encoding an HindIII site, 3× FLAG tag, a stop codon (TGA), and an EcoRV site were introduced between the HindIII and EcoRV sites of the newly created plasmid. The resulting plasmid (pShuttle-CMV-SREBP-1c) was linearized with PmeI and transformed into Escherichia coli BJ5183-AD-1. Recombinant adenovirus plasmid was digested with PacI and used to transfect AD-293 cells to prepare adenovirus vector stock expressing histidine and FLAG-tagged SREBP-1c (Ad-HisSREBP-1cFLAG).
Ad-HisSREBP-2 was created as follows. The cDNA insert from pTK-SREBP-2 (ATCC) containing the full coding sequence of human SREBP-2 was inserted into the pShuttle-CMV vector described above in place of the full-length SREBP-1c fragment. The resulting plasmid was linearized with PmeI and transformed into E. coli BJ5183-AD-1. Recombinant Ad plasmid was digested with PacI and used to transfect AD-293 cells to prepare vector designed to express histidine-tagged SREBP-2 adenovirus stock (Ad-HisSREBP-2). Recombinant Ad-HisSREBP-1cFLAG, Ad-HisSREBP-2, and control Ad-LacZ viruses were propagated in HEK-293 cells and purified by CsCl density gradient centrifugation. Recombinant SREBP and control adenovirus stocks were stored in PBS containing 10% glycerol at –80 °C until use. Hepatocytes were infected at 10–30 multiplicities of infection for 16–24 h.
Expression and Purification of GST-Sar1a and GST-SREBP-1c Recombinant Proteins—Bacterial expression plasmid of GST-Sar1a was constructed by inserting cDNA encoding human Sar1a (OriGene Technologies, Inc., Rockville, MD) into pGEX-4T-2 (GE Healthcare). SREBP-1c was subcloned from the above-described pShuttle-CMV-SREBP-1c into pGEX-4T-2. Recombinant glutathione S-transferase (GST)-Sar1a and GST-SREBP-1c proteins were expressed in E. coli and purified by using glutathione agarose (GE Healthcare) according to the manufacturer's protocols.
Primary Hepatocyte Culture—Hepatocytes were obtained from livers of male Sprague-Dawley rats (Harlan Laboratories, Indianapolis, IN) by collagenase perfusion as described previously (24). Cells were suspended in Dulbecco's modified Eagle's medium (Invitrogen) containing 5% fetal bovine serum (Sigma), 10 mm glucose, 1 μm dexamethasone, and 100 nm insulin. Each 60-mm culture dish, coated with rat tail collagen (Collaborative Biochemical Products, Bedford, MA), was seeded with 3 × 106 cells; after 4 h, nonadherent cells were removed, and adherent cells were infected with Ad-HisSREBP-1cFLAG, Ad-HisSREBP-2, or Ad-LacZ-viruses at 10–30 multiplicities of infection for 12–18 h in Dulbecco's modified Eagle's medium without serum or hormones and subsequently treated with insulin (100 nm) and dibutyryl cyclic AMP (100 μm).
Preparation of Total Cell Extracts and Nuclear Extracts—Rat hepatocytes or McA-RH7777 rat hepatoma cells (catalog number CRL-1601, ATCC, Manassas, VA) were homogenized in cell lysis buffer (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) containing protease and phosphatase inhibitors and then centrifuged at 10,000 × g for 10 min at 4 °C to obtain supernatant for use as total cell extracts. Nuclear extracts were prepared by a modification of a published procedure (25). In brief, hepatocytes from five 60-mm plates were washed twice with PBS and collected by centrifugation at 1,000 × g for 5 min. Pelleted cells were then suspended by passing through a 25-gauge needle 10 times in 1 ml of lysis buffer (20 mm Hepes, pH 7.9, 10 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, 0.5% Nonidet P40) and protease inhibitor mixture (Sigma), and the cell suspension was kept for 10 min on ice. Nuclei were pelleted by centrifugation (500 × g) for 10 min at 4 °C and washed once in the lysis buffer. The pelleted nuclei were resuspended in a hypertonic buffer (20 mm Hepes, pH 7.9, 0.42 m NaCl, 1.5 mm MgCl2, 2.5% glycerol, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 25 μg/ml ALLN, and protease inhibitor mixture), incubated for 30 min at 4 °C, and centrifuged at 25,000 × g for 30 min at 4 °C; the supernatant containing nuclear proteins was collected.
Preparation of Microsomal Membranes—Microsomal membranes were prepared as described (26) with minor modifications. Briefly, hepatocytes were washed three times with ice-cold PBS. The cells were scraped off the dish with a rubber policeman into 10 mm Hepes-KOH, pH 7.2, 250 mm sorbitol, 10 mm KOAc, 1.5 mm Mg(OAc)2 plus protease inhibitors and then broken with 20 passes through a 22-gauge syringe. The broken cells were pelleted at 500 × g for 10 min at 4 °C and the supernatant recentrifuged at 16,000 × g for 20 min at 4 °C. The pellet containing ER and pre-Golgi and Golgi membranes was washed with 2.5 m urea in 10 mm Hepes, pH 7.2, 250 mm sorbitol, 10 mm KOAc, 1.5 mm Mg(OAc)2 plus protease inhibitors for 30 min on ice and then centrifuged at 16,000 × g for 20 min at 4 °C. The membranes were resuspended in 20 mm Hepes, pH 7.2, 250 mm sorbitol, 50 mm KOAc, 1 mm Mg(OAc)2 plus protease inhibitors and used in the binding assays.
Purification of the Sec23-24 Complex from Rat Liver Cytosol—Sec23/24 proteins were isolated and purified by a modification of a procedure described previously (27). Briefly, rat livers were homogenized in a buffer containing 20 mm Hepes-KOH, pH 7.2, 150 mm KOAc, 250 mm sorbitol, 1 mm DTT, 1 mm EGTA, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride (buffer A) at 4 °C. The homogenate was centrifuged at 1,000 × g for 10 min, and the supernatant obtained was recentrifuged at 12,500 × g for 20 min. The supernatant was collected and further centrifuged at 186,000 × g for 1 h. The final supernatant, designated as cytosol, was concentrated in Amicon filter YM10 (Amicon, Beverly, MA). The concentrated cytosol was loaded onto a gel filtration column (Sephacryl S-300; GE Healthcare) previously equilibrated with buffer B (buffer A plus protease inhibitor mixture). The column was eluted at 0.4 ml/min, and the immunoreactive fractions were pooled, concentrated by centrifugation in Amicon Filter-YM10, and loaded onto an anion exchange column (5-ml Econo column, Bio-Rad). The column was eluted with buffer B with a nonlinear salt gradient from 0.15 to 1 m KOAc at a flow rate of 1 ml/min. The immunoreactive fractions were pooled, dialyzed against buffer B, and concentrated on an Amicon YM10 membrane. The concentrated Sec23/24 fraction was used in the binding assays. For Akt/PKB immunodepletion studies, 100 μl of cytosol (1 mg of protein) isolated from primary hepatocytes was incubated with 20 μl of a 1:1 slurry of immobilized Akt-Sepharose beads (Cell Signaling Technology, Beverly, MA) for 3 h at 4 °C. The antibody-protein complexes were isolated by centrifugation, and immunodepletion of Akt from the cytosol was confirmed by immunoblot analysis. Akt-depleted cytosol was used in the GST pulldown assays.
GST Pulldown Assay—GST pulldown experiments were performed as described (21, 28). Microsomal membranes (120–160 μg of protein) prepared as described above were incubated with 10 μg of GST-Sar1 and 10 μg of Sec23/24 complex in a reaction mixture containing 20 mm Hepes, pH 7.2, 250 mm sorbitol, 1 mm Mg(OAc)2, 50 mm KOAc, 5 mm EGTA, 0.1 mm GMP-PNP, and ATP-regenerating system. Reactions were incubated at 30 °C for 10–30 min. The binding reaction was terminated by transferring the samples to ice and then centrifuging at 20,000 × g for 10 min at 4 °C to obtain rapidly sedimenting pellet. Pellets were solubilized in 50 mm Hepes-KOH, pH 7.2, 2.5 mg/ml bovine serum albumin, 150 mm KCl, and 1 mm MgCl2 containing 0.5% digitonin. Insoluble material was removed by centrifugation (4 °C, 20,000 × g for 5 min), and the supernatant was diluted and incubated with glutathione-Sepharose beads for 30 min at 4 °C to pull down GST-Sar1. The beads were pelleted by centrifugation, and the supernatant (unbound) was mixed with 5× SDS loading buffer and analyzed by immunoblotting. The beads were washed, and bound material was solubilized and denatured in 2× SDS loading buffer for 5 min at 95 °C. Samples were separated by SDS-PAGE and immunoblotted with antibodies for GST (Sar1), Sec23, SCAP, or His (full-length SREBP-1c or SREBP-2). For phosphatase treatment assays, microsomal membranes were pretreated with alkaline phosphatase (0.4 IU) from calf intestine (Roche Applied Science) for 30 min at 37 °C. The phosphatase reactions were stopped by adding 0.01 volumes of 200 mm EGTA and incubating for 5 min on ice.
SDS-PAGE and Immunoblot Analysis—Protein concentrations of total cell extracts and nuclear extracts were measured with a BCA kit (Pierce), and equal amounts of protein were subjected to SDS-PAGE. Proteins were transferred to nitrocellulose membranes (Amersham Biosciences), and immunoblot analysis was carried out with ECL plus Western blotting detection kit (Amersham Biosciences) according to the manufacturer's instructions. Images were captured with a PhosphorImager (Bio-Rad) and were quantified with the National Institutes of Health ImageJ software. The following primary antibodies were used: anti-FLAG (F3040), and anti-phosphoserine (P3430) were from Sigma; anti-His (SC-8036), anti-SREBP1 (SC-13551), anti-SCAP (SC-9675), anti-Sec23 (SC-12107), anti-histone H1 (SC-8030), anti-β-actin (SC-1616), anti-Akt (SC-5298), and anti-GST (SC-138) were from Santa Cruz Biotechnology; and anti-phospho-GSK-3β (9336) and GSK-3β (9315) were from Cell Signaling Technology.
Immunoprecipitation—After appropriate treatments, hepatocytes were rinsed with cold PBS and lysed in 400 μl of lysis buffer (PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 100 μg/ml phenylmethylsulfonyl fluoride, 100 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mm sodium orthovanadate) on ice for 20 min. The cell extracts were scraped into 1.5-ml Eppendorf tubes and cleared by centrifugation at 12,500 × g for 20 min at 4 °C. Extracts containing 500 μg of protein from control and treated hepatocytes were immunoprecipitated with 2 μg of either anti-SREBP1 or anti-SCAP antibodies overnight at 4 °C. The immunoprecipitated proteins were collected by incubation with 40 μl of 50% (w/v) protein A/G-Sepharose beads, followed by serial washings with lysis buffer and PBS. The A/G-Sepharose beads containing immunocomplexed protein were suspended in 40 μl of the Laemmli sample buffer, heated for 10 min in boiling water, resolved by SDS-PAGE, and subjected to Western blot analysis.
32P Labeling of Proteins and Autoradiography—Following infection with Ad-HisSREBP-1cFLAG for 24 h, hepatocytes were preincubated for 2 h at 37 °C in 2 ml of phosphate-free Dulbecco's modified Eagle's medium (Sigma) containing 0.5 mCi/ml [32P]orthophosphate (PerkinElmer Life Sciences) before incubation in the absence or presence of insulin for 1 h at 37 °C. 32P-Labeled SREBP-1c was immunoprecipitated (500 μg of extracts of hepatocyte protein) (see above). Immunoprecipitated proteins were fractionated on 7.5% SDS-PAGE and electrotransferred to a polyvinylidene difluoride membrane to reduce the background in autoradiograms. Membranes were dried, and autoradiography was performed at –80 °C 4–6 days with Hyperfilm-MP (Amersham Biosciences). The autoradiographs were quantified as described above, and results were expressed as percentage ± S.E. change in the optical density of bands before and after insulin treatment.
Phosphoamino Acid Analysis—Briefly, the 32P-labeled SREBP-1c was immunoprecipitated from insulin-treated hepatocytes and subjected to SDS-PAGE as described above. The SREBP-1c was electrotransferred to a polyvinylidene difluoride membrane. The band corresponding to 32P-labeled SREBP-1c was excised and subjected to acid hydrolysis for 90 min in 5.7 n HCl at 110 °C, and the samples were concentrated to dryness in a SpeedVac. The samples from acid hydrolysis were dissolved in 10 μl of pH 1.9 buffer (88% formic acid:acetic acid:water, 50:156:1794) containing 3 μg each of phosphoamino acid standards (phosphothreonine, phosphoserine, and phosphotyrosine). The samples deposited on 20 × 20-cm cellulose thin layer chromatography plates (EM Science) were subjected to ascending chromatography in butanol:acetic acid:ethanol:water (1:1:1:1). Standard phosphoamino acids were located by spraying 0.2% ninhydrin in acetone. The [32P]phosphoamino acids from SREBP-1c were located by autoradiography and identified using the standards. The incorporations 32P-radiolabel into phosphoserine, phosphothreonine, and phosphotyrosine in the full-length and nSREBP-1c were determined from phosphoamino acid spots scraped from TLC plates followed by measurement of radioactivity in a scintillation spectrometer.
In Vitro Kinase Assay with PKB/Akt—One μg of SREBP-1c or GSK3β expressed and purified from E. coli was incubated with recombinant active Akt (400 ng; Upstate Biotechnology, Inc., Lake Placid, NY) in 30 μl of kinase buffer containing 25 mm HEPES, 25 mm β-glycerophosphate, 25 mm MgCl2, 2 mm dithiothreitol, and 0.1 mm sodium orthovanadate. The reaction was initiated by adding 10 μCi of [γ-32P]ATP followed by incubation for 2 h at 30 °C. The reaction was stopped by adding Laemmli SDS sample dilution buffer. Proteins were separated on 7.5% SDS-PAGE and transferred to nitrocellulose membrane, and radioactive proteins were detected by x-ray film autoradiography.
Isolation of RNA and Real Time PCR—Total RNA was isolated from primary rat hepatocytes by using RNeasy mini kit (Qiagen Inc., Valencia, CA). Twenty microliters of RNA were initially treated with DNase I (Ambion, Austin, EX) at 37 °C for 1 h. The DNase I enzyme activity was stopped by adding DNase inactivation reagent to the sample and quantified by absorbance at 260 nm. Equal amounts of DNA-free RNA were used for the first strand cDNA synthesis by using SuperScript III first strand synthesis kit (Invitrogen). Up to 1 μg of RNA was mixed with 1 μl of 10 mm dNTP mix and 1 μl of random hexamers (50 ng/μl). Each sample was incubated at 70 °C for 10 min and then placed on ice for at least 2 min. Next, 2 μl of 10× RT buffer, 4.5 μl of 25 mm MgCl2, 2 μl of 0.1 m DTT, and 1 μl of RNaseOUT recombinant ribonuclease inhibitor was added to each tube. After being incubated at room temperature for 2 min, 1 μl of SuperScript III RT was added to each tube. The tubes were sequentially incubated at room temperature for 10 min, at 42 °C for 1 h, and at 70 °C for 15 min. RNase H was added to each tube and incubated at 37 °C for 20 min. Synthesized cDNA was subsequently mixed with 2× SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) and a set of following forward and reverse primers and subjected to real time PCR quantification. The primers for SREBP-1c were 5′-ggagccatggattgcacatt-3′ (forward) and 5′-aggaaggcttccagagagga-3′ (reverse). The primers for 18 S were 5′-cggctaccacatccaaggaa-3′ (forward) and 5′-ttttcgtcactacctccccg-3′ (reverse). The parameters for real time PCR were as follows: 95 °C for 11 min followed by 40 cycles of 95 °C for 30 s and 60 °C for 1 min and for the melt curve 100 cycles of 10 s from 60 to 100 °C in 0.4 °C increments. All reactions were performed in triplicate. The relative amounts of mRNAs were calculated by using the comparative Ct method.
Statistics—All experiments were repeated at least three times, and data are presented as mean ± S.E. The treatment effects were analyzed by unpaired Student's t test by using Prism 4.0 (GraphPad Software, San Diego). p values <0.05 were considered to be statistically significant.
RESULTS
Insulin Stimulates and Bt2cAMP Inhibits Processing of Nascent SREBP-1c in Rat Hepatocytes—Insulin stimulates gene expression in part by increasing the abundance of nuclear SREBP-1c. In rat hepatocytes and in intact liver, insulin induces SREBP-1c mRNA and nascent protein, whereas glucagon via cyclic AMP opposes the effect of insulin. To determine whether, in addition to increasing the expression of nascent SREBP-1c, insulin also stimulates SREBP-1c proteolytic processing, we determined the effect of insulin on generation of nSREBP-1c from constitutively expressed recombinant full-length SREBP-1c in primary rat hepatocytes infected with either Ad-LacZ (control) or Ad-HisSREBP-1cFLAG for 24 h. The exogenously expressed SREBP-1c was quantified from Western blots of cell lysates probed with anti-His or anti-SREBP1 antibodies. The expression of His-tagged full-length SREBP-1c could be readily detected in cell lysates from the infected hepatocytes (Fig. 1A). There was no detectable expression of SREBP-1c in hepatocytes infected with Ad-LacZ. Furthermore, the levels of endogenous SREBP-1c were too low to be detected in the whole cell extracts. Reprobing the Western blots with antibodies against SREBP-1c further corroborated the expression data obtained with antibodies to His tag. These blots were also probed for β-actin to assess protein loading in various lanes (Fig. 1A).
FIGURE 1.
Exogenous expression of nascent SREBP-1c by recombinant adenovirus vectors and proteolytic processing in primary hepatocytes. A, primary hepatocytes were infected with Ad-LacZ or Ad-HisSREBP-1cFLAG for 24 h. Cell lysates (20 μg of protein) were resolved by SDS-PAGE and immunoblotted with His, SREBP1-specific antibodies. Blots were stripped and reprobed with β-actin-specific antibodies to assess protein loading in all samples. B, hepatocytes infected with Ad-HisSREBP-1cFLAG were incubated in medium containing insulin (100 nm), Bt2cAMP (db-cAMP) (100 μm), insulin (100 nm) + Bt2cAMP (100 μm), or without additions. ALLN (25 μg/ml) was added during incubation, and cells were harvested after 1 h of incubation. Nuclear protein extracts (50 μg) were fractionated by SDS-PAGE, and immunoblots were developed using anti-His or anti-histone antibodies. Microsomal fraction (20 μg) was subjected to SDS-PAGE, blotted to nitrocellulose membranes, and probed using His-specific antibodies to assess the levels of full-length SREBP-1c (bottom panel). C, densitometric quantification of nSREBP-1c bands from three independent blots is shown in B; the data are expressed as a percentage of control optical density of nSREBP-1c-specific band. *, p < 0.05 versus control; **, p < 0.05 versus insulin treatment. D, primary hepatocytes infected with Ad-HisSREBP-1cFLAG were treated with or without insulin (100 nm) for the indicated times, and RNA was isolated. The mRNA abundance of SREBP-1c was measured by real time PCR as described under “Materials and Methods” using 18 S rRNA as the control. The data are expressed as fold induction of control SREBP-1c mRNA abundance. E, primary hepatocytes were infected with recombinant Ad-lacZ or Ad-HisSREBP-2 for 24 h. Cell lysates were resolved by SDS-PAGE and immunoblotted with anti-His, anti-SREBP2, or β-actin antibodies. F, hepatocytes infected with Ad-His-SREBP-2 were incubated in medium containing insulin (100 nm), Bt2cAMP (100 μm), insulin (100 nm) + Bt2cAMP (100 μm), or without additions. ALLN (25 μg/ml) was added during incubation, and cells harvested after 1 h of incubation. Immunoblot of nuclear protein extracts (50 μg) were developed with anti-His or anti-histone H1 antibodies. Representative blots are shown.
To examine the ability of insulin to stimulate SREBP-1c processing, hepatocytes were infected overnight with Ad-HisSREBP-1cFLAG and then incubated in the presence or absence of insulin (100 nm), Bt2cAMP (100 μm), or both agents for 1 h. The nuclear SREBP-1c levels were measured using anti-His antibodies; equivalency of nuclear protein loading was assessed by reprobing the Western blots with anti-histone H1 antibodies. The proteasomal inhibitor ALLN (25 μg/ml) was added during the 1-h incubation period to prevent degradation of newly generated nSREBP-1c. Insulin increased the accumulation of nSREBP-1c in nuclear extracts by 2.62 ± 0.21-fold. The insulin-mediated enhanced proteolysis of the nascent SREBP-1c was completely prevented by concurrent treatment with Bt2cAMP. Treatment of hepatocytes expressing full-length SREBP-1c by dibutyryl-cAMP alone had a modest effect to reduce nSREBP-1c below control levels; however, the major effect of dibutyryl cAMP was to antagonize the effect of insulin (Fig. 1, B and C). The full-length His-SREBP-1c was exclusively contained in the microsomal fraction (Fig. 1B, bottom panel).
Insulin is known to potently stimulate transcription of srebp-1c gene, and it is conceivable but highly unlikely that (i) CMV promoter-driven transcription of SREBP-1c is affected by insulin and (ii) the temporal changes in the rates of transcription of endogenous SREBP-1c gene occur rapidly with in 1 h of insulin treatment. To investigate that the ability of insulin to stimulate processing of epitope-tagged nascent SREBP-1c was not because of its stimulatory effects on the expression of epitope-tagged SREBP-1c from the CMV promoter incorporated in adenoviral vector, we measured SREBP-1c mRNA levels. Hepatocytes infected with Ad-His-SREBP-1cFLAG were incubated with and without insulin for 1–3 h, and RNA was isolated. Equal amounts of RNA from control and insulin-treated hepatocytes were analyzed by real time PCR. Insulin treatment did not affect the expression of SREBP-1c mRNA over a 3-h period (Fig. 1D).
The observations outlined in Fig. 1, B and C, do not reveal whether the effect of insulin to stimulate proteolytic processing of nascent SREBP-1c is specific for this lipogenic regulator, or whether the effect also extends to the SREBP-2 isoform whose processing is regulated primarily by sterol balance. Both SREBP-1c and SREBP-2 are assembled in the ER in association with the chaperone protein SCAP and are held in the ER by the ER retention proteins Insig1 or -2. As nuclear levels of SREBP-1c and SREBP-2 are differentially regulated by lipid and sterol balance, respectively, selective regulation of processing is one potential mechanism to confer specificity of regulation. Therefore, we experimentally investigated this question using adenoviral vector to constitutively express histidine-tagged SREBP-2 (Ad-HisSREBP-2). We measured the efficacy of expression of full-length SREBP-2 in hepatocytes infected with Ad-HisSREBP-2 by Western blot analysis of cell extracts using anti-His or anti-SREBP-2 antibodies. The expression of exogenous full-length SREBP-2 in the infected hepatocytes was readily observed (Fig. 1E). There was no detectable expression of endogenous SREBP-2 in uninfected hepatocytes or cells infected with Ad-LacZ virus. To examine whether the proteolytic cleavage of the nascent SREBP-2 was affected under these conditions, hepatocytes were infected overnight with Ad-HisSREBP-2 and incubated for 1 h in the presence or absence of insulin, Bt2cAMP, or with both agents. The steady state levels of nuclear SREBP-2 were measured using anti-His antibody. The parallel Western blot analysis of histone H1 levels was used as a normalization control. As shown in Fig. 1F, neither insulin nor Bt2cAMP significantly altered the levels of nSREBP-2. These findings indicate that insulin selectively enhances processing of SREBP-1c but not SREBP-2.
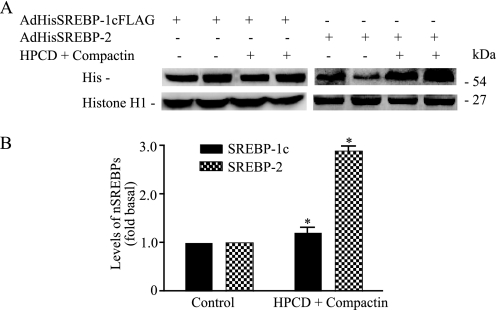
The selectivity of the effect insulin on SREBP-1c and SREBP-2 processing was further examined. We experimentally assessed whether SREBP-1c processing, like that of SREBP-2, is responsive to sterol balance. Hepatocytes were infected with either AdHisSREBP-1cFLAG or AdHisSREBP-2 and then incubated with or without 1% (w/v) hydroxypropyl-β-cyclodextrin, an agent known to remove sterols from intact cells, and the hydroxymethylglutaryl-CoA reductase inhibitor compactin (50 μm) for 3 h to deplete the cells of sterols. The levels of nSREBP-1c and -2 were measured using anti-His antibody. The parallel blots were probed for anti-histone H1 for normalization. Sterol depletion effectively increased the proteolytic processing of SREBP-2 by 2.89 ± 0.13-fold. In contrast, sterol depletion had no effect on SREBP-1c processing (1.26 ± 0.64-fold; Fig. 2, A and B). Altogether, these data indicate that insulin rapidly and selectively stimulates processing of SREBP-1c by a mechanism that is independent of sterol balance.
FIGURE 2.
Sterol depletion of primary hepatocytes does not affect processing of SREBP-1c. A, hepatocytes infected with either Ad-HisSREBP-1cFLAG or Ad-HisSREBP-2 were incubated with and without 1% (w/v) hydroxypropyl-β-cyclodextrin (HPCD) and 50 μm compactin for 3 h. ALLN (25 μg/ml) was added for the last 1 h of incubation. Nuclear protein extracts (50 μg) were electroblotted to nitrocellulose membranes and developed using antibodies specific for His or histone H1. B, densitometric quantifications of nSREBP bands (a representative blot shown in A is expressed as a fold change over control optical densities of nSREBP-1c bands). *, p < 0.05 versus control.
Insulin Increases Binding of SCAP/SREBP-1c to COPII Proteins in Rat Hepatocytes—To undergo sequential proteolysis by S1P and S2P, nascent SREBPs must first be transported from the ER to Golgi in COPII vesicles. The COPII coat consists of five proteins as follows: the small GTPase Sar1 and two heterodimeric protein complexes, Sec23/24 and Sec13/31. Formation of COPII vesicles is initiated by Sar1 binding to the ER membrane, where it is activated by the exchange of bound GDP for GTP. Then Sar1-GTP sequentially recruits Sec23/24 and Sec13/31 to the ER membrane, facilitating vesicle formation. Sterol-mediated regulation of transport of SREBP-1a and -2 is initiated by binding of SCAP/SREBP to COPII proteins that subsequently bud from the ER. We reasoned that ER to Golgi transport may also serve as a control point for proteolytic processing of SREBP-1c. Therefore, we experimentally determined whether insulin alters the affinity of one or more of the COPII vesicular proteins for the SCAP-SREBP-1c complex.
To discern whether insulin promotes binding of the SCAP-SREBP-1c complex to COPII proteins, we performed a glutathione S-transferase (GST) pulldown assay according to the published protocols (21, 27, 28). We prepared microsomes from hepatocytes expressing AdHis-SREBP-1cFLAG in the presence or absence of insulin treatment for 1 h, and we removed prebound COPII proteins by washing with urea. The urea-washed microsomes were then incubated with recombinant human wild-type GST-tagged Sar1 (GST-Sar1) in a reaction supplemented with ATP and GMP-PNP (a nonhydrolyzable analog of GTP) and/or purified COPII proteins (Sec23/24 complex), and we incubated these reactants at 30 °C for 15 min. To terminate the reaction, membranes were re-isolated by centrifugation and washed, and bound proteins were solubilized in a detergent-containing buffer. Proteins bound to GST-Sar1 were recovered on glutathione-Sepharose beads, separated by SDS-PAGE, transferred to nitrocellulose, and then probed with antibodies to the Sec23, SCAP, or GST tag on Sar1 or His tag on SREBP-1c. As illustrated in Fig. 3A (bottom panel, 2nd lane), all of the GST-Sar1 was found in the bound fraction when a reaction was supplemented with ATP/GMP-PNP, indicating complete recovery of the GST-Sar1 on the glutathione beads. This finding is consistent with previous data showing that ATP is required for binding of Sar1 to microsomal membranes (29). A fraction of Sec23 was copelleted when the incubations were performed in the presence of the Sec23/24 complex and ATP and GMP-PNP (Fig. 3A, 3rd panel, 3rd lane). This is consistent with the earlier observations that recruitment of COPII proteins to membranes requires an ATP-regenerating system and GTP or GMP-PNP. Preincubation of the hepatocytes with insulin had no effect on the GST-Sar1 pulldown of Sec23, indicating that binding of Sar1 to Sec23/24 is not regulated by insulin (Fig. 3A, 3rd panel, 4th lane). In contrast, we observed a marked increase in SCAP and SREBP-1c binding to the Sar1-Sec23/24 complex in microsomes derived from insulin-treated hepatocytes (Fig. 3A, 1st and 2nd panels). The amount of SCAP and SREBP-1c binding to the Sar1-Sec23/24 complex increased 2.34 ± 0.24- and 2.42 ± 0.56-fold, respectively, in microsomes isolated from insulin-treated hepatocytes (Fig. 3B). To determine the specificity of this effect, we next determined whether insulin also enhances binding of the SREBP-2 isoform to COPII proteins. In contrast to the enhanced binding of SREBP-1c observed in Fig. 3A, SREBP-2 binding to Sar1-Sec23/24 was not increased in microsomes isolated from insulin-treated hepatocytes expressing recombinant HisSREBP-2 (Fig. 3C).
FIGURE 3.
Insulin selectively increases binding of SCAP and SREBP-1c to COPII proteins. A, hepatocytes infected with Ad-HisSREBP1cFLAG were incubated for 1 h at 37 °C in the absence or presence of insulin (100 nm), and microsomes were assayed for SCAP and SREBP-1c binding to COPII proteins by using the GST-Sar1 pulldown assay as described under “Materials and Methods.” Microsomal membranes (160 μg) were incubated with GST-Sar1 (10 μg) in the presence or absence of Sec23/24 proteins (10 μg) or ATP/GMP-PNP and incubated for 15 min at 30 °C. Microsomes were pelleted by centrifugation, solubilized, and incubated with glutathione-Sepharose beads. The resulting bound and unbound fractions were separated by SDS-PAGE and immunoblotted with antibodies specific against GST (Sar1), Sec23, SCAP, or His (SREBP-1c). Representative blots are shown. B, amount of bound SCAP or SREBP-1c was quantitated by densitometry. Data are expressed as a percentage change over control optical density values of SCAP or SREBP-1 bands. *, p < 0.05 versus control. C, hepatocytes infected with Ad-HisSREBP-2 were treated with and without insulin for 1 h. Microsomes prepared were assayed for SREBP-2 binding to COPII proteins by GST-Sar1 pulldown assay as described under “Materials and Methods.” Representative blots are shown. D, hepatocytes were incubated with or without insulin (100 nm) for 1 h, and microsomes were isolated. Microsomal membranes (500 μg) were immunoprecipitated (IP) with anti-SCAP or preimmune IgG, fractionated by SDS-PAGE, and immunoblotted to detect Sec24 or SCAP. Representative blots are shown. E, following incubation of hepatocytes with or without insulin for 1 h, microsomes were prepared. Microsomal membranes (500 μg) were immunoprecipitated using anti-Sec23 or preimmune IgG and analyzed by immunoblotting using SCAP, SREBP-1c, or Sec23 antibodies.
To confirm the effect of insulin on membrane binding of COPII protein in situ, we further analyzed the association of Sec24 with SCAP from control and insulin-treated hepatocytes by immunoprecipitation of SCAP followed by immunoblotting to detect the presence of Sec24. As shown in Fig. 3, in hepatocytes treated with insulin the amount of Sec24 associated with SCAP was significantly increased (Fig. 3D). Conversely, when we immunoprecipitated Sec23 and probed for the presence of SCAP or SREBP-1c, we found that both SCAP and SREBP-1c were coimmunoprecipitated with Sec23, and this association was increased more than 2.5-fold in hepatocytes treated with insulin (Fig. 3E). Reprobing with anti-SCAP or anti-Sec23 antibodies confirmed that equal amounts of protein had been immunoprecipitated and that increased association was not the result of induction of either protein by insulin (Fig. 3, D and E, bottom panels). These data indicate that increased processing of SREBP1c in the presence of insulin results from increased recruitment of the SCAP/SREBP-1c by the COPII coat of nascent ER vesicles and that this effect of insulin is specific for the SREBP-1c isoform.
Insulin-stimulates Phosphorylation of SREBP-1c in Rat Hepatocytes—The rapid onset of the effect of insulin on proteolytic processing of SREBP-1c suggested a post-translational mechanism for this effect rather than a requirement for protein synthesis. To exclude the possibility that insulin-stimulated SREBP-1c processing resulted from de novo synthesis of an ER-export protein relegated to full-length SREBP-1c, we incubated hepatocytes with cycloheximide for 30 or 120 min before adding insulin (100 nm), followed by a subsequent 1-h incubation. As illustrated in Fig. 4, cycloheximide (2 μg/ml) pretreatment had no effect on insulin-stimulated SREBP-1c processing. The processing of SREBP-1c in insulin-treated hepatocytes was 265 ± 31% (Fig. 4, lane 2). Insulin treatment resulted in an enhanced generation of nSREBP-1c by 241 ± 21 (Fig. 4, lane 3) and 240 ± 17 (lane 4) (mean ± S.E.) in the presence of cycloheximide for 30 or 120 min, respectively. Cycloheximide treatment by itself did not significantly alter processing (Fig. 4, lane 5).
FIGURE 4.
Insulin-mediated stimulation of SREBP-1c processing does not require de novo synthesis. Hepatocytes infected with Ad-HisSREBP-1cFLAG were preincubated with and without cycloheximide for 30 min (lane 3) or 120 min (lane 4) and treated with or without insulin for 1 h. Nuclear protein extracts (50 μg) were prepared and analyzed by immunoblotting using anti-His antibodies to detect nSREBP-1c. Parallel blots were similarly probed with histone H1-specific antibodies to determine protein loading. Representative blots are shown.
Insulin exerts many effects via altering phosphorylation of proteins. The rapid onset and absence of a requirement for protein synthesis is entirely consistent with a mechanism for enhanced SREBP-1c processing following insulin treatment that involves post-translational modification of one or more of the participating proteins. We therefore hypothesized that phosphorylation of either full-length SREBP-1c and/or its chaperone protein SCAP could result in altered conformation of the SREBP-1c-SCAP complex with resulting increased affinity toward the COPII Sec23/24 complex. Phosphorylation of SCAP has not been previously reported, and in our initial experiments we were unable to identify phosphorylation of SCAP. To further extend these observations, we immunoprecipitated SCAP from lysates of 32P-labeled hepatocytes after 1 h of treatment with insulin and found that immunoprecipitated SCAP was not significantly phosphorylated (data not shown). On the other hand, the NH2-terminal nuclear fragment of SREBP-1c contains consensus sequences that may be phosphorylated by a number of kinases (30–32). Although it has been demonstrated that the NH2-terminal fragment of SREBP-1c is phosphorylated in vivo, it is not known whether it is phosphorylated in the ER or in the Golgi. If phosphorylation of nascent SREBP-1c mediates its enhanced transport from the ER to Golgi in response to insulin treatment, it must involve the full-length, ER-bound SREBP-1c. Because the effect of insulin on phosphorylation of full-length SREBP-1c has not been demonstrated previously, we infected rat hepatocytes with COOH-terminal FLAG-tagged SREBP-1c (Ad-HisSREBP-1cFLAG). SREBP-1c was immunoprecipitated with anti-FLAG antibody from lysates of 32P-labeled hepatocytes after 1 h of incubation at 37 °C in the absence or presence of insulin. The immunoprecipitated samples were then subjected either to immunoblotting with FLAG antibodies or were fractionated by SDS-PAGE followed by autoradiography. As shown in Fig. 5A, nascent SREBP-1c is a phosphoprotein, and insulin treatment leads to its increased phosphorylation. The incorporation of 32P more than doubled in insulin-treated samples (Fig. 5B). Insulin treatment led to increased phosphorylation of the full-length SREBP-1c at least in part resulting from its serine phosphorylation; insulin increased the phosphoserine immunoreactivity of the SREBP-1c by 2.18 ± 0.19-fold (Fig. 5, C and D). Conversely, the amount of SREBP-1c found in the immunoprecipitates of anti-phosphoserine agarose was increased by 2.06 ± 0.22-fold by insulin (Fig. 5, E and F). The enhanced phosphorylation of the full-length SREBP-1c potentially suggests a novel mechanism by which insulin promotes movement of the nascent SREBP-1c from the ER to Golgi where enhanced proteolysis releases a 60-kDa fragment, the transcriptionally active nSREBP-1c.
FIGURE 5.
Insulin increases the phosphorylation of full-length SREBP-1c and its phosphoserine immunoreactivity in rat primary hepatocytes. A, hepatocytes infected with Ad-HisSREBP1cFLAG were metabolically labeled with [32P]orthophosphate for 3 h at 37 °C and then incubated in the absence or presence of insulin (100 nm) for 1 h. Cell lysates (500 μg) were immunoprecipitated (IP) with anti-FLAG antibody and were immunoblotted using FLAG antibody (left panel); immunoblots were subjected to autoradiography (right panel). B, densitometric quantification of phosphorylated SREBP-1c shown in A (right panel); the data are expressed as a percentage of control optical density. *, p < 0.05 versus control. C, hepatocytes infected with Ad-HisSREBP1cFLAG were incubated in the absence or presence of insulin (100 nm). Cell extracts (500 μg) were immunoprecipitated with anti-FLAG antibody and immunoblotted with FLAG antibody (left panel) or phosphoserine antibody (right panel). D, densitometric quantification of phosphorylated SREBP-1c (a representative blot shown in C) was carried out; the data are expressed as a percentage of control optical density. *, p < 0.05 versus control. E, hepatocytes infected with Ad-HisSREBP1cFLAG were incubated in the absence or presence of insulin (100 nm). Cell extracts (500μg) were immunoprecipitated with anti-FLAG (left panel) or anti-phosphoserine-agarose (right panel) and immunoblotted with FLAG antibody (full-length SREBP-1c). F, densitometric quantification of phosphorylated SREBP-1c shown in E (right panel); the data are expressed as a percentage of control optical density. *, p < 0.05 versus control. G, hepatocytes labeled with [32P]orthophosphate were treated with and without insulin for 1 h. Cell lysates (500 μg) were immunoprecipitated with anti-FLAG (full-length SREBP-1c; left panel) or anti-His (nSREBP-1c; right panel) and separated on SDS-PAGE. The 32P-labeled SREBP-1c bands excised from separate phosphorylation assays were pooled and subjected to acid hydrolysis and phosphoamino acid analysis as described under “Materials and Methods.” Locations of ninhydrin-stained standard phosphoamino acids (pSer, phosphoserine; pThr, phosphothreonine; pTyr, phosphotyrosine) are indicated. H, 32P radiolabel incorporated into phosphoserine spots of SREBP-1c as shown in G were determined by scintillation counting as described under “Materials and Methods.” Data are expressed as a percentage of control optical density. *, p < 0.05 versus control.
Insulin activates a number of downstream kinases (33–35) that phosphorylate their targets at serine, threonine, or tyrosine residues. Therefore, we assessed the identities of phosphorylated amino acids in endogenous SREBP-1c; we immunoprecipitated the full-length SREBP-1c from 32P-loaded hepatocytes and subjected the immunoprecipitated protein to phosphoamino acid analysis. Precursor SREBP-1c immunoprecipitated from untreated hepatocytes contained only serine and tyrosine phosphorylation (Fig. 5G, left panel). In contrast, SREBP-1c isolated from insulin-treated hepatocytes not only had increased serine phosphorylation (3.4 ± 0.76-fold) but also was enriched in phosphothreonine (Fig. 5, G, left panel, and H). These results indicate that serines are the primary phosphorylated amino acids in full-length SREBP-1c and insulin enhances both serine and threonine phosphorylation. To determine whether the NH2-terminal segment of full-length SREBP-1c is also phosphorylated in situ, we immunoprecipitated the NH2-terminal fragment of SREBP-1c (60 kDa, nSREBP-1c) from 32P-labeled hepatocytes by using antibodies directed toward the NH2 terminus (anti-His). The immunoprecipitated NH2-terminal segment of SREBP-1c was fractionated by SDS-PAGE and subjected to phosphoamino acid analysis. The 32P content of serine spots from control and insulin-treated hepatocytes was measured. Insulin treatment also led to a consistent but lesser increase in serine phosphorylation of nSREBP-1c (Fig. 5G, right panel). Quantification of the autoradiographs (Fig. 5H, right panel) revealed that insulin treatment enhanced phosphorylation of nSREBP-1c by 2.3 ± 0.34-fold. These data suggest that phosphorylation sites exist in both NH2-terminal and COOH-terminal fragments of full-length SREBP-1c. However, our attempts to demonstrate the phosphorylation of COOH-terminal fragment directly were not successful. Under similar conditions, the fractionation of the immunoprecipitated COOH-terminal fragment of SREBP-1c failed to detect the band corresponding to the size of the COOH terminus (∼60–70 kDa; data not shown). We interpret this observation to indicate that unlike the nSREBP-1c-fragment the cytosolic COOH-terminal domain is highly susceptible to enzymatic degradation.
These results demonstrate that nascent SREBP-1c is phosphorylated under basal conditions and that phosphorylation increases in response to insulin treatment in primary rat hepatocytes. Because insulin-induced increase in SREBP-1c phosphorylation correlated with the magnitude of its proteolysis by S1P and S2P, we hypothesized that phosphorylation of the full-length SREBP-1c may promote its movement from the ER to Golgi where enhanced proteolysis occurs under these conditions. We experimentally assessed if phosphorylation of nascent SREBP-1c by insulin was necessary for its increased association with COPII proteins. Using a GST-Sar1 pulldown assay, we tested the ability of alkaline phosphatase treatment to prevent binding of SCAP/SREBP-1c to the Sec23/24 complex. For this purpose, the microsomal membranes derived from control and insulin-treated hepatocytes were preincubated with alkaline phosphatase as described under “Materials and Methods.” As shown in Fig. 6, A and B, SCAP and SREBP-1c binding to the Sar1-Sec23/Sec24 complex was 2-fold higher in membranes isolated from insulin-treated hepatocytes. The ability of insulin to promote SCAP/SREBP-1c binding was completely abolished when membranes were incubated with alkaline phosphatase (Fig. 6, A and B). Taken together, these data strongly support our contention that enhanced ER to Golgi transport of SREBP-1c was facilitated by its phosphorylation in response to insulin.
FIGURE 6.
Phosphatase treatment inhibits the ability of insulin to stimulate binding of SCAP/SREBP-1c to COPII proteins. A, hepatocytes infected with Ad-HisSREBP-1cFLAG were incubated in the presence or absence of insulin for 1 h. Microsomes were preincubated with or without alkaline phosphatase (0.4 IU) and assayed for SCAP and SREBP-1c binding to COPII proteins by using GST-Sar1 pulldown assay as described under “Materials and Methods.” Representative blots are shown. B, densitometric quantification of bound SCAP and SREBP-1c from three independent experiments are shown in A; the data are expressed as a percentage of control optical density of SCAP and SREBP-1c bands. *, p < 0.05 versus control. **, p < 0.05 versus insulin treatment.
The physiological relevance of phosphorylation of the full-length SREBP-1c was further evaluated by comparing the time course of the insulin-induced phosphorylation of nascent SREBP-1c and the appearance of nSREBP-1c in rat hepatocytes. The phosphorylation of newly synthesized full-length SREBP-1c was revealed by autoradiography after immunoprecipitation of 32P-labeled nascent SREBP-1c with the FLAG antibody. Concomitantly, we quantified the levels of nSREBP-1c by Western blot analysis followed by densitometry. As shown in Fig. 7, phosphorylation of nascent SREBP-1c was rapid and apparently preceded the appearance of nSREBP-1c. These data strongly support the hypothesis that increased phosphorylation of nascent SREBP-1c in response to insulin results in stimulation of SREBP-1c/SCAP binding to COPII proteins and concomitant acceleration of ER to Golgi transport of SREBP-1c.
FIGURE 7.
The insulin-induced phosphorylation of the nascent SREBP-1c and stimulation of the SREBP-1c processing follow a similar time course. Hepatocytes infected with Ad-HisSREBP-1cFLAG were treated with or without insulin for various times (10–60 min). The phosphorylation of the nascent SREBP-1c and SREBP-1c processing was determined as described under “Materials and Methods.” Top, representative autoradiogram and Western blot are shown. Bottom, data from three independent experiments are expressed as fold change over basal phosphorylation of nascent SREBP-1c (open squares) and as fold change over basal levels of nSREBP-1c (closed squares) and were plotted on the same graph.
Inhibitors of Phosphoinositide 3-Kinase Block Both Insulin-stimulated Phosphorylation of the Full-length SREBP-1c and Its Proteolytic Processing—To examine whether phosphorylation of nascent SREBP-1c mediates the effect of insulin to promote ER to Golgi transport and subsequent proteolytic processing of SREBP-1c, we next determined the effect of inhibitors of insulin-mediated protein phosphorylation. We reasoned that if phosphorylation of nascent SREBP-1c mediates the effect of insulin to enhance processing, then preventing SREBP-1c phosphorylation will also prevent its controlled proteolysis by S1P and S2P. Activation of the insulin receptor initiates a number of signal transduction pathways that include activation of PI3K that is critical for the acute and chronic actions of insulin in the liver and in hepatocytes in culture (35–37). Previous studies have shown that PI3K is essential for the effects of insulin on SREBP-1c expression and synthesis (17, 38, 39). However, a role of PI3K pathway in insulin regulation of SREBP-1c phosphorylation and processing remains to be determined. Therefore, we sought to determine whether the PI3K pathway mediates the effect of insulin to promote both SREBP-1c phosphorylation and processing by using two specific, cell-permeable inhibitors of PI3K. Rat hepatocytes were infected with Ad-HisSREBP-1cFLAG and then incubated for 1 h with insulin or with control medium in the presence or absence of LY294002 (10 μm) or wortmannin (250 nm). Proteins solubilized from the membrane fraction from hepatocytes were subjected to immunoprecipitation with anti-FLAG antibody. The resulting immunoprecipitates containing ER-associated full-length SREBP-1c were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The blots were probed with anti-phosphoserine antibody. As shown in Fig. 8, both LY294002 and wortmannin effectively blocked phosphorylation of full-length SREBP-1c by insulin. Neither inhibitor of PI3K affected the steady state levels of the total nascent FLAG-tagged SREBP-1c (Fig. 8, A and B, bottom panels).
FIGURE 8.
Inhibitors of phosphoinositide 3-kinase prevent insulin-induced phosphorylation of full-length SREBP-1c. A, hepatocytes infected with Ad-HisSREBP-1cFLAG were incubated with or without LY294002 (10 μm) for 30 min before addition of insulin. Microsomal membranes (500 μg) were immunoprecipitated (IP) with anti-FLAG antibodies to detect full-length SREBP-1c; and immunoblots were then probed with phosphoserine (upper panel) or FLAG-specific antibody (lower panel). B, hepatocytes infected with Ad-HisSREBP-1cFLAG were incubated with or without wortmannin (250 nm) for 30 min before adding insulin. Microsomal membranes (500 μg) were immunoprecipitated with anti-FLAG (full-length SREBP-1c) and immunoblotted using phosphoserine antibody (upper panel) or FLAG antibody (lower panel). Representative blots are shown.
We next determined the effect of the phosphoinositide 3-kinase inhibitors LY294002 or wortmannin on the ability of insulin to stimulate proteolytic cleavage of SREBP-1c. Rat hepatocytes were infected with Ad-HisSREBP-1cFLAG and then incubated for 1 h with and without insulin after preincubation with or without LY294002 or wortmannin. Proteins from nuclear extracts were resolved by SDS-PAGE and transferred to nitrocellulose membranes, and blots were probed with anti-His antibody to detect the nuclear fragment of SREBP-1c, nSREBP-1c. As shown in Fig. 9, insulin treatment stimulated SREBP-1c processing, and inhibition of PI3K by both LY294002 and wortmannin effectively prevented insulin-stimulated processing. Quantification of these Western blots indicated that insulin treatment led to a 2.5–3-fold increase in the formation of nSREBP-1c that was completely neutralized by inhibitors of PI3K. These results suggest that PI3K-mediated phosphorylation is required to stimulate SREBP-1c processing by insulin.
FIGURE 9.
Stimulation of SREBP-1c processing by insulin is prevented by inhibitors of PI3K. A, hepatocytes infected with Ad-HisSREBP-1cFLAG were treated with or without insulin (100 nm) in the presence or absence of LY294002 (10 μm) for 1 h, and nuclear extracts were prepared. Nuclear protein extracts (50 μg) were separated by SDS-PAGE and immunoblotted for anti-His (nSREBP-1c) or anti-histone H1 antibodies. B, hepatocytes infected with Ad-HisSREBP-1cFLAG were treated with or without insulin (100 nm) in the presence or absence of wortmannin (250 nm) for 1 h. Nuclear protein extracts (50 μg) were separated by SDS-PAGE and immunoblotted for anti-His or anti-histone H1 antibodies. Representative blots for both experiments are shown in A and B.
We further confirmed these results in McARH7777 rat hepatoma cells that were cotransfected with dominant negative (pdelta p85) and constitutively active (p110) subunits of PI3K with the HisSREBP-1cFLAG construct. Cotransfection of dominant-negative p85 reduced the phosphoserine content of the precursor SREBP-1c by 45% (Fig. 10A). On the other hand, the phosphoserine content of the nascent SREBP-1c was enhanced when the constitutively active p110 subunit of PI3K was coexpressed by 160% (Fig. 10A). Cotransfection of the dominant negative p85 subunit also resulted in an ∼2-fold decrease in SREBP-1c processing, whereas cotransfection of the constitutively active p110 subunit of PI3K resulted in a similar increase in SREBP-1c processing (Fig. 10B). These data corroborate the results of chemical inhibitors of the PI3K pathway and, together with the phosphatase treatment studies, indicate a tight link between phosphorylation of the nascent SREBP-1c, its ER to Golgi transport, and enhanced proteolytic processing.
FIGURE 10.
Constitutively active p110 subunit of PI3K increases and dominant negative p85 subunit decreases both phosphorylation and processing of SREBP-1c. A, rat hepatoma cells were cotransfected with pHisSREBP-1cFLAG and p110 or delta-p85 expression vectors and membranes were prepared. Microsomal membrane proteins (500 μg) were immunoprecipitated (IP) with anti-FLAG and immunoblotted for anti-phosphoserine antibody (upper panel) or FLAG antibody (lower panel). B, rat hepatoma cells were cotransfected with pHisSREBP-1cFLAG and p110 or delta-p85 expression vectors, and nuclear extracts were prepared. Nuclear protein extracts (50 μg) were separated by SDS-PAGE and immunoblotted for anti-His or anti-histone H1 antibodies. Representative blots are shown.
Effects of Serine/Threonine Kinase PKB/Akt on SREBP-1c Phosphorylation and Activation by Insulin—The PKB/Akt enzyme has been suggested to be a major downstream mediator of PI3K in the insulin signaling pathway (40, 41) and has been shown to be involved in the stimulation of SREBP-1c mRNA expression and synthesis in the liver in response to insulin (39, 42, 43). To further investigate the signaling pathway(s) that mediate insulin-induced proteolytic processing of SREBP-1c and to provide additional support for a mechanistic link with phosphorylation of SREBP-1c, we next determined whether phosphorylation of SREBP-1c by PKB/Akt is involved in its proteolytic activation by insulin. Our initial approach was to assess the ability of selective PKB/Akt inhibitors (Akt inhibitors II and III) to prevent insulin-dependent phosphorylation and proteolytic processing of SREBP-1c (44, 45). We incubated hepatocytes expressing FLAG-tagged SREBP-1c with or without PKB inhibitors and then maintained these cultures in insulin-supplemented (100 nm) or insulin-free medium for an additional 1 h. Immunoprecipitated FLAG-tagged SREBP-1c was sequentially probed with antibodies against phosphorylated serine and FLAG. We demonstrated that insulin increased serine phosphorylation of nascent SREBP-1c by ∼2-fold (Fig. 11A, left panel, 2nd lane). The insulin-dependent increase in SREBP-1c phosphorylation was abolished by pretreatment of cells with either inhibitors of PKB/Akt (Fig. 11A, upper left panel, 3rd and 4th lanes). To investigate whether the stimulatory effects of insulin on SREBP-1c proteolytic activation is attributable to its stimulatory effect of PKB-dependent phosphorylation, we investigated the effects of PKB inhibitors on proteolytic activation by insulin. Hepatocytes expressing His-tagged SREBP-1c were preincubated with and without PKB inhibitors and then treated with and without insulin for 1 h. Consistent with what was observed for phosphorylation of the nascent SREBP-1c, we discovered that insulin treatment led to a significant increase in accumulation of nuclear SREBP-1c, and both inhibitors of PKB/Akt inhibited the insulin-induced generation of nuclear SREBP-1c (Fig. 11A, upper right panel). The efficacy of these Akt inhibitors in primary hepatocyte cultures was confirmed by assessing the ability of these inhibitors to prevent Akt-dependent insulin-mediated phosphorylation of GSK-3β, a known Akt target protein. As expected, insulin treatment increased phosphorylation of GSK-3β, and this effect was effectively prevented by both Akt inhibitors (Fig. 11A, lower panel). These data are consistent with the interpretation that Akt-dependent phosphorylation of SREBP-1c by insulin is required for its proteolytic activation.
FIGURE 11.
Effects of PKB on SREBP-1c phosphorylation and activation by insulin. A, hepatocytes infected with Ad-HisSREBP-1cFLAG were incubated with or without Akt inhibitor II (10 μm) or III (10 μm) for 30 min before adding insulin. Microsomal membranes (500 μg) were immunoprecipitated (IP) with anti-FLAG (full-length SREBP-1c) and immunoblotted for phosphoserine antibody or FLAG antibody (upper left panel). Hepatocytes infected with Ad-His-SREBP-1cFLAG were preincubated with or without Akt inhibitor II or III for 30 min and then treated with insulin for 1 h. Nuclear protein extracts (50 μg) were separated by SDS-PAGE and immunoblotted for anti-His (nSREBP-1c) or anti-histone H1 antibodies (upper right panel). Hepatocytes were treated with or without insulin (100 nm) in the presence and absence of Akt inhibitor II (10 μm) or III (10 μm), and cell extracts were prepared. An equal amount of protein (50 μg) from control and each treatment was analyzed by Western blotting for phosphorylated GSK-3β-specific antibodies. The blots were reprobed with anti-GSK-3β antibodies to assess the levels of total GSK-3β (lower panel). B, hepatocytes were incubated with or without insulin (100 nm) in the presence or absence of LY294002 (10 μm) or Akt inhibitor II (10 μm), and microsomes were prepared. Microsomal membrane proteins were immunoprecipitated (500 μg) with anti-SREBP-1c or preimmune IgG, fractionated by SDS-PAGE, and immunoblotted for Sec23 (left panel). Hepatocytes were incubated with or without insulin (100 nm) in the presence or absence of LY294002 (10 μm) or Akt inhibitor II (10 μm), and microsomes were prepared. Microsomal membrane proteins (500 μg) were immunoprecipitated with anti-Sec23 or preimmune IgG, fractionated by SDS-PAGE, and immunoreacted with anti-SREBP-1c antibodies (right panel). C, hepatocytes infected with Ad-HisSREBP-1cFLAG were incubated with or without insulin (100 nm) for 1 h, and cytosol and native membranes (control-treated) were prepared. Cytosol immunodepleted of PKB/Akt or treated with control IgG was incubated with urea-washed native membranes and assayed by GST-Sar1 pulldown assay as described under “Materials and Methods.” D, hepatocytes infected with Ad-HisSREBP-1cFLAG were incubated with or without insulin (100 nm) for 1 h, and cytosol and native membranes (control-treated) were prepared. Cytosol immunodepleted of PKB/Akt or treated with control IgG was incubated with urea-washed native membranes for 1 h at 37°C and an ATP-regenerating system containing 50 μCi of [γ-32P]ATP. Membranes were reisolated, solubilized, fractionated by SDS-PAGE, and subjected to autoradiography as described under “Materials and Methods.” E, recombinant SREBP-1c, recombinant active PKB, or recombinant SREBP-1c and active PKB were incubated in vitro in the presence of [γ-32P]ATP. Proteins were separated by SDS-PAGE, and autoradiographed. Alternatively, proteins were detected by immunoblotting using either SREBP-1 antibody or Akt antibody (left panel). Recombinant wild-type GSK-3β or its nonphosphorylatable mutant (S9A) and active PKB were incubated in the presence of [γ-32P]ATP. Proteins were separated by SDS-PAGE and autoradiographed or immunoblotted using GSK-3β or Akt antibody (right panel).
To further elucidate the potential mechanistic relationship between PKB-dependent SREBP-1c phosphorylation and its increased association with COPII proteins, the binding of SREBP-1c to the COPII complex was assessed by coimmunoprecipitation of these proteins from hepatocytes treated with PI3K/PKB inhibitors. It is evident that insulin treatment increased the amount of COPII protein (Sec23) associated with SREBP-1c, and inhibition of either PI3K or PKB/Akt significantly inhibited the ability of insulin to stimulate binding of SREBP-1c to Sec23 (Fig. 11B, left panel). Conversely, when we immunoprecipitated Sec23 and carried out immunoblot analysis for SREBP-1c, as expected, insulin treatment increased the amount of SREBP-1c binding to this COPII protein, and both PI3K and PKB inhibitors inhibited insulin-increased association (Fig. 11B, right panel).
To further extend and corroborate the role of PKB-dependent SREBP-1c phosphorylation in binding of COPII proteins, we assessed the effect of immunodepletion of PKB on the ability of insulin to promote SREBP-1c/Sec23 association in vitro using a GST-Sar1 pulldown assay. For this assay, PKB in the cytosol derived from insulin-treated hepatocytes was depleted using antibody bound to Sepharose beads and incubated with microsomes isolated from hepatocytes maintained in insulinfree medium. As shown in (Fig. 11C), SREBP-1c binding to COPII proteins was increased in cytosol isolated from insulin-treated hepatocytes. The effect of insulin to promote SREBP-1c binding was inhibited when Akt-depleted cytosol was used. By contrast, control IgG-treated cytosol had no effect, as expected (Fig. 11C). We also directly determined the effect of PKB/Akt immunodepletion on phosphorylation of nascent SREBP-1c under these conditions and its association with COPII proteins by using [γ-32P]ATP in GST-Sar1 pulldown experiments. After the binding reaction, the membranes were re-isolated, solubilized, and fractionated by SDS-PAGE, and 32P-labeled SREBP-1c was detected by autoradiography. Consistent with the GST-Sar1 pulldown experiments, phosphorylation experiments revealed that SREBP-1c phosphorylation was increased when cytosol isolated from insulin-treated hepatocytes was incubated with native microsomes in a GST-Sar1 binding reaction. However, when cytosol depleted in PKB/Akt was used, the insulin-induced phosphorylation of SREBP-1c was reduced. By contrast, control IgG-treated cytosol had no effect (Fig. 11D).
To determine whether SREBP-1c could be directly phosphorylated by PKB/Akt, we performed a cell-free kinase assay by coincubating bacterially expressed recombinant SREBP-1c and active Akt. The autoradiogram of reaction mixtures subjected to SDS-PAGE shows that Akt directly phosphorylates the full-length SREBP-1c; as expected, incubation of either recombinant SREBP-1c or Akt alone in the presence of [32P]ATP did not result in phosphorylation (Fig. 11E, left panel). Akt also effectively phosphorylates its well known substrate, the wild-type GSK-3β; in contrast the mutated form of GSK-3β (S9A) is refractory to phosphorylation by Akt (Fig. 11E, right panel). These data strongly support the hypothesis that PKB/Akt-dependent SREBP-1c phosphorylation is required for its enhanced association with COPII proteins by insulin and thus enhanced generation of active nuclear SREBP-1c.
DISCUSSION
SREBP-1c is the primary regulator of insulin-induced hepatic lipogenesis. Insulin potently induces expression of SREBP-1c mRNA and its encoded protein (10, 16, 17, 46, 47) that must be transported to the Golgi for regulated proteolysis to yield the transcriptionally active N-terminal fragment nSREBP. Using adenoviral vectors to constitutively express epitope-tagged full-length SREBPs in primary hepatocyte cultures, we have demonstrated that insulin selectively enhances the ER to Golgi transport and controlled proteolytic cleavage of nascent SREBP-1c, resulting in increased nuclear content of the transcriptionally active NH2-terminal fragment. Increased insulin-stimulated SREBP-1c processing is preceded by its phosphorylation on serine and threonine residues that resulted in enhanced binding of SCAP/SREBP-1c to COPII proteins.
Although regulation of SREBP-1c by nutritional and hormonal factors has been attributed primarily to transcriptional mechanisms, there is emerging indirect evidence to suggest that SREBP-1c may also be regulated at the level of its posttranslational processing (13, 23). Based on their observations of rapid appearance of nSREBP-1c with administration of insulin following LXR-dependent induction of nascent SREBP-1c in hepatocytes, Hegarty et al. (23) proposed that insulin enhances proteolytic processing of nascent full-length SREBP-1c. This study directly confirms this hypothesis and further links the effect of insulin to enhanced association of the SREBP-1c-SCAP complex with COPII vesicle proteins and resulting enhanced ER to Golgi transport via Akt/PKB-dependent phosphorylation of nascent SREBP-1c. To determine the effect of insulin on post-translational processing of nascent SREBP-1c independent of the known effect of insulin to promote synthesis of the nascent protein, primary hepatocytes were transduced to constitutively overexpress precursor SREBP-1c tagged with an NH2-terminal His epitope. Degradation of the resulting nuclear His-SREBP-1c fragment was prevented by treatment with the proteosomal inhibitor ALLN. Thus, appearance of the His-SREBP-1c nuclear protein directly reflected proteolytic processing of nascent recombinant SREBP-1c. In this regard, it is important to note that the behavior of a recombinant tagged protein may not completely mimic that of the native protein. In this case, however, the recombinant SREBP-1c behaved as one would expect of the native protein, i.e. it intercalated into the endoplasmic reticulum, associated with SCAP and COPII proteins, and underwent ER to Golgi transport and proteolytic cleavage. In contrast to the finding of enhanced proteolytic processing of SREBP-1c in response to insulin treatment, processing of exogenously expressed full-length SREBP-2 was enhanced by sterol depletion but not by insulin. Thus, SREBP-2 and SREBP-1c exhibit divergent regulation by sterol and insulin in primary hepatocytes. This is consistent with previous in vivo studies that showed distinct regulation of SREBP-2 and SREBP-1c by altered sterol balance and fasting-refeeding (13, 48–50). The failure of insulin to stimulate proteolytic cleavage of full-length SREBP-2 is also consistent with the reported lack of effect of insulin depletion or of insulin replacement on levels of nSREBP-2 in livers of streptozotocin-diabetic rats (16). Furthermore, increased levels of nuclear SREBP-1c but not SREBP-2 were shown to be associated with fatty livers in mouse models of hyperinsulinemia (12). This selectivity has been attributed to differential transcriptional response of the promoter regions of the different SREBP isoforms to nutritional and hormonal stimuli (51). This study demonstrates that the selective response of SREBP-1c to nutritional and hormonal stimuli also reflects differences in post-translational processing. We conclude that insulin selectively enhances SREBP-1c processing and thereby fatty acid biosynthesis. This allows for independent regulation of fatty acid and cholesterol biosynthesis in response to varying dietary intake of carbohydrate, fat, and cholesterol respectively. Our findings do not rule out the possibility of cross-talk between the two pathways under certain dietary and hormonal conditions, however.
There are several possible mechanisms by which insulin could enhance SREBP-1c processing. The binding of the SCAP-SREBP complex to COPII proteins is a key event in the export of the nascent SREBPs from the ER. Our data show that insulin treatment increased binding of SCAP-SREBP-1c with Sar1-Sec23/24 complex, and the kinetics of this process were concordant with enhanced processing of SREBP-1c. This increase was not because of higher levels of SCAP or of Sec24 protein in membranes of insulin-treated hepatocytes but rather indicate increased affinity of the SCAP-SREBP-1c complex toward COPII proteins and decreased affinity for the ER retention protein Insig. Insulin treatment reduces expression of Insig-2a, the liver-specific isoform of Insig-2, in rat hepatocytes (52, 53), which could result in increased release of SCAP-SREBP-1c complexes for transport to the Golgi and proteolytic cleavage. This work demonstrates that the acute effect of insulin to stimulate processing of SREBP-1c occurs prior to a measurable reduction in levels of Insig-2 mRNA (53), implying that mechanisms other than reduction in Insig-2a expression alone are responsible for the acute effect of insulin. This explanation is consistent with our data showing that cycloheximide did not prevent insulin-stimulated SREBP-1c processing. On the other hand, we cannot rule out an additional effect of reduced Insig-2a expression in longer term regulation of SREBP-1c processing under conditions of hyperinsulinemia.
Previous studies have indicated that the NH2-terminal transcriptionally active fragment of SREBP-1c could be phosphorylated in vitro, and phosphorylation increased its stability and transcriptional activity (30–32). For insulin to regulate ER to Golgi transport of nascent SREBP-1c via this mechanism, full-length SREBP-1c would also have to be subject to phosphorylation in the ER. We have demonstrated that full-length SREBP-1c is, in fact, a phosphoprotein and furthermore that insulin enhances the serine/threonine phosphorylation of full-length SREBP-1c. The demonstration of phosphorylation of nascent SREBP-1c in the ER is a novel finding and may have important implications for regulating SREBP-1c processing by insulin. For example, if phosphorylation of SREBP-1c by insulin specifically alters the conformation of the SREBP-1c-SCAP complex to allow its transport from ER to Golgi, this might explain dissimilar regulation of SREBP-2-SCAP by insulin. In support of the hypothesis that insulin-mediated phosphorylation of nascent SREBP-1c is an obligatory step in enhanced ER to Golgi transport and proteolytic processing, we demonstrated that inhibitors of both the key insulin signaling pathway PI3K and its downstream kinase PKB/Akt effectively prevented both insulin-induced serine phosphorylation of nascent SREBP-1c and its proteolytic processing. The enhanced association of SREBP-1c-SCAP complex with COPII proteins was also prevented by inhibitors of PI3K and PKB/Akt. Thus the relationship between phosphorylation of SREBP-1c precursor and its processing could be demonstrated regardless of whether we employed chemical inhibitors, molecular inhibitors, or immunodepletion methods. The proposed mechanistic scenario was bolstered by the observation that enzymatic dephosphorylation of SREBP-1c in microsomes of insulin-treated hepatocytes effectively reversed the effect of insulin to promote the association of SCAP-SREBP-1c with COPII proteins.
In summary, we have presented several lines of evidence in support of the hypothesis that insulin enhances ER to Golgi transport of the SREBP-1c-SCAP complex via phosphorylation of SREBP-1c as follows. 1) The temporal kinetics and magnitude of phosphorylation of full-length SREBP-1c and its processing were similar. 2) Inhibitors of both PI3K and Akt/PKB concomitantly prevented insulin-mediated phosphorylation and processing of SREBP-1c. 3) Immuno-depletion of PKB from cytosol inhibited the enhanced phosphorylation and association of SCAP/SREBP-1c to COPII proteins by insulin. 4) Alkaline phosphatase treatment of microsomal membranes abolished the increased binding of SCAP/SREBP-1c to COPII proteins in response to insulin treatment. Therefore, the process of insulin-induced phosphorylation of SREBP-1c and enhanced ER to Golgi transport and proteolytic processing appear to be tightly coupled. Although our observations strongly support the hypothesis that insulin enhances ER to Golgi transport of SREBP-1c via phosphorylation of nascent SREBP-1c, the identities of the specific amino acids of nascent SREBP-1c that are phosphorylated by insulin remains to be defined. The question of the specific kinase(s) that mediate insulin-dependent serine phosphorylation of the nascent SREBP-1c remains partially answered. We have demonstrated that phosphorylation of nascent SREBP-1c is dependent upon at least one of the downstream kinases, PKB/Akt, and that SREBP-1c is phosphorylated by PKB in vitro. However, this does not eliminate the possibility that insulin-mediated phosphorylation of full-length SREBP-1c is mediated by kinases downstream to PKB. Based on the kinases known to mediate insulin-induced phosphorylation and the analysis of potential phosphorylation sites of SREBP-1c, atypical protein kinase C and mammalian target of rapamycin are candidate kinases for increased phosphorylation in response to insulin (37, 39, 54). Alternatively, insulin may inhibit a serine phosphatase and prevent dephosphorylation of full-length SREBP-1c (55). Furthermore, although SCAP does not appear to be a phosphoprotein, these studies do not exclude the possibility that increased affinity of the SCAP-SREBP-1c complex with COPII proteins may be indirectly regulated by phosphorylation of another yet to be identified protein(s). We conclude that the coordinated effect of insulin on synthesis and post-translational processing of nascent SREBP-1c allows for rapid delivery of transcriptionally active nSREBP-1c to the nucleus in response to changes in nutritional and hormonal status. Furthermore, enhanced posttranslational processing in response to chronic hyperinsulinemia may also serve as an additional mechanism for the observed dysregulation of hepatic lipogenesis and consequent overproduction of triglyceride-rich lipoproteins, dyslipidemia, and hepatic steatosis observed in hyperinsulinemic states, including obesity and type II diabetes mellitus.
Acknowledgments
We thank Dr. William E. Balch (Department of Cell and Molecular Biology, The Scripps Research Institute, La Jolla, CA) for generously providing Sec24 antiserum. We also thank Poonam Kumar for expert technical assistance. We gratefully acknowledge the suggestions of Dr. Kafait U. Malik for experiments related to the role of Akt/PKB in SREBP-1c post-translational processing. We thank Dr. David Armbruster for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant DK075504 (to M. B. E., R. R., and C. R. Y.). This work was also supported by a postdoctoral fellowship grant, American Heart Association (to C. R. Y.), grant-in-aid, American Heart Association, Southeast Affiliate (to M. B. E. and C. R. Y.), and the Office of Research and Development, Department of Veterans Affairs (to M. B. E.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SREBP, sterol regulatory element-binding protein; ER, endoplasmic reticulum; COPII, coatomer protein II; PKB, protein kinase B; PI3K, phosphoinositide 3-kinase; SCAP, SREBP cleavage activating protein; DTT, dithiothreitol; GST, glutathione S-transferase; PBS, phosphate-buffered saline; CMV, cytomegalovirus; ALLN, acetylleucyl-leucyl-norleucinal; Bt2cAMP, dibutyryl cyclic AMP; S1P, site 1 protease; S2P, site 1 protease; GMP-PNP, 5′-guanylyl imidodiphosphate.
References
- 1.Edwards, P. A., Tabor, D., Kast, H. R., and Venkateswaran, A. (2000) Biochim. Biophys. Acta 1529 103–113 [DOI] [PubMed] [Google Scholar]
- 2.Eberle, D., Hegarty, B., Bossard, P., Ferre, P., and Foufelle, F. (2004) Biochimie (Paris) 86 839–848 [DOI] [PubMed] [Google Scholar]
- 3.McPherson, R., and Gauthier, A. (2004) Biochem. Cell Biol. 82 201–211 [DOI] [PubMed] [Google Scholar]
- 4.Brown, M. S., and Goldstein, J. L. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 11041–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espenshade, P. J., and Hughes, A. L. (2007) Annu. Rev. Genet. 41 401–427 [DOI] [PubMed] [Google Scholar]
- 6.Espenshade, P. J. (2006) J. Cell Sci. 119 973–976 [DOI] [PubMed] [Google Scholar]
- 7.Raghow, R., Yellaturu, C., Deng, X., Park, E. A., and Elam, M. B. (2008) Trends Endocrinol. Metab. 19 65–73 [DOI] [PubMed] [Google Scholar]
- 8.Hua, X., Wu, J., Goldstein, J. L., Brown, M. S., and Hobbs, H. H. (1995) Genomics 25 667–673 [DOI] [PubMed] [Google Scholar]
- 9.Shimomura, I., Shimano, H., Horton, J. D., Goldstein, J. L., and Brown, M. S. (1997) J. Clin. Investig. 99 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foretz, M., Guichard, C., Ferre, P., and Foufelle, F. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 12737–12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koo, S. H., Dutcher, A. K., and Towle, H. C. (2001) J. Biol. Chem. 276 9437–9445 [DOI] [PubMed] [Google Scholar]
- 12.Shimomura, I., Bashmakov, Y., and Horton, J. D. (1999) J. Biol. Chem. 274 30028–30032 [DOI] [PubMed] [Google Scholar]
- 13.Horton, J. D., Bashmakov, Y., Shimomura, I., and Shimano, H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5987–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, J. B., Sarraf, P., Wright, M., Yao, K. M., Mueller, E., Solanes, G., Lowell, B. B., and Spiegelman, B. M. (1998) J. Clin. Investig. 101 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elam, M. B., Wilcox, H. G., Cagen, L. M., Deng, X., Raghow, R., Kumar, P., Heimberg, M., and Russell, J. C. (2001) J. Lipid Res. 42 2039–2048 [PubMed] [Google Scholar]
- 16.Shimomura, I., Bashmakov, Y., Ikemoto, S., Horton, J. D., Brown, M. S., and Goldstein, J. L. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 13656–13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azzout-Marniche, D., Becard, D., Guichard, C., Foretz, M., Ferre, P., and Foufelle, F. (2000) Biochem. J. 350 389–393 [PMC free article] [PubMed] [Google Scholar]
- 18.Deng, X., Yellaturu, C., Cagen, L., Wilcox, H. G., Park, E. A., Raghow, R., and Elam, M. B. (2007) J. Biol. Chem. 282 17517–17529 [DOI] [PubMed] [Google Scholar]
- 19.Yabe, D., Brown, M. S., and Goldstein, J. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams, C. M., Goldstein, J. L., and Brown, M. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10647–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun, L. P., Li, L., Goldstein, J. L., and Brown, M. S. (2005) J. Biol. Chem. 280 26483–26490 [DOI] [PubMed] [Google Scholar]
- 22.Goldstein, J. L., DeBose-Boyd, R. A., and Brown, M. S. (2006) Cell 124 35–46 [DOI] [PubMed] [Google Scholar]
- 23.Hegarty, B. D., Bobard, A., Hainault, I., Ferre, P., Bossard, P., and Foufelle, F. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thorngate, F. E., Raghow, R., Wilcox, H. G., Werner, C. S., Heimberg, M., and Elam, M. B. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 5392–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dignam, J. D., Lebovitz, R. M., and Roeder, R. G. (1983) Nucleic Acids Res. 11 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowe, T., Aridor, M., McCaffery, J. M., Plutner, H., Nuoffer, C., and Balch, W. E. (1996) J. Cell Biol. 135 895–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aridor, M., Weissman, J., Bannykh, S., Nuoffer, C., and Balch, W. E. (1998) J. Cell Biol. 141 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuehn, M. J., Herrmann, J. M., and Schekman, R. (1998) Nature 391 187–190 [DOI] [PubMed] [Google Scholar]
- 29.Aridor, M., and Balch, W. E. (2000) J. Biol. Chem. 275 35673–35676 [DOI] [PubMed] [Google Scholar]
- 30.Lu, M., and Shyy, J. Y. (2006) Am. J. Physiol. 290 C1477–C1486 [DOI] [PubMed] [Google Scholar]
- 31.Roth, G., Kotzka, J., Kremer, L., Lehr, S., Lohaus, C., Meyer, H. E., Krone, W., and Muller-Wieland, D. (2000) J. Biol. Chem. 275 33302–33307 [DOI] [PubMed] [Google Scholar]
- 32.Sundqvist, A., Bengoechea-Alonso, M. T., Ye, X., Lukiyanchuk, V., Jin, J., Harper, J. W., and Ericsson, J. (2005) Cell Metab. 1 379–391 [DOI] [PubMed] [Google Scholar]
- 33.White, M. F., and Kahn, C. R. (1994) J. Biol. Chem. 269 1–4 [PubMed] [Google Scholar]
- 34.Czech, M. P., Klarlund, J. K., Yagaloff, K. A., Bradford, A. P., and Lewis, R. E. (1988) J. Biol. Chem. 263 11017–11020 [PubMed] [Google Scholar]
- 35.Taniguchi, C. M., Emanuelli, B., and Kahn, C. R. (2006) Nat. Rev. Mol. Cell Biol. 7 85–96 [DOI] [PubMed] [Google Scholar]
- 36.Tobe, K., Kadowaki, T., Hara, K., Gotoh, Y., Kosako, H., Matsuda, S., Tamemoto, H., Ueki, K., Akanuma, Y., and Nishida, E. (1992) J. Biol. Chem. 267 21089–21097 [PubMed] [Google Scholar]
- 37.Peak, M., Rochford, J. J., Borthwick, A. C., Yeaman, S. J., and Agius, L. (1998) Diabetologia 41 16–25 [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto, M., Ogawa, W., Teshigawara, K., Inoue, H., Miyake, K., Sakaue, H., and Kasuga, M. (2002) Diabetes 51 1672–1680 [DOI] [PubMed] [Google Scholar]
- 39.Fleischmann, M., and Iynedjian, P. B. (2000) Biochem. J. 349 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franke, T. F., Yang, S. I., Chan, T. O., Datta, K., Kazlauskas, A., Morrison, D. K., Kaplan, D. R., and Tsichlis, P. N. (1995) Cell 81 727–736 [DOI] [PubMed] [Google Scholar]
- 41.Burgering, B. M., and Coffer, P. J. (1995) Nature 376 599–602 [DOI] [PubMed] [Google Scholar]
- 42.Ono, H., Shimano, H., Katagiri, H., Yahagi, N., Sakoda, H., Onishi, Y., Anai, M., Ogihara, T., Fujishiro, M., Viana, A. Y., Fukushima, Y., Abe, M., Shojima, N., Kikuchi, M., Yamada, N., Oka, Y., and Asano, T. (2003) Diabetes 52 2905–2913 [DOI] [PubMed] [Google Scholar]
- 43.Ribaux, P. G., and Iynedjian, P. B. (2003) Biochem. J. 376 697–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chrysis, D., Zaman, F., Chagin, A. S., Takigawa, M., and Savendahl, L. (2005) Endocrinology 146 1391–1397 [DOI] [PubMed] [Google Scholar]
- 45.Kierbel, A., Gassama-Diagne, A., Mostov, K., and Engel, J. N. (2005) Mol. Biol. Cell 16 2577–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foretz, M., Pacot, C., Dugail, I., Lemarchand, P., Guichard, C., Le Liepvre, X., Berthelier-Lubrano, C., Spiegelman, B., Kim, J. B., Ferre, P., and Foufelle, F. (1999) Mol. Cell. Biol. 19 3760–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cagen, L. M., Deng, X., Wilcox, H. G., Park, E. A., Raghow, R., and Elam, M. B. (2005) Biochem. J. 385 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown, M. S., and Goldstein, J. L. (1997) Cell 89 331–340 [DOI] [PubMed] [Google Scholar]
- 49.Foufelle, F., and Ferre, P. (2002) Biochem. J. 366 377–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng, Z., Otani, H., Brown, M. S., and Goldstein, J. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002) J. Clin. Investig. 109 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yabe, D., Komuro, R., Liang, G., Goldstein, J. L., and Brown, M. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3155–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yellaturu, C. R., Deng, X., Cagen, L. M., Wilcox, H. G., Park, E. A., Raghow, R., and Elam, M. B. (2005) Biochem. Biophys. Res. Commun. 332 174–180 [DOI] [PubMed] [Google Scholar]
- 54.Taniguchi, C. M., Kondo, T., Sajan, M., Luo, J., Bronson, R., Asano, T., Farese, R., Cantley, L. C., and Kahn, C. R. (2006) Cell Metab. 3 343–353 [DOI] [PubMed] [Google Scholar]
- 55.Srinivasan, M., and Begum, N. (1994) J. Biol. Chem. 269 12514–12520 [PubMed] [Google Scholar]