Abstract
NRC/NCoA6 plays an important role in mediating the effects of ligand-bound nuclear hormone receptors as well as other transcription factors. NRC interacting factor 1 (NIF-1) was cloned as a novel factor that interacts in vivo with NRC. Although NIF-1 does not directly interact with nuclear hormone receptors, it enhances activation by nuclear hormone receptors presumably through its interaction with NRC. To further understand the cellular and biological function of NIF-1, we identified NIF-1-associated proteins by in-solution proteolysis followed by mass spectrometry. The identified components revealed factors involved in histone methylation and cell cycle control and include Ash2L, RbBP5, WDR5, HCF-1, DBC-1, and EMSY. Although the NIF-1 complex contains Ash2L, RbBP5, and WDR5, suggesting that the complex might methylate histone H3-Lys-4, we found that the complex contains a H3 methyltransferase activity that modifies a residue other than H3-Lys-4. The identified components form at least two distinctly sized NIF-1 complexes. DBC-1 and EMSY were identified as integral components of an NIF-1 complex of ∼1.5 MDa and were found to play an important role in the regulation of nuclear receptor-mediated transcription. Stimulation of the Sox9 and HoxA1 genes by retinoic acid receptor-α was found to require both DBC-1 and EMSY in addition to NIF-1 for maximal transcriptional activation. Interestingly, NRC was not identified as a component of the NIF-1 complex, suggesting that NIF-1 and NRC do not exist as stable in vitro purified complexes, although the separate NIF-1 and NRC complexes appear to functionally interact in the cell.
A number of crucial insights into the multilayered regulation of transcription have been uncovered through study of nuclear hormone receptor-mediated gene expression. Nuclear hormone receptors are hormone- and ligand-dependent transcription factors that control the coordinated expression of gene networks in numerous physiological, developmental, and metabolic processes (1). Dysfunction of nuclear receptor signaling leads to a number of proliferative, reproductive, and metabolic diseases such as cancer, infertility, obesity, and diabetes (2). The biological functions of nuclear hormone receptors rely on coactivators that represent a diverse group of proteins that enhance nuclear receptor-mediated transcription (3). Extensive studies on the in vivo functions of nuclear receptors have led to the identification and characterization of ∼200 coactivators, all of which have been catalogued on-line at the Nuclear Receptor Signaling Atlas. Coactivators, in general, are known to act by: (i) bridging factors to recruit additional cofactors to DNA-bound nuclear receptors, e.g. p160/SRC proteins (4, 5); (ii) exhibiting various enzymatic activities such as methylation, acetylation, and others to modulate chromatin (6–8); and (iii) interfacing between DNA-bound nuclear receptors and the basal transcriptional machinery, e.g. TRAP/DRIP complex (9).
Nuclear receptor coregulator (NRC)3 also referred to as ASC-2, TRBP, PRIP, and RAP250, was identified through yeast two-hybrid screens as a ligand-dependent interacting factor of a number of nuclear hormone receptors (10–14). The NCBI has also annotated this factor as NCoA6 (nuclear receptor coactivator 6) (NCBI accession number NM_014071). NRC has emerged as an important coactivator not only for nuclear receptors, but also for a number of other well known transcription factors such as c-Fos, c-Jun, CREB, NF-κB, ATF-2, heat shock factors, E2F-1, Rb, and p53 (10, 12, 15–18). The importance of NRC as an essential coregulator is reflected by the finding that NRC null mice are embryonic lethal (19, 20). Various genetic and biochemical studies with NRC–/– and NRC+/– knockout mice have revealed an essential role for NRC in growth, development, cell survival, wound healing, and maintenance of energy homoeostasis (15, 18, 20–22).
Biochemical purification and analysis of NRC-containing complexes suggests that NRC complexes are involved in histone methylation, DNA repair, and nuclear receptor-mediated transcription. An NRC-containing ASCOM complex of ∼2 MDa includes components of the histone methyltransferase machinery, including the Trithorax-related proteins ALL-1/MLL1, ALR-1 (MLL2), ALR-2, HALR (MLL3), Ash2, retinoblastoma-binding protein RbBP5, WDR5 and α/β-tubulins (23, 24). Recent studies have identified NRC as a component of the ALR/MLL2 and PTIP complexes, which are clearly distinct from the previously characterized ASCOM complex (25–27). Both ALR/MLL2 and PTIP complexes contain SET/MLL methyltransferase proteins and have a histone H3 lysine 4-specific methyltransferase activity. The PTIP (Pax transactivation domain-interacting protein) complex is thought to play an additional role in DNA damage response (25).
We have previously reported the cloning and characterization of NIF-1 (NRC-interacting factor 1) (28). NIF-1 is a novel factor that has been shown to interact with NRC in vivo. The expression of NIF-1 enhances the activity of nuclear hormone receptors as well as other transcription factors such as c-Fos and c-Jun (28). Because NIF-1 does not directly interact with nuclear receptors, this ability of NIF-1 to enhance receptor function was proposed to occur through NRC (28). NIF-1 has also recently been shown to associate with components of the CCR4-NOT complex and plays an important role in the regulation of retinoic acid receptor-α (RARα) target genes (29). Surprisingly, NIF-1 has not been identified as a component of any of the NRC-containing complexes described above.
To understand how NIF-1 enhances the activity of nuclear receptors and other transcription factors, we developed methodology to purify NIF-1 complexes from cells and identify their components by mass spectrometry. In this study, we report the purification and characterization of novel NIF-1 complexes that include the core components of SET/MLL complexes (Ash2L, RbBP5, and WDR5) as well as two unique components, deleted in breast cancer-1 (DBC-1) and EMSY. DBC-1 and EMSY exist in a NIF-1 complex that is distinct from the complex containing Ash2L, RbBP5, and WDR5. The purified NIF-1 complex containing Ash2L, RbBP5, and WDR5 does not contain any of the SET/MLL proteins. Interestingly, the isolated NIF-1 complex contains a H3 methyltransferase activity that methylates one or more residues other than H3-Lys-4 (H3K4). Lastly, siRNA studies provide new information on DBC-1 and EMSY and indicate that, along with NIF-1, they play an important role in mediating transcriptional activation by nuclear hormone receptors.
EXPERIMENTAL PROCEDURES
Cell Culture and Plasmids—The 293T packaging cell line (ψA) and the HeLa S3 suspension cell line were a kind gift from Dr. Jeffrey Ye. The ψA and HeLa S3 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum supplemented with glutamine and antibiotics. pLPC is a retroviral expression vector with a Puromycin resistance marker that expresses N-terminal FLAG and HA tags (a gift from Dr. Jeffrey Ye). pLPC-NIF-1 was constructed by introducing the full-length NIF-1 insert into the HindIII-EcoRI sites of the parent pLPC vector. The HindIII site of the original pLPC vector was brought in-frame by an initial digestion and bluntend re-ligation at the BamHI site.
Retroviral Transfection and Cell Culture—The ψA cells were seeded in 10-cm dishes at 5 × 106 cells per dish. The cells were transfected with either the pLPC vector or pLPC-NIF-1 by the calcium phosphate precipitation method. The retroviral supernatant was collected 36–48 h post transfection and filtered through a 0.45-μm sterile filter. HeLa S3 cells were infected with the retroviral supernatant and subsequently selected through resistance to Puromycin (1.5 μg/ml). Single colonies of pLPC-NIF-1 HeLa S3 cells were isolated by serial dilution and screened for the expression of FLAG-HA-NIF-1. A clone expressing a full-length NIF-1 with both FLAG and HA epitope tags was used for the purification of the NIF-1 complex. HeLa S3 cells were routinely maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum supplemented with glutamine and antibiotics. For NIF-1 complex purification, HeLa S3 Vector- and NIF-1 cells were expanded in suspension in Eagle's minimum essential medium, Joklik modification.
Preparation of Nuclear Extracts—Nuclear extracts were obtained from HeLa S3 cells expressing NIF-1 and from control cells transformed with just the pLPC vector using a modified Dignam procedure (30). Cells were collected from ∼8 liters of cells, washed with ice-cold phosphate-buffered saline followed by hypotonic buffer (10 mm Tris-HCl, pH 7.3, 10 mm KCl, 1.5 mm MgCl2, 10 mm β-ME, 0.2 mm PMSF). The cell pellet was swollen with an equal volume of hypotonic buffer and then homogenized 15 times with pestle “A” (Kontes) followed by centrifugation. The nuclear pellet was then resuspended in low salt buffer (20 mm Tris-HCl, pH 7.3, 20 mm KCl, 1.5 mm MgCl2, 0.2 mm EDTA, pH 8.0, 25% glycerol, 10 mm β-ME, 0.2 mm PMSF). The nuclear pellet was homogenized six times and then, 0.5 vol of high salt buffer (20 mm Tris-HCl, pH 7.3, 1.2 m KCl, 1.5 mm MgCl2, 0.2 mm EDTA, pH 8.0, 25% glycerol, 10 mm β-ME, 0.2 mm PMSF) was added dropwise over 30 min. The nuclear extract was centrifuged at 13,000 rpm (SS-34 rotor) for 20 min, and the lipid layer was removed before dialysis (Dialysis buffer BC100: 20 mm Tris-HCl, pH 7.3, 100 mm KCl, 20% glycerol, 0.2 mm EDTA, 10 mm β-ME, 0.2 mm PMSF).
Affinity Purification of NIF-1 Protein Complexes—60 mg of dialyzed nuclear extract protein from pLPC-vector (control) and pLPC-NIF-1 HeLa S3 cultures was first bound to FLAG-M2 antibody-agarose affinity matrix for 4 h. The bound beads were washed eight times with binding buffer (20 mm Tris-HCl, pH 8, 0.1 m KCl, 5 mm MgCl2, 10% glycerol, 0.1% Tween 20, 10 mm β-ME, 0.2 mm PMSF), transferring the FLAG beads to new sterile microcentrifuge tubes with each wash. NIF-1 and the associated proteins were then eluted thrice with 0.5 mg/ml 3×-FLAG peptide (Sigma) for 45 min at 4 °C. The eluted proteins were subsequently bound by incubation overnight to HA antibody-affinity matrix (Roche Applied Science). After extensively washing the HA beads (five times), the associated proteins were eluted twice with 1.2 mg/ml HA peptide (Covance) for 1 h at room temperature. The purified fractions were either analyzed by silver staining or further processed for MS. For analysis by MS, the Tween 20 in the binding buffer was replaced with 1% n-octylglucoside after the HA-affinity immunoprecipitation. The HA-bound proteins were eluted with the HA peptide reconstituted in the n-octylglucoside buffer. The purified fractions were finally ultrafiltered by centrifugation sequentially five times to concentrate the protein and to simultaneously exchange the n-octylglucoside-containing buffer for one containing RapiGest™ (Waters, Milford, MA) (0.1% RapiGest™, 20 mm Tris-HCl, pH 8.0, 0.1 m KCl, 5 mm MgCl2).
MS Analysis—30 μl of purified protein complex was mixed with an equal volume of 100 mm ammonium bicarbonate solution. The mixed solution was denatured at 95 °C for 5 min. After cooling down, 2 μl (0.2 μg/μl) of MS grade trypsin (Promega, Madison, WI) was added into the solution, and the solution was incubated at 37 °C for 12 h. 7 μl 10% trifluoroacetic acid solution was then added into the protein digests which were incubated again at 37 °C for 30 min to decompose the RapiGest™ (Waters). The protein digests were centrifuged at 10,000 rpm for 10 min, and the supernatant fluid under the thin by-product film of RapiGest™ was collected and cleaned-up using a μ-C18 Zip-Tip. The eluted peptides were dried under vacuum and resuspended in 5 μl of 0.1% formic acid. 5 μl of the peptide mixture was analyzed by using nanoflow LC/electrospray ionization-MS/MS with the configuration of a nanoAcquity UPLC mass spectrometer (Waters) coupled directly to an LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific Inc., Waltham, MA). A C18 pre-column was used to load the sample to a 75-μm × 15-cm fused silica C18 BEH analytical column (Waters). A gradient of 1–40% acetonitrile in 0.1% formic acid was delivered over 120 min at a flow rate of 200 nl/min through a fused silica distal end-coated tip nano-electrospray needle (New Objective, Woburn, MA). The data acquisition involved MS survey scans using Orbitrap analyzer and up to five automatic data-dependent MS/MS acquisitions using the linear ion trap. The raw MS data were subsequently processed using Bioworks software (ThermoFisher Scientific), which generated .dta files from each MS/MS spectrum. The .dta files were used to search the NCBI non-redundant protein data base using the Mascot search engine (Version 2.1.0, Matrix Science, Boston, MA). The search parameters included peptide mass tolerance of up to 10 ppm, MS/MS mass tolerance of up to 0.6 Da, and variable oxidation of methionines with up to one missed tryptic cleavage allowed.
Antibodies and Coimmunoprecipitation—The antibodies against NIF-1, NRC, DBC-1, EMSY, MLL1-C, hSET1, and SirT1 were purchased from Bethyl Laboratories. Antibodies against RbBP5, WDR5, and Ash2L were a gift from Danny Reinberg's laboratory (New York University School of Medicine). Antibodies against MLL3, MLL4, and Menin were a kind gift from Dr. Kai Ge (NIDDK, National Institutes of Health, Bethesda, MD). The anti-UTX antibody was generously provided by Dr. Yang Shi (Harvard University). The antibody against Sox9 was purchased from Chemicon International. The anti-HA and anti-FLAG M2 antibodies were purchased from Roche Applied Sciences and Sigma-Aldrich, respectively. The anti-β-actin antibody was from Abcam. The antibodies against HCF-1, HP-1β, and Sin3A were a gift from Dr. Angus Wilson, Dr. Naoko Tanese, and Dr. Gregory David (New York University School of Medicine), respectively. For coimmunoprecipitation, nuclear extract from the NIF-1-expressing HeLa S3 cells was initially incubated with FLAG-affinity matrix. NIF-1 and other associated proteins were eluted off the beads using a FLAG peptide, and the eluates were then immunoprecipitated with either control IgG or with antibodies against Ash2L, DBC-1, or EMSY. 2 μg of antibody was used for each immunoprecipitation. The associated proteins were finally analyzed by Western blotting using antibodies against HA, Ash2L, DBC-1, EMSY, and RbBP5.
Gel Filtration Chromatography—For the size-dependent fractionation of the NIF-1 complex, a 2.4-ml Superose 6 column was used with an exclusion volume of 0.9 ml. The flow rate of the column was 25 μl/min, and the elution profile of the column was determined by an initial fractionation of molecular weight standards (buffer: 20 mm Tris-HCl, pH 7.4, 0.2 m KCl, 10% glycerol, 0.05% Nonidet P-40, 0.2 mm EDTA, 10 mm β-ME). For the fractionation, 100 μl of the purified NIF-1 complex from 3.5 liters of HeLa S3 cells was injected into the column. A total of 60 fractions was collected and analyzed for the distribution profile of NIF-1 and its associated proteins.
siRNA Transfection—siRNAs against DBC-1 (siDBC-1), NIF-1 (siNIF-1) and a non-targeting control (siControl) were purchased from Dharmacon. The siRNA targeted against EMSY and SirT1 was from Qiagen. MCF-7 cells were transfected with the siRNAs (final concentration, 40 nm) using Hiperfect siRNA transfection reagent from Qiagen. 42 h later the cells were incubated with all-trans-retinoic acid (RA) for 24 h, and the cells were harvested for analysis of Sox9 and HoxA1 expression (66 h after the siRNA transfection). Whole cell lysates were prepared in lysis buffer (50 mm KCl, pH 7.4, 250 mm KCl, 1 mm dithiothreitol, 0.25% Nonidet P-40, 7× protease inhibitor tablets (Roche Applied Science)), and equal amounts of protein were analyzed by SDS-PAGE followed by Western blotting with an anti-Sox9 antibody. The knockdown of DBC-1, EMSY, and NIF-1 was also analyzed by Western blotting.
Reverse Transcription-PCR Analysis—Total RNA was extracted using TRIzol® (Invitrogen). For quantitative PCR, 1 μg of total RNA was reverse-transcribed using Superscript III® Reverse Transcriptase. 1 μl of cDNA was used for real-time PCR using SYBR Green (Sigma) on a Roche Applied Science LightCycler® 2.0 System. The values were normalized to an internal glyceraldehyde-3-phosphate dehydrogenase control. The following primers were used: HoxA1, 5′-CCAACTTCACTACCAAGCAGC-3′ and 5′-GACTTCTCTGAGGATTCCTCG-3′; Sox9, 5′-CTCAAAGGCTACGACTGGACG-3′ and 5′-CTCGTTCAGAAGTCTCCAGAGC-3′; and glyceraldehyde-3-phosphate dehydrogenase, 5′-CCTCAACGACCACTTTGTCA-3′ and 5′-CCCTGTTGCTGTAGCCAAAT-3′.
Histone Methylation Assays—Recombinant histone octamers were prepared by one of the investigators (Patrick Trojer) at Danny Reinberg's laboratory at the New York University School of Medicine. The histone methylation assays were performed as described previously (31, 32). The Vector control samples or the NIF-1-purified complex(es) were incubated at 30 °C for 1 h in buffer containing 50 mm Tris-HCl, pH 8.5, 5 mm MgCl2, 4 mm dithiothreitol, 1 μm 3H-labeled S-adenosylmethionine (Amersham Biosciences). 2 μg of histone octamer was used as substrates, and the total volume of the reaction mixture was adjusted to 25 μl. The reaction was stopped by addition of SDS loading buffer and fractionated by 15% SDS-PAGE. The separated histones were transferred to a polyvinylidene difluoride membrane and visualized by Coomassie staining. The membrane was sprayed with EN3HANCE (PerkinElmer Life Sciences) and exposed to Kodak BioMax film.
Yeast Two-hybrid Assay and Cloning of DBC-1/pEGΔPL—Full-length human DBC-1 cDNA was released from the DBC-1/pCS2 expression plasmid (40), a gift from Dr. Tom Boyer at the University of Texas Health Science Center, San Antonio. The NcoI and NcoI-XhoI fragments released from DBC-1/pCS2 were cloned in a two-part ligation with the LexA vector pEGΔPL, cut with NcoI and XhoI. The two-hybrid expression construct DBC-1/pEGΔPL was confirmed by sequencing and also verified for correct orientation and insert size. The following NIF-1 two-hybrid constructs as B42-fusions in pJG4–5ΔPL have been described earlier (28): full-length NIF-1 (NIF-1/pJG4–5ΔPL), NIF-2/pJG4–5ΔPL (an isoform of NIF-1 that lacks the DE region and zinc fingers 1–4), NIFb/pJG4–5ΔPL (contains N-terminal amino acids 42–644), and NIFf/pJG4–5ΔPL (contains C-terminal amino acids 1041–1342, including zinc fingers 5 and 6 and a leucine-zipper like region). A two-hybrid assay using X-gal intensity on plates was carried out as described earlier (28).
RESULTS
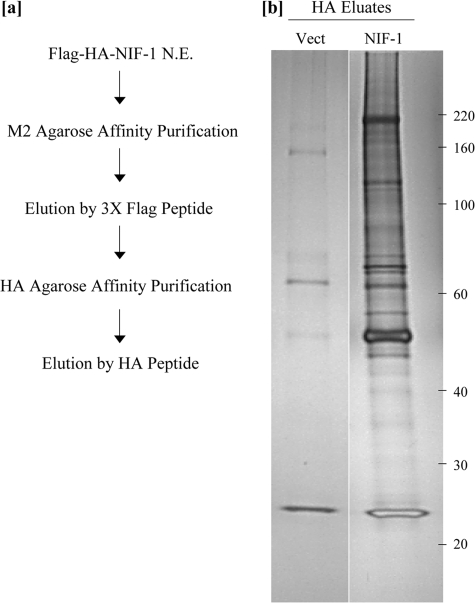
Affinity Purification and Characterization of NIF-1 Complexes—To understand the role of NIF-1 in transcriptional activation, we purified NIF-1 protein complexes by a sequential affinity-purification method. A HeLa S3 cell line stably expressing FLAG-HA-tagged full-length NIF-1 was established using retrovirus-mediated gene transfer, as described under “Experimental Procedures.” A control HeLa S3 cell line was established with a retroviral control vector that only expresses the FLAG and HA tags. Nuclear extracts, prepared from both the NIF-1 stable cell line and the control cell line using a modified Dignam procedure (30), were used for the isolation of NIF-1-associated proteins. The scheme for the two-step purification scheme of NIF-1 complexes is depicted in Fig. 1a. The nuclear extracts were first incubated with FLAG-M2 antibody-agarose affinity matrix. The bound beads were washed extensively, and NIF-1 and associated proteins were then eluted off the beads with a competitive FLAG peptide. The eluted proteins were subsequently bound to HA-antibody-affinity matrix, washed, and then eluted twice with an HA peptide. The HA eluates were analyzed by SDS-PAGE electrophoresis followed by silver staining. Fig. 1b is a representative silver-stained gel comparing the pooled HA peptide eluates from the NIF-1-expressing cells and the control stable HeLa S3 cell lines, which were transformed with just the FLAG-HA vector that was used to express NIF-1. The NIF-1 HA eluates clearly reveal a number of candidate proteins that specifically associate with NIF-1.
FIGURE 1.
Two-step sequential purification scheme for the NIF-1 complex(es). a, 60 mg of nuclear extract protein from pLPC-Vector and pLPC-NIF-1 cells was initially dialyzed to a final salt concentration of 0.1 m KCl. The dialyzed nuclear extract was affinity-purified using an anti-FLAG affinity matrix, and the bound proteins were subsequently eluted from the beads using 3×-FLAG peptide. The eluates were further purified by HA-affinity matrix and, after extensive washing, eluted with a competitive HA peptide. b, the affinity-purified Vector- and NIF-1-complex was resolved in 4–15% SDS-polyacrylamide gels, and the associated proteins were visualized by silver staining. The molecular weight markers are as shown on the right.
The identification of proteins by MS of gel-excised bands is a well established technique. However, this method was unsuccessful in identifying most of the components of the NIF-1 complex. As an alternative approach, an in-solution digestion method was pursued to identify the candidate NIF-1-associated proteins. This technique circumvented the gel electrophoretic step and identified over 15 proteins that are unique to the NIF-1-purified complex (Table 1). A Mascot protein score of 33 was applied as a cut-off for 95% confident identification. In addition to NIF-1 (∼200 kDa), the one or more complexes include Host Cell Factor 1 (HCF-1) (∼200 kDa), retinoblastoma binding protein RbBP5 (∼70 kDa), WD-40 repeat protein WDR5 (∼35 kDa), RuvB-like 2 (Tip49b), RbBP7/RbAp46, deleted in breast cancer 1 (DBC-1, ∼120 kDa), EMSY (∼180 kDa), Ki-67 (∼350 kDa), Matrin 3, nuclear mitotic apparatus protein (NuMA), DEAD-box protein 5 (DDX5), DEAD-box protein 48 (DDX48), α- and β-tubulins, Hsp70, and a number of heterogeneous nuclear ribonucleoproteins.
TABLE 1.
Components of the NIF-1 complex identified by MS and their confidence scores
| Protein results | Accession no. | Score | |
|---|---|---|---|
| 1 | α/β-Tubulin | gi 29788785 | 2072 |
| 2 | Matrin 3 | gi 21626466 | 790 |
| 3 | NIF-1/ZNF 335 | gi 17921989 | 698 |
| 4 | Heat shock protein (Hsp 70) | gi 5729877 | 434 |
| 5 | Host cell factor C1 (HCF-1) | gi 4885403 | 343 |
| 6 | p30 DBC-1 | gi 24432106 | 227 |
| 7 | Ki-67 antigen | gi 19923217 | 217 |
| 8 | EMSY | gi 19923559 | 210 |
| 9 | Nuclear mitotic apparatus protein 1 (NuMA) | gi 71361682 | 205 |
| 10 | RuvB-like 2 | gi 5730023 | 151 |
| 11 | Retinoblastoma binding protein 5 (RbBP5) | gi 53759148 | 137 |
| 12 | Histone 2 A | gi 4504239 | 120 |
| 13 | DEAD-box polypeptide 5 (DDX5) | gi 4758138 | 98 |
| 14 | WD-40 repeat protein 5 (WDR5) | gi 16554627 | 90 |
| 15 | DEAD-box polypeptide 48 (DDX48) | gi 7661920 | 84 |
| 16 | LOC284058 | gi 32698714 | 61 |
| 17 | Retinoblastoma binding protein 7 (RbBP7) | gi 4506439 | 52 |
HCF-1 was originally identified as a host cell factor required for herpes simplex virus transcription (33). It has been shown to associate with multiple histone modifying activities, including the MLL/SET1 H3K4 methyltransferases, Sin3A histone deacetylases, and the MLL1-MOF histone acetyltransferases (34–37). Ash2L, along with RbBP5 and WDR5, form the core components of all the H3K4-specific methyltransferase complexes that have been characterized such as MLL1, PTIP, ASCOM, and SET complexes (23, 25, 26, 35, 36, 38). The biological function of DBC-1 is relatively unknown, with a putative role in apoptosis of ERα-positive breast cancer cells (39, 40). Recently DBC-1 was reported to act as an inhibitor of the Class III deacetylase, SirT1 (41, 42). EMSY was identified as a BRCA2-interacting partner and is amplified in 13% of sporadic breast cancer and 17% of high grade ovarian cancer (43). Ki-67 is a well known proliferation marker whose function is not yet fully understood (44). DBC-1, EMSY, and Ki-67 have not been identified previously as components of a multiprotein complex. DDX5, DDX48, and Matrin 3 have been attributed a role in mRNA splicing (45–48). A recent study indicated that Matrin 3 is a component of an ALR/MLL2-methyltransferase complex (26).
NIF-1 Complexes Are Distinct from Other Known Coactivator Complexes—The mass spectrometric findings indicate that NIF-1 complexes share some core components with other coactivator complexes, including those described for MLL1·MOF, PTIP, and ALR/MLL2 complexes (25, 26, 36). However, a comparative analysis of the components of NIF-1 and other characterized complexes clearly indicated a distinct cohort of proteins unique to NIF-1 complexes.
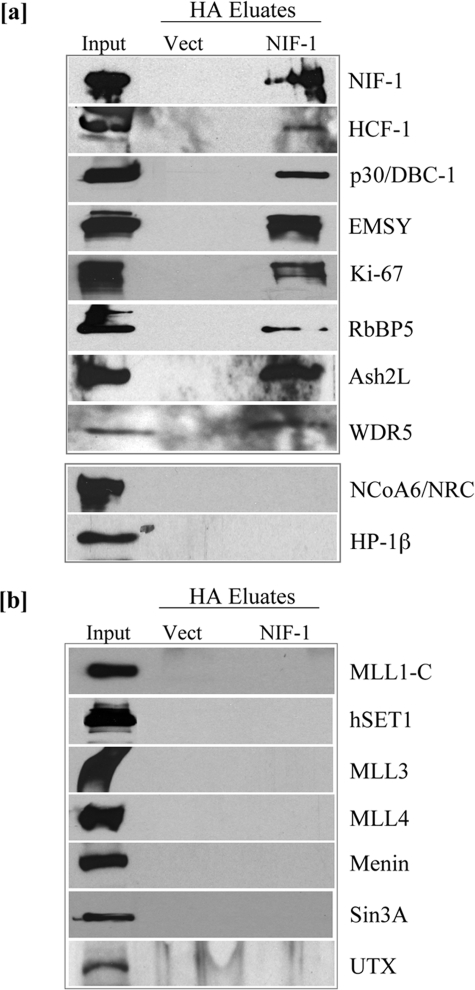
A number of the proteins identified by MS were confirmed by Western blotting as components of NIF-1 complexes. As shown in Fig. 2a, HCF-1, RbBP5, WDR5, DBC-1, EMSY, and Ki-67 were specifically detected in the NIF-1-purified fractions. Although Ash2L was not detected in the NIF-1 complex by MS, its presence was confirmed by Western blotting. The presence of Matrin 3, Hsp70, and α/β-tubulins was confirmed by repeated identification through MS. These results suggest that HCF-1, Ash2L, RbBP5, WDR5, DBC-1, EMSY, Ki-67, Matrin 3, Hsp70, and α/β-tubulins are integral components of one or more novel NIF-1 complexes in the cell.
FIGURE 2.
Western blotting confirmation of components of the purified NIF-1 complex. a, NIF-1-associated proteins were analyzed by Western blot with antibodies indicated on the right. Equivalent amounts of the Vector-purified fractions were used to confirm specificity. 5 μg of nuclear extract was used as Input. b, the purified NIF-1 complex does not include a number of known MLL/SET methyltransferase components. NIF-1-associated proteins were analyzed by Western blot with antibodies indicated on the right.5 μgof nuclear extract was used as Input.
Although NIF-1 was originally identified as an NRC-interacting factor (28), NRC was not identified as a component of the NIF-1 complex by either MS or Western blotting (Fig. 2a). HP-1β, which has been shown to interact in vitro with EMSY (43), was also not detected in the NIF-1 complex (Fig. 2a). The presence of the core components of histone methyltransferase complexes, Ash2L, RbBP5, and WDR5, raised the possibility of a histone methyltransferase component in the purified NIF-1 sample. The two-step affinity-purified NIF-1 fractions were therefore assessed for the presence of known methyltransferases such as SET1, MLL1, MLL3, and MLL4. As shown in Fig. 2b, none of these known methyltransferases were detected in the NIF-1 complex. The NIF-1-purified fractions were also analyzed for other factors identified as components of the MLL/SET methyltransferase complexes. The tumor suppressor protein Menin, shown to be a component of the MLL1·MOF methyltransferase complex, was not present in the NIF-1 complex (35, 49). The recently characterized H3K27 demethylase UTX, an integral member of the ALR/MLL2 and PTIP complexes, was also not detected in the NIF-1-purified fractions (26, 50, 51). Sin3A, a component of both the HCF-1 complex (34) and the ALL-1 supercomplex (52), was also not found as a component of the NIF-1 complex (Fig. 2b). Although Ash2L, RbBP5, and WDR5, the “core components” of histone methyltransferase complexes, are integral components of NIF-1 complexes, none of the known H3K4-specific methyltransferases were detected, perhaps because of the stringent high salt conditions used to purify NIF-1-associated proteins.
NIF-1 Forms at Least Two Distinct Sub-complexes—The components of the NIF-1 complexes revealed factors involved in histone methylation, cell cycle control, and splicing suggesting the possibility of several functionally distinct groups of NIF-1-associated proteins. Further biochemical evidence in support of the observed composition of the NIF-1 complex was obtained by size-dependent gel filtration chromatography. The distribution of the NIF-1-associated proteins after enrichment by FLAG- and HA-affinity purification across a Superose 6 gel filtration column was analyzed by immunoblotting. A 2.4-ml Superose 6-SMART® system with an exclusion size of 2 MDa was utilized to study the elution profile of the NIF-1 complex. The void volume elutes at fraction 17, thyroglobulin dimer (1.3 MDa) at fraction 23, thyroglobulin monomer (667 kDa) at fraction 27, apoferritin (440 kDa) at fraction 31 and alcohol dehydrogenase (150 kDa) at fraction 35 (Fig. 3a). NIF-1 protein was distributed from fractions 19 to 31, peaking in fractions 25–29. The elution profile of DBC-1 and EMSY demonstrated a discrete distribution from fractions 19 to 25. Neither of the two proteins was detected in the lower molecular weight fractions. The elution profile of RbBP5, Ash2L, and WDR5 was also analyzed, and all three proteins were distributed over fractions 27–35. A small fraction of RbBP5 was detected in fraction 25; however Ash2L and WDR5 are distinctly notable for their absence in the high molecular weight fractions (Fig. 3a).
FIGURE 3.
NIF-1 exists in at least two distinct molecular weight complexes and interacts with DBC-1. a, NIF-1-associated proteins purified as described were fractionated in a Superose 6 gel filtration column in buffer containing 0.2 m KCl. The collected fractions were analyzed by Western blotting with antibodies indicated on the right. Positions of protein standards are indicated on the top. The two distinct groups of NIF-1-associated proteins are detected. A high molecular weight complex with a peak of ∼1.5 MDa contains NIF-1, DBC-1, and EMSY. b, evidence for association between the two distinct sub-groups of NIF-1-associated proteins. 20 mg of nuclear extract from NIF-1-expressing HeLa S3 cells was partially purified using FLAG-affinity matrix and eluted with competing 3×-FLAG peptide. The FLAG eluates were immunoprecipitated with either rabbit IgG (control) or with antibodies against Ash2L, DBC-1, or EMSY indicated at the top. The immunoprecipitates were subjected to Western blotting with the antibodies indicated on the right. c, various deletion constructs of NIF-1 as B42 fusions (described under “Experimental Procedures”) were tested for interaction with full-length LexA-DBC-1 in two-hybrid assays as assessed by an X-gal colony response. LexA (pEGΔPL) and B42 (pJG4–5ΔPL) vectors were used as control plasmids and showed no response. The results were reproduced and verified twice. As expected, no interaction was found on dextrose plates. Those indicated by “++++” showed a strong X-gal colony response on Gal/Raf plates within 12–20 h of incubation. The interaction assay with NIF-1b indicated as negative (–) showed no interaction as judged by color reaction after several weeks of incubation even though NIF-1b is expressed as assessed by Western blotting.
The size-dependent fractionation of the NIF-1 complex revealed the presence of at least two distinct NIF-1 sub-complexes. DBC-1 and EMSY co-migrate with NIF-1 as part of a higher molecular mass complex (1–2 MDa with a peak at 1.5 MDa). From the determined size of ∼1.5 MDa, this sub-complex appears to consist of other unidentified protein components. It must be noted here that the DBC-1·EMSY·NIF-1 complex appears to be quite distinct from the previously reported DBC-1·SirT1 complex (41, 42). The conserved histone methyltransferase core proteins, Ash2L, RbBP5, and WDR5, along with a fraction of NIF-1, form a separate sub-complex. Notwithstanding the mutually exclusive distributions of DBC-1·EMSY and Ash2L·RbBP5·WDR5, these results provide further confirmation of the composition and integrity of NIF-1 complexes.
The presence of at least two functionally distinct groups of NIF-1-associated proteins does not necessarily exclude the possibility of an association between the two sub-complexes. The continuous distribution of NIF-1 and the slight overlap between RbBP5 and DBC-1·EMSY in the size fractionation of the NIF-1 complex may translate into an association between the two sub-complexes. To test this hypothesis, a partially purified NIF-1 complex was subjected to a reciprocal immunoprecipitation. Nuclear extracts from the NIF-1-expressing cells were affinity-purified by FLAG-affinity matrix followed by an elution by FLAG peptide. The FLAG eluates were then immunoprecipitated either with control IgG or with antibodies against Ash2L, DBC-1, or EMSY. As shown in Fig. 3b, the antibody against EMSY immunoprecipitated DBC-1, consistent with the comigration of the two factors (Fig. 3a). However, immunoprecipitation with EMSY antibody also detected Ash2L and RbBP5, suggesting a functional interaction between the two NIF-1 sub-complexes. Conversely, an immunoprecipitation with Ash2L detected DBC-1 (although not as efficiently as with EMSY antibody) in addition to RbBP5 (Fig. 3b).
Because Fig. 3a indicated that NIF-1 forms a high molecular weight complex with DBC-1, we sought to determine if this interaction was with NIF-1 or through another protein in the complex. To assess this, we examined the interaction of DBC-1 with NIF-1 and various deletion mutants of NIF-1 in a yeast two-hybrid assay (Fig. 3c). These results support the notion that DBC-1 interacts with NIF-1 through the most C-terminal ∼200 amino acids of NIF-1 (amino acids 1041–1342) containing zinc fingers 5 and 6 and a leucine-zipper-like region.
Role of DBC-1 and EMSY in Nuclear Hormone Receptor-mediated Transcription—Previous studies have elucidated the role of NIF-1 in nuclear hormone receptor-mediated transcription and the regulation of RARα target genes by RA (15, 29). Retinoids induce senescence-like growth arrest in MCF-7 cells through endogenous RARα (53); this effect is associated with the induction of several oncogenic and tumor suppressor genes (54). Sox9 and HoxA1 are two such RA-responsive RARα target genes (53, 55). In fact, we previously reported that induction of Sox9 by ligand-bound RARα plays an important role in RA-mediated inhibition of breast cancer cell growth (53). siRNA-mediated knockdown of NIF-1 resulted in a marked decrease in the RA-mediated stimulation of both Sox9 and HoxA1, indicating a critical role of NIF-1 in the regulation of endogenous RARα target genes (29). The size fractionation of the NIF-1 complex indicates that both DBC-1 and EMSY are closely associated with NIF-1 as components of a distinct subcomplex (Fig. 3a). Consequently, we hypothesized that DBC-1 and EMSY, in concert with NIF-1, may be involved in the regulation of endogenous nuclear receptor target genes.
The effect of knockdown of DBC-1 and EMSY on the stimulation of Sox9 and HoxA1 by RA was assessed. MCF-7 cells were transfected with siRNAs designed specifically against DBC-1 and EMSY mRNA. The cells were incubated with RA, and the effect of knockdown of DBC-1 and EMSY on RAR-mediated stimulation of Sox9 was analyzed by Western blotting. siRNA-mediated silencing of DBC-1 and EMSY expression specifically attenuated stimulation of Sox9 by RA but had no detectable effect on β-actin (Fig. 4a). The observed effect on Sox9 expression by the knockdown of DBC-1 and EMSY might reflect a nonspecific, global decrease in transcription. Thus, we also analyzed the effect of siRNA-mediated knockdown of DBC-1 and EMSY on Menin and hSET1, both less abundant and less stable than β-actin. As indicated in Fig. 4b, knockdown of DBC-1 and EMSY did not affect the expression of either Menin or hSET1, supporting the notion that the effect found with Sox9 does not reflect a global effect of the knockdowns on transcription.
FIGURE 4.
DBC-1 and EMSY are involved in stimulation of Sox9, an RAR target gene. MCF-7 cells were transfected with either an siRNA against DBC-1 (a) or an siRNA against EMSY along with a control siRNA at a concentration of 40 nm. 42 h later the cells were incubated with 1 μm RA for 24 h before whole cell lysates were prepared and immunoblotted with an anti-Sox9 antibody. The extent of knockdown of both DBC-1 and EMSY was also analyzed. β-Actin was used as a loading control. b, siRNA knockdown does not alter the levels of hSRT1 or Menin, which are less abundant and less stable than β-actin. This indicates that the effect of knockdown of DBC-1 and EMSY on stimulation of Sox9 by RA does not reflect a general effect on gene expression.
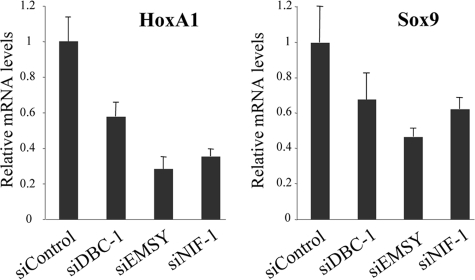
An additional RA-responsive gene, HoxA1, was analyzed to further investigate the relation between DBC-1-, EMSY-, NIF-1-, and RARα-mediated transcription. MCF-7 cells were transfected with either siRNAs against DBC-1, EMSY, or NIF-1 or a corresponding control siRNA. The HoxA1 mRNA levels were initially analyzed by semi-quantitative reverse transcription-PCR to determine the relative important of DBC-1, EMSY, and NIF-1 (data not shown). These results were further confirmed and quantified by real-time PCR analysis (Fig. 5, left panel). The siRNA-mediated knockdown of DBC-1 and EMSY resulted in a significant reduction in RA-mediated stimulation of HoxA1 expression. NIF-1 has been previously shown to be important for the RA-dependent induction of HoxA1 (29) and was therefore used as a positive control. The endogenous Sox9 mRNA level was also analyzed, and the effect of knockdown of DBC-1 and EMSY was validated by both semi-quantitative reverse transcription-PCR (data not shown) and real-time PCR analysis (Fig. 5, right panel). These results therefore imply that DBC-1 and EMSY, as components of a NIF-1 complex, play an important role in the RA-mediated stimulation of Sox9 and HoxA1 by RARα.
FIGURE 5.
Real-time PCR analysis of Hox1 and Sox9. MCF-7 cells were transfected with siRNAs against DBC-1, EMSY, and NIF-1 at a concentration of 40 nm as indicated. A control siRNA was included in the study. The cells were incubated with 1 μm RA for 24 h before the cells were processed. Total RNA (1 μg) was reverse-transcribed, and 1 μl of each cDNA sample was used for the quantitative PCR analysis. All samples were assayed in triplicate. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal normalizing control. The quantification was done using the comparative Ct method. Error bars represent ± S.D.
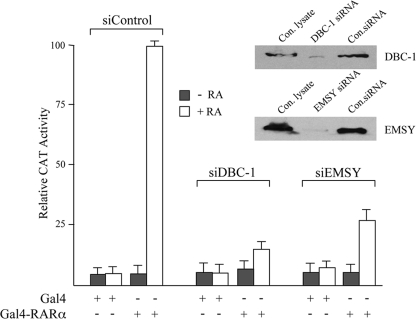
To further exclude the possibility that DBC-1 and EMSY act indirectly through RARα or selectively on the Sox9 and HoxA1 genes, transient transfection studies were carried out with the corresponding siRNAs using a Gal4-RARα vector and a chloramphenicol acetyltransferase reporter gene (pMC110). The Gal4-RARα vector contains only the RARα ligand-binding domain. Thus, such a study could provide further support that the RA-mediated change in the receptor ligand-binding domain is important for the effects of DBC-1 and EMSY. Fig. 6 shows that the knockdown of DBC-1 and EMSY resulted in a 4- to 5-fold reduction in the RA-mediated stimulation of the chloramphenicol acetyltransferase reporter by Gal4-RARα. Western blotting confirmed that the siRNAs decreased the levels of DBC-1 and EMSY about 5- to 10-fold.
FIGURE 6.
Knockdown of DBC-1 and EMSY affects ligand-dependent stimulation by Gal4-RARα in MCF-7 cells. MCF-7 cells were transfected with siRNAs against DBC-1 or EMSY along with a control siRNA at a concentration of 40 nm. 48 h later, the cells were transfected with the reporter pMC110 (100 ng, all lanes) and either Gal4-RARα (200 ng) or equimolar amounts of the Gal4 vector pSG424 as indicated. The cells were incubated with or without RA (0.25 μm) and assayed for chloramphenicol acetyltransferase (CAT) activity 72 h post-siRNA transfection as described previously (28). The maximum chloramphenicol acetyltransferase activity designated as 100 was obtained with Gal4-RARα in the presence of RA with samples transfected with control siRNA. All transfections were carried out in triplicate, and error bars represent ± S.D. The figure also indicates the knockdown of DBC-1 and EMSY as confirmed by Western blotting using equal amounts of protein extracts (35 μg). In addition to the control siRNA (Con.siRNA), Western blotting was also performed with lysates from cells that did not receive siRNA (Con.lysate).
Effect of DBC-1 on RAR-mediated Stimulation of Sox9 Is Independent of SirT1—Studies with endogenous nuclear receptor target genes suggest that DBC-1 and EMSY play important roles in the regulation of Sox9 and HoxA1. However, the one or more mechanisms by which either DBC-1 or EMSY regulate gene expression have not yet been elucidated. DBC-1 has recently been reported to be a negative regulator of SirT1, an NAD+-dependent deacetylase (41, 42). Thus, it remained possible that the attenuation of Sox9 expression after knockdown of DBC-1 could involve deacetylation by SirT1. To address this question, the effect of nicotinamide, a SirT1 inhibitor on the effect of DBC-1 silencing, was analyzed (56). MCF-7 cells were transfected with either a control siRNA or an siRNA designed specifically against DBC-1 mRNA. The cells were treated with or without nicotinamide and then incubated with RA. Endogenous Sox9 protein levels were subsequently analyzed (Fig. 7). Knockdown of DBC-1 markedly decreased stimulation of Sox9 by RA. However, nicotinamide did not reverse the decrease in Sox9 stimulation, suggesting that DBC-1 affects the regulation of Sox9 through a SirT1-independent mechanism (Fig. 7a). The role of SirT1 in RA-induced Sox9 expression could not be analyzed directly, because the viability and proliferation of MCF-7 cells was severely affected by the siRNA-mediated knockdown of SirT1 (data not shown). We also examined whether SirT1 is a component of one of the NIF-1 complexes by Western blotting. As shown in Fig. 7b, SirT1 was not detected as a component of the NIF-1 complex. Furthermore, the previously reported DBC-1/SIRT1 complex elutes at ∼440 kDa, quite distinct from the NIF-1/DBC-1·EMSY complex observed in this study (∼1.5 MDa) (41).
FIGURE 7.
Effect of DBC-1 on stimulation of Sox9 expression is SirT1-independent. a, MCF-7 cells were transfected with either an siRNA against DBC-1 or a control siRNA at a concentration of 40 nm. The cells were treated with 50 nm nicotinamide, a concentration that we have found fully inhibits SirT1 in MCF-7 cells. Following preincubation with nicotinamide, the cells were incubated with RA as indicated for 24 h before whole cell lysates were prepared and immunoblotted with an anti-Sox9 antibody. The extent of knockdown of DBC-1 was also analyzed using an antibody against DBC-1. b, SirT1 is not a component of the NIF-1 complex. Purified NIF-1 and Vector fractions were analyzed by Western blotting with an antibody against SirT1. 5 μg of nuclear extract was used as Input.
NIF-1 Complexes Exhibit an H3-specific Methyltransferase Activity—The size-dependent fractionation of the NIF-1 complex indicated the presence of a lower molecular weight NIF-1 sub-complex consisting of NIF-1, Ash2L, RbBP5, and WDR5 (Fig. 3a). A comprehensive analysis of the NIF-1 complex by MS and Western blotting did not identify any of the known MLL/SET family of methyltransferases (Fig. 2b). Nevertheless, the presence of Ash2L, RbBP5, and WDR5 in the NIF-1 complex alludes to a potential methyltransferase activity. An in vitro histone methyltransferase assay would therefore definitively resolve this issue.
The methyltransferase activity of the purified NIF-1 complex was assayed using histone octamers as substrates. The NIF-1 complex showed a strong H3-specific methyltransferase activity on purified octamers (Fig. 8). However, the absence of the MLL/SET proteins in the NIF-1 complex suggests that the activity may not be specific for H3K4. When the methyltransferase activity of the NIF-1 complex was assayed with recombinant histone octamers carrying a H3K4A mutation, there was no detectable decrease in the H3-specific methylation (Fig. 8). This indicates that, although the NIF-1 complex has a H3-specific methyltransferase activity on histone octamers, the methyltransferase activity is not H3K4-specific. Through Western blotting we excluded CARM1 (PRMT4) as the possible methyltransferase, because CARM1 is known to methylate H3R17, which is associated with transcriptional activation (data not shown). The identification of the methylated residue on H3 and the identity of the enzymatic component in the NIF-1 complex is a subject for additional study.
FIGURE 8.
One or more NIF-1 complexes contain methyltransferase activity that does not methylate H3K4. 5 and 10 μl (out of 1 ml of purified NIF-1 complex) were incubated with 2 μg of WT and recombinant H3K4A histone octamers and the methyl donor [3H]S-adenosylmethionine. The samples were resolved in 15% SDS-polyacrylamide gels, transferred to a polyvinylidene difluoride membrane, and stained with Coomassie Blue (lower panel). The polyvinylidene difluoride membrane was sprayed with EN3HANCE and analyzed by autoradiography (upper panel). The vector-purified fraction was also analyzed with 10 μl of the final preparation. Total nuclear extract (NE) served as a positive control for the in vitro methyltransferase assay.
DISCUSSION
NRC has emerged as a multifunctional coregulator of transcription, with an essential, non-redundant role in growth and development (18). However, NRC-interacting factor, NIF-1, has not been found to associate with any of the coactivator complexes containing NRC. Although NIF-1 has been characterized to enhance ligand-dependent activation by nuclear hormone receptors (28), little is known about its mode of action. In an attempt to elucidate the biological role of NIF-1, a sequential affinity purification approach was utilized to purify NIF-1 associated factors. Identification of the NIF-1-associated proteins by MS of silver- or Coomassie blue-stained gel bands was found to be inefficient. Therefore, we developed a more sensitive “in-solution” MS method to identify components of the NIF-1 complex(es). This approach established that NIF-1 is present within a novel, multiprotein complex that is quite distinct from other known coactivator complexes. Biochemical purification of the NIF-1 complex identified a number of interesting factors, including HCF-1, Ash2L, RbBP5, WDR5, DBC-1, EMSY, Ki-67, Matrin 3, α/β-tubulins, and Hsp70 and a number of heterogeneous nuclear ribonucleoproteins.
The one or more NIF-1 complexes display some similarity to other histone methyltransferase complexes. Ash2L, RbBP5, and WDR5 are considered to be the “core components” of known H3K4-specific MLL/SET methyltransferase complexes (38, 57, 58). HCF-1 has also been identified as an integral component of several histone-modifying complexes (34–37). However, the similarity between NIF-1 and MLL/SET complexes does not extend to their enzymatic activity. Although the NIF-1 complexes exhibit an H3-specific methyltransferase activity on histone octamers, analysis with an H3K4A mutant clearly indicates that the modification on H3 is not on Lys-4. Consistent with this observation, none of the known MLL/SET family of methyltransferases that methylate H3K4 were identified as components of the NIF-1 complex(es). The residue on H3 modified by the NIF-1 complex has not yet been identified. Methylation of other residues, which are marks for activation such as H3-Lys-36 or H3-Lys-79, is an attractive possibility (59, 60), given the functional correlation between NIF-1 and transcriptional activation (28, 29). However, an H3-Arg methyltransferase activity of the NIF-1 complex is also a possibility.
While sharing some factors with previously characterized complexes, the one or more NIF-1 complexes contain several unique components. DBC-1 and EMSY have not been previously associated with any known nuclear receptor coactivator/corepressor complex. However, they have been confirmed to be integral components of the purified NIF-1 complex (Fig. 2). DBC-1 was originally identified as a tumor suppressor gene found to be frequently deleted in breast cancer (39). The biological function of DBC-1 is relatively unknown, with a putative role in apoptosis of ERα-positive breast cancer cells (39, 40). DBC-1 has also been identified as a ligand-independent ERα-interacting protein and is thought to stabilize the expression of ERα (40). Recent studies reported that DBC-1 may function as a negative regulator of SirT1 (41, 42). EMSY was initially identified as a BRCA2-interacting partner and is thought to silence the activation potential of BRCA2 (43, 61). EMSY is amplified in 13% of breast cancers and 17% of ovarian cancers and has been associated with poor survival (43). A potential role for EMSY in chromatin modulation has been proposed through its association with HP-1β (43, 62), although little further is known regarding its function.
The identification and characterization of NIF-1 complexes provides insight into the physiological roles of DBC-1 and EMSY. Biochemical fractionation of the purified NIF-1 complex indicated that DBC-1 and EMSY associate with NIF-1 to form a complex of ∼1.5 MDa. NIF-1 functions as a nuclear hormone receptor coactivator and has been shown to enhance RA-dependent transcriptional activation by RARα (28). Studies with endogenous NIF-1 have revealed a critical role in the regulation of Sox9 and HoxA1, two endogenous RARα target genes (29). Consistent with their association with NIF-1, both DBC-1 and EMSY play an important role in transcriptional activation mediated by RARα. Knockdown of DBC-1 and EMSY resulted in a decrease in RA stimulation of both Sox9 and HoxA1. However, there was no effect on the expression of either β-actin or the less stable Menin or hSET1 proteins, indicating that the effect of siRNA knockdown of DBC-1 and EMSY does not represent a global decrease in transcription. Furthermore, the effect of DBC-1 on the regulation of Sox9 and HoxA1 was found to be SirT1-independent (41, 42), again suggesting a more specific mechanism of transcriptional modulation.
It should be pointed out that the siRNA-mediated silencing of EMSY had a more potent effect on Sox9 and HoxA1 expression, as compared with DBC-1. However, this may be explained by the different extent of knockdown of DBC-1 and EMSY. As shown previously, knockdown of NIF-1 expression also resulted in a decrease in Sox9 and HoxA1 mRNA levels (29). It is interesting to note that EMSY appears to have at least a similar if not greater effect on the expression of endogenous RARα target genes compared with NIF-1.
Although NIF-1 enhances ligand-dependent activation of RARα, no direct interaction has been observed between nuclear receptors and NIF-1 (15, 18, 28). It was postulated that NIF-1 is recruited to the receptor via association with NRC, which has been shown to bind directly to liganded RARα (28). However, NRC was not identified as a component of the NIF-1 complexes, suggesting a different mode of recruitment to the receptor target genes. In this context, DBC-1 contains several LXXLL motifs that may be involved in receptor interaction. Interestingly, DBC-1 has been previously shown to interact with ERα, albeit in a ligand-independent manner (40). Nevertheless, this raises the possibility that DBC-1 could function as a physical bridge between nuclear receptors and the NIF-1 complex. An alternate scenario for the interaction between nuclear receptors and the NIF-1 complex includes an association between NRC and components of the NIF-1 complex under more physiological conditions than those used to purify the NIF-1 complex.
In summary, characterization of NIF-1 protein complexes revealed factors involved in histone methylation, transcription, cell cycle control, and splicing, suggesting the presence of several discrete groups of NIF-1-associated proteins. Size-dependent fractionation of the NIF-1 complex did indeed uncover at least two distinct NIF-1 sub-complexes. The methyltransferase core proteins, Ash2L, RbBP5, and WDR5, form a physically separate complex with NIF-1 as compared with DBC-1 and EMSY (Fig. 3a). Although not all of the remaining components of the NIF-1/DBC-1·EMSY sub-complex have been identified, SirT1 does not appear to be a candidate protein irrespective of its reported association with DBC-1. It is tempting to speculate that the two groups of NIF-1-associated proteins are functionally distinct with varied functions: histone methylation and nuclear receptor-mediated transcription. However, the physical association between the two NIF-1 sub-complexes (Fig. 3b) may indicate a more collaborative function. Biochemical purification of various protein complexes is generally achieved by using stringent salt extraction procedures, which may result in loss of some otherwise integral components of the complex. Thus, it remains possible that the activity of NIF-1/DBC-1·EMSY in nuclear receptor function involves both a yet to be identified methyltransferase as well as recruitment of the second NIF-1 sub-complex containing Ash2L, RbBP5, and WDR5, which contain SET/MLL proteins that associate with NIF-1 protein complexes under the physiological conditions found in cells.
Acknowledgments
We thank those that provided us with antibodies as listed in the report and Jeffrey Ye who provided us with the pLPC vector and for helpful suggestions. We thank Tom Boyer for providing us with the DBC-1/pCS2 plasmid. We also thank Danny Reinberg for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant DK16636 (to H. H. S.). This work was also supported by an NIH Shared Instrumentation Grant 1S10 RR017990-01 and NCI Cancer Institute Core Grant P30CA016087-239025 (to T. A. N.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NRC, nuclear receptor coregulator; PTIP, Pax transactivation domain-interacting protein; X-gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; NIF-1, NRC interacting factor 1; DBC-1, deleted in breast cancer-1; H3K4, H3-Lys-4; siRNA, small interference RNA; HA, hemagglutinin; β-ME, β-mercaptoethanol; HCF-1, host cell factor 1; PMSF, phenylmethylsulfonyl fluoride; MS, mass spectrometry; MS/MS, tandem MS; RA, retinoic cid; Ash2L, Absent, Small or Homeotic-like; RbBP5, retinoblastoma-binding protein 5; WDR5, WD repeat domain 5; RbBP7, retinoblastoma-binding protein 7; Ki-67, antigen identified by monoclonal antibody Ki-67; ERα, estrogen receptor-α.
References
- 1.McKenna, N. J., and O'Malley, B. W. (2002) Cell 108 465–474 [DOI] [PubMed] [Google Scholar]
- 2.Lonard, D. M., Lanz, R. B., and O'Malley, B. W. (2007) Endocr. Rev. 28 575–587 [DOI] [PubMed] [Google Scholar]
- 3.Lonard, D. M., and O'Malley, B. W. (2007) Mol. Cell 27 691–700 [DOI] [PubMed] [Google Scholar]
- 4.Demarest, S. J., Martinez-Yamout, M., Chung, J., Chen, H., Xu, W., Dyson, H. J., Evans, R. M., and Wright, P. E. (2002) Nature 415 549–553 [DOI] [PubMed] [Google Scholar]
- 5.Xu, J., and Li, Q. (2003) Mol. Endocrinol. 17 1681–1692 [DOI] [PubMed] [Google Scholar]
- 6.Tsai, C. C., and Fondell, J. D. (2004) Vitam. Horm. 68 93–122 [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides, T. (2007) Cell 128 693–705 [DOI] [PubMed] [Google Scholar]
- 8.Trotter, K. W., and Archer, T. K. (2008) Nucl. Recept. Signal. 6 e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belakavadi, M., and Fondell, J. D. (2006) Rev. Physiol. Biochem. Pharmacol. 156 23–43 [DOI] [PubMed] [Google Scholar]
- 10.Mahajan, M. A., and Samuels, H. H. (2000) Mol. Cell Biol. 20 5048–5063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, S. K., Anzick, S. L., Choi, J. E., Bubendorf, L., Guan, X. Y., Jung, Y. K., Kallioniemi, O. P., Kononen, J., Trent, J. M., Azorsa, D., Jhun, B. H., Cheong, J. H., Lee, Y. C., Meltzer, P. S., and Lee, J. W. (1999) J. Biol. Chem. 274 34283–34293 [DOI] [PubMed] [Google Scholar]
- 12.Ko, L., Cardona, G. R., and Chin, W. W. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6212–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu, Y., Kan, L., Qi, C., Kanwar, Y. S., Yeldandi, A. V., Rao, M. S., and Reddy, J. K. (2000) J. Biol. Chem. 275 13510–13516 [DOI] [PubMed] [Google Scholar]
- 14.Caira, F., Antonson, P., Pelto-Huikko, M., Treuter, E., and Gustafsson, J. A. (2000) J. Biol. Chem. 275 5308–5317 [DOI] [PubMed] [Google Scholar]
- 15.Mahajan, M. A., and Samuels, H. H. (2005) Endocr. Rev. 26 583–597 [DOI] [PubMed] [Google Scholar]
- 16.Hong, S., Choi, H. M., Park, M. J., Kim, Y. H., Choi, Y. H., Kim, H. H., Choi, Y. H., and Cheong, J. (2004) J. Biol. Chem. 279 16996–17003 [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. K., Na, S. Y., Jung, S. Y., Choi, J. E., Jhun, B. H., Cheong, J., Meltzer, P. S., Lee, Y. C., and Lee, J. W. (2000) Mol. Endocrinol. 14 915–925 [DOI] [PubMed] [Google Scholar]
- 18.Mahajan, M. A., and Samuels, H. H. (2008) Nucl. Recept. Signal 6 e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonson, P., Schuster, G. U., Wang, L., Rozell, B., Holter, E., Flodby, P., Treuter, E., Holmgren, L., and Gustafsson, J. A. (2003) Mol. Cell Biol. 23 1260–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahajan, M. A., Das, S., Zhu, H., Tomic-Canic, M., and Samuels, H. H. (2004) Mol. Cell Biol. 24 4994–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi, C., Kashireddy, P., Zhu, Y. T., Rao, S. M., and Zhu, Y. J. (2004) J. Biol. Chem. 279 33696–33701 [DOI] [PubMed] [Google Scholar]
- 22.Yeom, S. Y., Kim, G. H., Kim, C. H., Jung, H. D., Kim, S. Y., Park, J. Y., Pak, Y. K., Rhee, D. K., Kuang, S. Q., Xu, J., Han, D. J., Song, D. K., Lee, J. W., Lee, K. U., and Kim, S. W. (2006) Mol. Cell Biol. 26 4553–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goo, Y. H., Sohn, Y. C., Kim, D. H., Kim, S. W., Kang, M. J., Jung, D. J., Kwak, E., Barlev, N. A., Berger, S. L., Chow, V. T., Roeder, R. G., Azorsa, D. O., Meltzer, P. S., Suh, P. G., Song, E. J., Lee, K. J., Lee, Y. C., and Lee, J. W. (2003) Mol. Cell Biol. 23 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S., Lee, D. K., Dou, Y., Lee, J., Lee, B., Kwak, E., Kong, Y. Y., Lee, S. K., Roeder, R. G., and Lee, J. W. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 15392–15397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho, Y. W., Hong, T., Hong, S., Guo, H., Yu, H., Kim, D., Guszczynski, T., Dressler, G. R., Copeland, T. D., Kalkum, M., and Ge, K. (2007) J. Biol. Chem. 282 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Issaeva, I., Zonis, Y., Rozovskaia, T., Orlovsky, K., Croce, C. M., Nakamura, T., Mazo, A., Eisenbach, L., and Canaani, E. (2007) Mol. Cell. Biol. 27 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel, S. R., Kim, D., Levitan, I., and Dressler, G. R. (2007) Dev. Cell 13 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahajan, M. A., Murray, A., and Samuels, H. H. (2002) Mol. Cell. Biol. 22 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garapaty, S., Mahajan, M. A., and Samuels, H. H. (2008) J. Biol. Chem. 283 6806–6816 [DOI] [PubMed] [Google Scholar]
- 30.Dignam, J. D., Lebovitz, R. M., and Roeder, R. G. (1983) Nucleic Acids Res. 11 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishioka, K., Rice, J. C., Sarma, K., Erdjument-Bromage, H., Werner, J., Wang, Y., Chuikov, S., Valenzuela, P., Tempst, P., Steward, R., Lis, J. T., Allis, C. D., and Reinberg, D. (2002) Mol. Cell 9 1201–1213 [DOI] [PubMed] [Google Scholar]
- 32.Sarma, K., Margueron, R., Ivanov, A., Pirrotta, V., and Reinberg, D. (2008) Mol. Cell. Biol. 28 2718–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristie, T. M., and Sharp, P. A. (1993) J. Biol. Chem. 268 6525–6534 [PubMed] [Google Scholar]
- 34.Wysocka, J., Myers, M. P., Laherty, C. D., Eisenman, R. N., and Herr, W. (2003) Genes Dev. 17 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoyama, A., Wang, Z., Wysocka, J., Sanyal, M., Aufiero, D. J., Kitabayashi, I., Herr, W., and Cleary, M. L. (2004) Mol. Cell. Biol. 24 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dou, Y., Milne, T. A., Tackett, A. J., Smith, E. R., Fukuda, A., Wysocka, J., Allis, C. D., Chait, B. T., Hess, J. L., and Roeder, R. G. (2005) Cell 121 873–885 [DOI] [PubMed] [Google Scholar]
- 37.Narayanan, A., Ruyechan, W. T., and Kristie, T. M. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 10835–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, J. H., Tate, C. M., You, J. S., and Skalnik, D. G. (2007) J. Biol. Chem. 282 13419–13428 [DOI] [PubMed] [Google Scholar]
- 39.Hamaguchi, M., Meth, J. L., von Klitzing, C., Wei, W., Esposito, D., Rodgers, L., Walsh, T., Welcsh, P., King, M. C., and Wigler, M. H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13647–13652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trauernicht, A. M., Kim, S. J., Kim, N. H., and Boyer, T. G. (2007) Mol. Endocrinol. 21 1526–1536 [DOI] [PubMed] [Google Scholar]
- 41.Zhao, W., Kruse, J. P., Tang, Y., Jung, S. Y., Qin, J., and Gu, W. (2008) Nature 451 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, J. E., Chen, J., and Lou, Z. (2008) Nature 451 583–586 [DOI] [PubMed] [Google Scholar]
- 43.Hughes-Davies, L., Huntsman, D., Ruas, M., Fuks, F., Bye, J., Chin, S. F., Milner, J., Brown, L. A., Hsu, F., Gilks, B., Nielsen, T., Schulzer, M., Chia, S., Ragaz, J., Cahn, A., Linger, L., Ozdag, H., Cattaneo, E., Jordanova, E. S., Schuuring, E., Yu, D. S., Venkitaraman, A., Ponder, B., Doherty, A., Aparicio, S., Bentley, D., Theillet, C., Ponting, C. P., Caldas, C., and Kouzarides, T. (2003) Cell 115 523–535 [DOI] [PubMed] [Google Scholar]
- 44.Endl, E., and Gerdes, J. (2000) Exp. Cell Res. 257 231–237 [DOI] [PubMed] [Google Scholar]
- 45.Liu, Z. R. (2002) Mol. Cell. Biol. 22 5443–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuller-Pace, F. V. (2006) Nucleic Acids Res. 34 4206–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Z., and Krainer, A. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11574–11579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeCerbo, J., and Carmichael, G. G. (2005) Curr. Opin. Cell Biol. 17 302–308 [DOI] [PubMed] [Google Scholar]
- 49.Caslini, C., Yang, Z., El-Osta, M., Milne, T. A., Slany, R. K., and Hess, J. L. (2007) Cancer Res. 67 7275–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, M. G., Villa, R., Trojer, P., Norman, J., Yan, K. P., Reinberg, D., Di Croce, L., and Shiekhattar, R. (2007) Science 318 447–450 [DOI] [PubMed] [Google Scholar]
- 51.Agger, K., Cloos, P. A., Christensen, J., Pasini, D., Rose, S., Rappsilber, J., Issaeva, I., Canaani, E., Salcini, A. E., and Helin, K. (2007) Nature 449 731–734 [DOI] [PubMed] [Google Scholar]
- 52.Nakamura, T., Mori, T., Tada, S., Krajewski, W., Rozovskaia, T., Wassell, R., Dubois, G., Mazo, A., Croce, C. M., and Canaani, E. (2002) Mol. Cell 10 1119–1128 [DOI] [PubMed] [Google Scholar]
- 53.Afonja, O., Raaka, B. M., Huang, A., Das, S., Zhao, X., Helmer, E., Juste, D., and Samuels, H. H. (2002) Oncogene 21 7850–7860 [DOI] [PubMed] [Google Scholar]
- 54.Chen, Y., Dokmanovic, M., Stein, W. D., Ardecky, R. J., and Roninson, I. B. (2006) Cancer Res. 66 8749–8761 [DOI] [PubMed] [Google Scholar]
- 55.Gillespie, R. F., and Gudas, L. J. (2007) J. Mol. Biol. 372 298–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee, H. I., Jang, S. Y., Kang, H. T., and Hwang, E. S. (2008) Biochem. Biophys. Res. Commun. 368 298–304 [DOI] [PubMed] [Google Scholar]
- 57.Crawford, B. D., and Hess, J. L. (2006) ACS Chem. Biol. 1 495–498 [DOI] [PubMed] [Google Scholar]
- 58.Dou, Y., Milne, T. A., Ruthenburg, A. J., Lee, S., Lee, J. W., Verdine, G. L., Allis, C. D., and Roeder, R. G. (2006) Nat. Struct. Mol. Biol. 13 713–719 [DOI] [PubMed] [Google Scholar]
- 59.Li, B., Carey, M., and Workman, J. L. (2007) Cell 128 707–719 [DOI] [PubMed] [Google Scholar]
- 60.Steger, D. J., Lefterova, M. I., Ying, L., Stonestrom, A. J., Schupp, M., Zhuo, D., Vakoc, A. L., Kim, J. E., Chen, J., Lazar, M. A., Blobel, G. A., and Vakoc, C. R. (2008) Mol. Cell. Biol. 28 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haber, D. A. (2003) Cell 115 507–508 [DOI] [PubMed] [Google Scholar]
- 62.Huang, Y., Myers, M. P., and Xu, R. M. (2006) Structure 14 703–712 [DOI] [PubMed] [Google Scholar]