Abstract
Follistatin is a transcriptional target and a modulator of activin action. Through an autocrine/paracrine loop, activin controls follistatin levels and thus regulates its own bioavailability. In gonadotropic αT3-1 cells, activin induces follistatin transcription primarily through the action of Smad3 at an intronic Smad-binding element (SBE1). Using a proteomics approach, we searched for endogenous αT3-1 proteins that participate in SBE1-mediated transcription. We identified FoxL2, a member of the forkhead family, as a candidate modulator of SBE1 function. Mutations of FoxL2 are associated with the blepharophimosis/ptosis/epicanthus inversus syndrome characterized with craniofacial defects and premature ovarian failure. FoxL2 localizes to α-glycoprotein subunit- and follicle-stimulating hormone β-positive cells of the adult mouse pituitary and is present in αT3-1 and LβT2 cells, but its pituitary actions remain largely unknown. We have determined that FoxL2 binds to a forkhead-binding element (FKHB) located just downstream of the SBE1 site of the follistatin gene and functions as a Smad3 partner to drive SBE1-mediated transcription in αT3-1 cells treated with activin. Chromatin immunoprecipitation assays confirm that endogenous FoxL2 and Smad3 are recruited to the intronic enhancer of the follistatin gene where the SBE1 and FKHB sites are located. Exogenous FoxL2 enhances SBE1-mediated transcription, and short hairpin RNA-mediated knockdown of endogenous FoxL2 protein compromises this effect in αT3-1 cells. FoxL2 directly associates with Smad3 but not Smad2 or Smad4. This association between Smad3 and FoxL2 is mediated by the MH2 domain of Smad3 and is dependent on an intact forkhead domain in FoxL2. Altogether, these observations highlight a novel role for FoxL2 and suggest that it may function as a transcriptional regulator and a coordinator of Smad3 targets.
The TGF-β3 family is represented by a group of evolutionarily conserved secreted proteins that control a broad spectrum of biological functions of embryonic and adult tissues (1, 2). Activin A and B comprising homodimers of the structurally related inhibin βA and βB subunits, respectively, are members of the TGF-β family (1). As such, they are present in most tissues, and both forms of activin are known to participate in the local control of a variety of essential developmental and homeostatic processes, including those involved in the control of reproduction (1, 3, 4). The varied actions of activins are in turn fine-tuned by cell-specific and context-dependent mechanisms of inactivation via binding proteins such as follistatin and FSTL3 (5, 6), receptor antagonists such as inhibin (7), or intracellular mechanisms that limit further signaling (2, 8, 9).
Follistatin is a single chain glycoprotein that binds activin with high affinity at a 2:1 molar ratio and interferes with its access to cell-surface receptors (10–12). Follistatin also binds and modulates the actions of several other members of the TGF-β family such as myostatin and certain bone morphogenetic proteins (13). First characterized as a product of the gonads with feedback FSH inhibitory actions on the pituitary, many studies have since demonstrated that follistatin is expressed and secreted locally by pituitary cells, as it is by many other tissues and organs (14–19). Genetic inactivation of the Fst in mice is associated with extensive defects and death soon after birth, reflecting the broad physiological importance of follistatin and its ability to target multiple TGF-β family ligands at relevant sites (20). The global overexpression of follistatin, on the other hand, is associated with reduced FSH levels, gonadal defects, and abnormalities of the skin and hair (21). The highly conserved human and rodent follistatin genes comprising six introns and exons produce two mRNA transcripts and the corresponding C-terminally extended FS315 or truncated FS288 proteins that are presumed to serve distinct physiological functions (22, 23).
Activin and follistatin have emerged as key players of the autocrine/paracrine milieu of the anterior pituitary involved in the control of the reproductive axis in rodents and primates (24). Activin B and follistatin expressed and secreted by pituitary gonadotropes exert local control on FSHβ and LHβ subunit expression in these cells and, working in conjunction with GnRH, inhibin, and steroid hormones, promote the cyclic variations of FSH/LH ratios required for normal reproduction (25). Intra-pituitary activin B exerts positive effects on FSH secretion from gonadotropes, and this local action of activin B is in turn controlled by the local “buffering” capacity of follistatin (26–29). Follistatin itself is a downstream target of activin signaling in pituitary gonadotropes, and the activin-induced rise in follistatin expression contributes to the local feedback control and preserves the critical level of activin signaling (27, 28, 30–35). Similar mechanisms are also critical for maintaining the functional integrity of other tissues, and associations between disrupted follistatin tone and altered proliferative, tumorigenic, or metastatic potential of differentiated endocrine and non-endocrine cells have been reported (36–39). Despite the established importance of this interplay between follistatin and activin, activin-dependent mechanisms that control follistatin expression in the pituitary or other tissues remain largely unknown.
Activin signals are transmitted by type II (ActRII or ActRIIB) and type I (ALK4) serine/threonine kinase receptors and the ALK4-dependent phosphorylation of the downstream Smad2 and/or Smad3 transcriptional effectors (2, 40). The phosphorylation of Smad2/3 at their C-terminal SSXS motifs enables them to form partnerships with Smad4/DPC4 and other cofactors and mediate transcriptional effects by binding to specific promoter elements on target genes and assembling co-activators and co-repressors (2, 8, 9). The interactions of Smads with co-factors are fundamental to their function, and the differential partnerships between P-Smad2 or P-Smad3 and cell-specific factors allows activin to generate a diverse set of transcriptional programs in many different cell types under different physiological conditions (2, 9).
We previously reported that Smad-mediated transcription of the rat Fst in activin-treated αT3-1 cells is dependent on a conserved Smad-binding element (SBE1) that localizes to an activin-responsive region of intron 1 (35). This SBE1 element preferentially binds Smad3 and mediates downstream effects of activin A. Luciferase reporter constructs that incorporate up to 2.86 kb of the upstream promoter of the rat Fst, but lack the intronic SBE1 enhancer, are unresponsive to activin signaling in either αT3-1 or LβT2 cells (29, 35, 41). By contrast, the same reporters display both basal and Smad-dependent inducible activity when transfected into HepG2 or HEK293T cells, indicating that the function of SBE1 is dispensable in these cell types. Similarly, the upstream promoter region of the human FST is sufficient for Smad-dependent activation in HepG2 cells treated with either activin or TGF-β (42). These observations raised the possibility that the differential modes of Smad-dependent regulation of follistatin expression are dictated through partnerships between Smads and other factors that are differentially expressed in a cell type-dependent fashion. We evaluated this hypothesis by searching for factors that cooperate with Smad3 at the intronic SBE1 site of the rat follistatin gene and mediate activin effects in αT3-1 cells. Using an oligonucleotide pulldown approach, we enriched endogenous αT3-1 proteins that assemble at or near the intronic SBE1 site and analyzed them by mass spectrometry. One of the proteins enriched by this strategy was FoxL2, a member of the forkhead family of transcription factors. In this study, we identify FoxL2 as a key Smad3 partner and evaluate its role in facilitating activin-dependent transcription of the follistatin gene in gonadotrope-derived αT3-1 cells.
EXPERIMENTAL PROCEDURES
Cell Lines—The mouse gonadotrope-derived αT3-1 (43) and the human embryonic kidney (HEK) 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 2 mm glutamine.
Plasmids and Reagents—The N-terminally Myc-tagged human (h) Smad2, -3, and -4 were subcloned into the pCS2+ expression vector as described previously (35). The hSmad2 and -3 cDNAs were provided by Dr. Rik Derynck, University of California, San Francisco, and the hSmad4/DPC4 cDNA was obtained from Dr. Scott Kern, The Johns Hopkins University School of Medicine, Baltimore. The myc-hSmad3-MH1 domain corresponding to amino acids 1–145 of hSmad3 was generated by PCR amplification using the myc-hSmad3 plasmid as a template (forward T7 primer, 5′-AATACGACTCACTATAGG; reverse primer, 5′-ACGTTCTAGATTACGTGTGGCGTGGCAC). The full-length myc-hSmad3 was also used as a template to generate myc-hSmad3-MH2 encompassing amino acids 220–425 of hSmad3 (forward primer incorporating the Myc tag, 5′-ACGTGGATCCACCATGGGAGAACAGAAACTGATCTCTGAAGAAGACCTGATGGACCTGCAGCCAGTTACC; reverse SP6 primer, 5′-ATTTAGGTGACACTATA). Both fragments were digested and directionally subcloned into the BamHI and XbaI sites of pCS2+ and verified by sequence analysis. The human FoxL2 cDNA clone was obtained from Open Biosystems (Huntsville, AL), and the entire coding region (AscI/SphI fragment) was subcloned into the pCS2+ expression vector at the StuI site. To obtain the mouse FoxL2, total RNA extracted from αT3-1 was reverse-transcribed, and a cDNA encoding mFoxL2 was amplified and subcloned into the BamHI/EcoRI sites of pCS2+ (forward primer, 5′-GGATCCACCATGATGGCCAGCTACCCCGAGCCC; reverse primer, 5′-GAATTCTCAGAGATCCAGACGCGAGTG). An N-terminal FLAG tag was then added by using an upstream primer that incorporates the corresponding nucleotides (forward primer, 5′-GGATCCACCATGGACTACAAAGACGACGACGACAAAATGATGGCCAGCTACCCCGAGCCC). C-terminally truncated forms of FoxL2 were generated using a PCR approach with upstream primers that incorporate the FLAG tag, as shown in Table 1.
TABLE 1.
Primers used to generate truncated forms of FLAG-tagged mFoxL2
| Forward primer for all C-terminal truncationsa | ||
|
5′-CGTTGGATCCACCATGGACTACAAAGACGACGACGACAAAATGATGGCCAGCTACCCCGAGCCC
|
||
| Reverse primers | ||
| FoxL2-(Δ268-375) | 5′-CGTTGAATTCTCACACGACGCCCGGAGGCAGCGC | |
| FoxL2-(Δ234-375) | 5′-CGTTGAATTCTCAGCCCGGGCCGGCGGCTGCAGC | |
| FoxL2-(Δ215-375) | 5′-CGTTGAATTCTCAGCAGGAGGCGTAGGGCATGGG | |
| FoxL2-(Δ161-375) | 5′-CGTTGAATTCTCACTTGCCGGGCTGGAAGTG | |
| FoxL2-(Δ133-375) | 5′-CGTTGAATTCTCAGTCCTCGCAGGCCGGGTCGAG | |
| Forward primersa | ||
| FoxL2-(Δ1-44) | 5′-CGTTGGATCCACCATGGACTACAAAGACGACGACGACAAACCAGACCCCGCGCAGAAGCCCCCG | |
| Reverse primer | ||
| FoxL2-(Δ1-44) | 5′-CGTTGAATTCTCAGAGATCCAGACGCGAGTG | |
Forward primers incorporate the sequence corresponding to the FLAG tag, and all primers include BamHI and EcoRI restriction sites for ligation into the pCS2+ mammalian expression vector.
The luciferase reporter plasmids that incorporate fragments of the rat Fst just upstream of the coding region of luciferase in the pGL2 basic vector (Promega, Madison, WI) have been characterized previously (35). The rFS(2.9)-luc and the rFS(0.3)-luc reporter plasmids incorporate the –2864/+136 and –312/+136 fragments of rat follistatin, respectively. In the rFS(2.9i)-luc or the rFS(0.3i)-luc plasmid, the entire first intron (+227/+2097) of the rat Fst is ligated to the 3′ end of either the –2864/+136 or the –312/+136 fragment via an engineered SpeI site. The rFS(0.3ex45)-luc reporter retains only the +1784/+1912 fragment of intron 1 of the rat follistatin gene just downstream of the –312/+136 fragment in rFS(0.3)-luc. The SBE1 mutant form of the rFS(0.3ex45)-luc reporter was generated by using a PCR approach (wild type, 5′...TTGT... 3′; SBE1 mutant, 5′... TTGTaatTGGGTCA... 3), as described previously (35). Mutations of the forkhead-binding site (FKHB) were also generated by a similar PCR approach to introduce those substitutions shown in Fig. 9. Recombinant human activin A was purified from the conditioned medium of stably transfected Chinese hamster ovary cells (Salk Institute, La Jolla, CA).
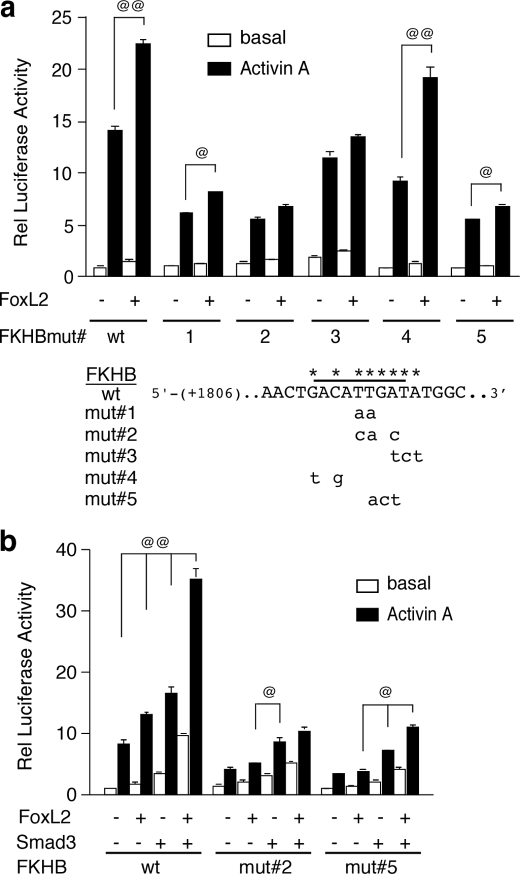
FIGURE 9.
FoxL2 effects on the follistatin gene are mediated via a regulatory element just downstream of SBE1. a, functional importance of a putative FKHB located just downstream of SBE1 was evaluated by mutating the indicated nucleotides within the context of the rFS(0.3ex45)-luc plasmid and comparing the activities of these mutant reporters to that of the wild type in αT3-1 cells co-transfected with mFoxL2 or vector and treated with vehicle or 1 nm activin A. b, FKHB mut#2 and mut#5 were further tested to determine their activities in αT3-1 cells co-transfected with mFoxL2 or Smad3 individually or together. Luciferase activity was measured as described above. Each panel shows a representative experiment performed in triplicate (@, p < 0.05 or @@, p < 0.001 in the presence or absence of co-transfected mFoxL2 or Smad3).
Transfections and Luciferase Reporter Assays—For transient transfection experiments, the αT3-1 cells were seeded in poly-l-lysine-coated 12-well tissue culture plates at a density of 3 × 105 cells/well in 2 ml of complete medium (DMEM, 10% FBS and 2 mm glutamine). The αT3-1 cells were transfected with the indicated plasmids for 6 h using the Superfect transfection reagent (Qiagen, Hilden, Germany). The cells were subsequently treated with vehicle or activin A in DMEM supplemented with 2% FBS and 2 mm glutamine. Luciferase reporter activity was measured 15 h later, as described (35). Where indicated, varying amounts of expression plasmids encoding Myc-tagged Smads, FLAG-tagged mFoxL2, or empty pCS2+ vector were co-transfected along with the reporter plasmids. Luciferase reporter activity of cell lysates was measured using d-luciferin luciferase substrate (Biosynth, Naperville, IL) with a Lumimark microplate luminometer (Bio-Rad) and internally normalized to the activity of the co-transfected cytomegalovirus-β-galactosidase plasmid, as described (35). Reported data correspond to luciferase/β-galactosidase ratios of each plasmid relative to the activity of the pGL2 basic vector. HEK293T cells were transiently transfected by plating 105 cells/well on poly-l-lysine-coated 24-well tissue culture plates in complete DMEM 24 h prior to transfection. The cells were transfected overnight with 200 ng of the appropriate reporter plasmid, 10 ng of β-galactosidase, and other plasmids, as indicated, using polyethyleneimine as the transfection reagent. The cells were washed 16–18 h later and treated for 18 h with either vehicle or activin A in complete DMEM. Luciferase reporter activity was measured as described above. The data were subjected to analysis of variance and Tukey's post hoc test.
Oligonucleotide Precipitation Assays—The oligonucleotide precipitation experiments were essentially as described previously (35). Briefly, αT3-1 cells (5 × 107/15-cm dish) were treated for 30 min with 1 nm activin A or vehicle in freshly supplemented growth medium. Samples were prepared by brief sonication in lysis buffer followed by a 10-min centrifugation at 12,000 × g at 4 °C. The supernatant obtained from the equivalent of 5 × 107 cells (∼4 mg) was incubated for 2 h at 4 °C with 2 μg of biotinylated double-stranded oligonucleotides, pre-coupled to streptavidin-agarose beads (Pierce), in the presence of 30 μg of poly(dI-dC) (Sigma). The agarose beads were washed three times by centrifugation, and endogenous αT3-1 proteins enriched by this method were recovered and subjected to Western analysis or processed for analysis by mass spectrometry. For Western analyses, the samples were resolved under reducing conditions by one-dimensional gel electrophoresis (4–12% SDS-NuPAGE gels, Invitrogen) with MOPS as the running buffer and then transferred to nitrocellulose membranes. Endogenous proteins corresponding to Smad2/3 or FoxL2 were detected using rabbit anti-hSmad2/3, as described previously (35), and a commercially available goat anti-m/hFoxL2 (Imgenex, San Diego), respectively, and the appropriate horseradish peroxidase-conjugated secondary antibodies (donkey anti-rabbit or rabbit anti-goat; Pierce). Immune complexes were visualized with SuperSignal West Pico chemiluminescence substrate (Pierce). The double-stranded biotinylated wild type or the equivalent mutant probe harboring a mutated SBE1 site corresponds to the SBE1-containing region of intron 1 of the rat Fst (35).
Identification of SBE1-associated Proteins by Mass Spectrometry—Proteins enriched by the oligonucleotide precipitation assays described above were processed and analyzed by two approaches. In one strategy, bound proteins were recovered off the biotinylated probes pre-coupled to streptavidin-agarose beads by boiling with sample loading buffer under reducing conditions. Proteins that co-purified with the wild type or mutant SBE1 oligonucleotides were separated by one-dimensional gel electrophoresis and visualized with Coomassie Blue. Mass-specific bands were excised from the stained gel. Gel slices were de-stained by treatment with 40% n-propyl alcohol and 50% acetonitrile and digested over 16 h at 37 °C with 100 ng of trypsin added in 10 μl of ammonium bicarbonate solution (20 mm). Peptides were extracted and dried down completely and then re-dissolved in 0.1% formic acid for ESI-MS/MS analysis on a Bruker Daltonics Esquire 3000 Plus mass spectrometer (Billerica, MA). MS analysis data were analyzed using the Mascot algorithm (Matrix Science, London, UK) on a local Mascot server (version 2.1.0) and searched against the latest NCBI nr protein data base. Peptides were required to be fully tryptic with one missed cleavage allowed. Results were filtered, combined, sorted, and displayed using Scaffold (Proteome Software, Portland, OR) to validate protein identifications derived from MS/MS sequencing results. Scaffold can verify peptide identifications assigned by Mascot using the X!Tandem data base searching program (44, 45). Scaffold then probabilistically validates these peptide identifications and derives corresponding protein probabilities using Protein-Prophet (44, 45). Only protein hits that met the Mascot algorithm dynamic significance threshold were considered. Alternatively, proteins were subjected to MudPIT analysis (46). For this strategy, protein complexes bound to the streptavidin-agarose coupled biotinylated oligonucleotides were washed in 400 μl of ammonium bicarbonate (20 mm) and then directly digested by the addition of 50 μl of ammonium bicarbonate buffer (20 mm) containing 2.5 μg of trypsin. Digestion was allowed to progress at 37 °C for 16 h. Supernatants containing peptides that resulted from the tryptic digests were recovered and lyophilized. The samples were then brought up in 0.1% formic acid for ESI-MS/MS analysis. Multidimensional protein identification technology (MudPIT) was performed on a Bruker Daltonics Esquire 3000 Plus mass spectrometer (Billerica, MA). A tandem capillary column packed with strong ion exchange and C18 reversed-phase material was developed using a 10-step pH gradient from 2.5 to 8, followed by a 40-min acetonitrile gradient (5–45%) for direct identification of proteins. Each experiment was performed three times using a gelbased LC/MS/MS approach or twice using the multidimensional protein identification technique (MudPIT) (46). Identified proteins had to be present in two of three individual experiments to be considered.
Chromatin Immunoprecipitation (ChIP)—Activin A or vehicle-treated αT3-1 cells were subjected to ChIP analyses as described previously (35). Briefly, following cross-linking with 1% formaldehyde, the nuclear fractions recovered from cell lysates were sonicated on ice (Misonix XL200 ultrasonic cell disruptor) and pre-cleared by incubation with protein A-Sepharose, salmon sperm DNA, 0.05% bovine serum albumin, and 0.3% normal rabbit or goat serum followed by centrifugation. Half of the material recovered from ∼107 αT3-1 cells was incubated overnight at 4 °C with 5 μl of normal rabbit IgG or protein A-purified rabbit anti-hSmad2/3 and protein-A-Sepharose, as described previously (35). Parallel samples were incubated with either normal goat IgG or goat anti-FoxL2 directed to a peptide at the C terminus of the protein (Imgenex, San Diego) and protein-A-Sepharose. After thoroughly washing the beads, the specifically bound complexes were eluted from the protein-A-Sepharose beads with 1% SDS, and cross-linking was reversed by an overnight incubation at 65 °C. The purified DNA samples obtained from QIAquick spin columns (Qiagen Inc., Valencia, CA) were then analyzed by real time PCR using the SYBR GREEN PCR master mix and the ABI PRISM 7700 sequence detector (PerkinElmer Life Sciences). Primer sets that surround the intronic SBE1 site were used to determine the abundance of the intervening fragment of the endogenous mouse follistatin gene (forward, 5′-AACAGTCTAGTAAAAGTCAATGCAAGCT, and reverse, 5′-TGCGCCCCAGCCATAT) relative to a fragment ∼5 kb upstream of the transcription start site (forward, 5′-AGATAGAGATCCCACCACAGAACAA, and reverse, 5′-GGATGGACTTGGGTGGTATCTGTA). The relative abundance of the amplified fragments in the treated and untreated samples was analyzed using the ΔΔCt method.
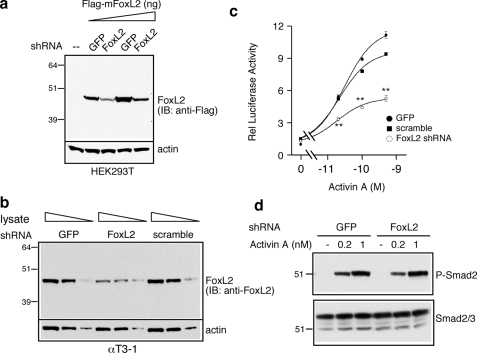
shRNA-mediated Knockdown of FoxL2—The strategy used to achieve shRNA-mediated knockdown was essentially as described previously (35). Several potential shRNA targets of the mouse FoxL2 were identified using the sfold software (47, 48). Of four shRNAs analyzed, the shRNA (5′-CCATGATGCATTGCTCATA) targeted to the coding region of the single-exon mouse FoxL2 was validated for its ability to knockdown FLAG-tagged mFoxL2 in transfected HEK293T cells and the endogenous FoxL2 protein expressed in αT3-1 cells. The shRNA cassettes were generated by PCR amplification from pSuper (Oligoengine, Seattle, WA) using a forward T3 primer and a reverse primer that incorporates an intervening nine-nucleotide hairpin loop (5′-TCTCTTGAA) between the sense and antisense target sequence (5′-CTGTCTAGACAAAAA (sense target) TCTCTTGAA (antisense target) GGGGATCTGTGGTCTCATACA-3′), in addition to the human H1 RNA polymerase III promoter sequence, as described (49). The resulting shRNA cassettes were inserted into the compatible NheI site in the 3′-long terminal repeat of the lentiviral internal ribosome entry site vector, p156RRLSINpptCMVGFPPRE, that incorporates cytomegalovirus-driven GFP as a marker (generously provided by Dr. Inder Verma, Salk Institute). The empty lentiviral internal ribosome entry site vector expressing only GFP and/or one that incorporates a scrambled sequence (5′-GGCATTACAGTATCGATCAGA) was used as control. The extent and specificity of FoxL2 knockdown by each shRNA were evaluated in HEK293T cells co-transfected with the FLAG-mFoxL2 expression plasmid. Cell extracts prepared 72 h after transfection were subjected to Western blot analysis using mAb anti-FLAG. To achieve shRNA-mediated knockdown of endogenous FoxL2 in αT3-1 cells, viral particles harboring FoxL2-targeted shRNA cassettes or the control vector were prepared using HEK293T as described previously (35). The αT3-1 cells were infected in the presence of 4 μg/ml Polybrene, and 3 days later, the extent of knockdown of endogenous mouse FoxL2 protein was assessed by Western blot analysis using goat anti-FoxL2 IgG, compared with actin levels detected by a mAb anti-actin. To achieve uniformity of knockdown for functional studies, αT3-1 cells were transduced similarly with lentivirus and then expanded under normal growth conditions. Approximately 107 cells expressing either GFP or scrambled shRNA as control or FoxL2 shRNA were subjected to fluorescence-activated cell sorting (FACS) on a FACSVantage SE DiVa (BD Biosciences) equipped with a 488-nm argon laser. Initial gating was based on forward scatter and side scatter to maximize recovery of live single cells. According to the fluorescence intensity histogram of each population of infected αT3-1 cells, the top 5% of GFP+ cells were sorted and collected for further analysis. Initial experiments had indicated that cells sorted based on GFP intensity, GFPlow, GFPmid, or GFPhigh, did not exhibit significant differences. The sorted GFP+ αT3-1 expressing either of the two control shRNAs or FoxL2-specific shRNA were expanded and evaluated for FoxL2 expression by Western blots and for activin responsiveness of the rFS(0.3ex45)-luc reporter.
Immunohistochemistry—Wild type mice were perfused with 4% paraformaldehyde, and the pituitary was removed and then frozen in embedding medium. Twenty μm sections were cut on a cryostat and mounted on slides. Slides were washed three times in KPBS and incubated overnight at 4 °C in KPBS containing 0.4% Triton X-100 and 2% normal donkey serum, along with primary antibodies at recommended dilutions as follows: goat anti-FoxL2 (1:500; Imgenex), rabbit anti-αGSU (1:1000; National Hormone and Peptide Program of NIDDK, National Institutes of Health), guinea pig anti-FSHβ (1:5000; National Hormone and Peptide Program) and rabbit anti-S100 (1:500; Sigma). After primary antibody incubation, slides were washed three times and exposed for 1 h at room temperature to the appropriate species-specific secondary antibodies conjugated to fluorophores. Donkey anti-goat antibodies were conjugated to Alexa-488 (Invitrogen). Donkey anti-guinea pig antibodies were conjugated to Cy3 (Jackson ImmunoResearch). Donkey anti-rabbit antibodies were conjugated to Cy3 (Jackson ImmunoResearch). All secondary antibodies were used at a dilution of 1:600. Slides were washed three times in KPBS and mounted with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole to visualize nuclei (Vector Laboratories, Burlingame, CA). Sections were imaged using a Leica TCS SP2 AOBS confocal microscope (Leica Microsystems, Inc, Bannockburn, IL).
RESULTS
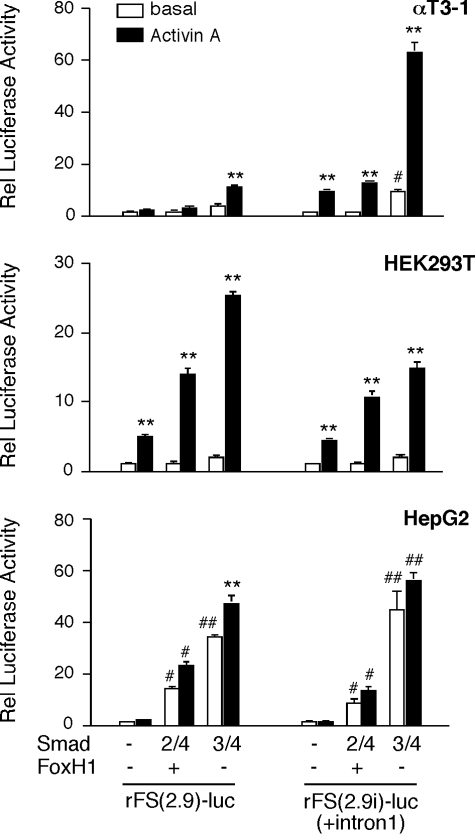
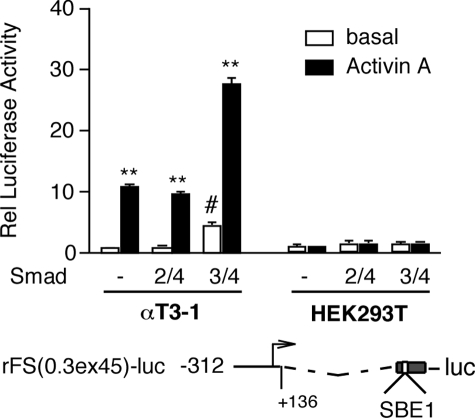
Activin Regulation of Follistatin Transcription Is Dependent on Cell Type-specific Mechanisms—We previously reported that an intronic Smad-binding element, SBE1, is required for the action of activin A on follistatin transcription in gonadotrope-derived αT3-1 cells (35). This conserved SBE1 site preferentially recruits Smad3 and mediates ALK4-dependent activin effects (35). An equivalent SBE1 site is conserved in the human FST, but its activity seems to be dispensable for activin or TGF-β action in HepG2 cells (42). To verify that these observations do not reflect species-specific mechanisms, we evaluated fragments of the rat Fst in αT3-1, HEK293T, and HepG2 cells. Consistent with our previous data, the rFS(2.9)-luc reporter lacking intron 1 was not activated in αT3-1 cells treated with activin A and was only slightly induced by co-transfected Smad3/4 but not the combination of Smad2/4 and FoxH1 (Fig. 1, upper panel). FoxH1 had no effect on reporter activity whether co-transfected with or without Smad2/4 (data not shown). Although Smad4 was routinely included in the transfection DNA mixture, it did not appreciably contribute to the effect of Smad3 alone, as we reported previously (35). In contrast to these results, the rFS(2.9i)-luc reporter that incorporates intron 1 was induced 9.5-fold in response to activin A in αT3-1 cells, and this activity was further enhanced by co-transfected Smad3/4 but not Smad2/4 (Fig. 1, upper panel). In HEK293T, activin A induced the two reporters to the same extent, and both were further activated by Smad3/4 and, to a lesser extent, by Smad2/4/FoxH1 (Fig. 1, middle panel). The inducible activity of these two reporter plasmids, rFS(2.9)-luc and rFS(2.9i)-luc, in HepG2 cells was more in line with those of HEK293T cells except that a maximum concentration of activin A (1 nm) produced a much more modest response (Fig. 1, bottom panel). To further assess the differential importance of the intronic SBE1, the activity of the rFS(0.3ex45)-luc reporter, which retains only 312 bp of the upstream promoter region and the 129 bp activin-responsive fragment of intron 1 that incorporates SBE1, was evaluated in αT3-1 and HEK293T cells. As expected, this reporter plasmid was responsive to activin A and Smad3/4 in αT3-1 but not in HEK293T cells (Fig. 2).
FIGURE 1.
The upstream promoter region or downstream regulatory elements of the rat Fst differentially mediate activin A (1 nm) effects in HEK293T or HepG2 cells compared with gonadotrope-derived αT3-1 cells. The indicated cell types were transiently co-transfected with either the rFS(2.9)-luc or rFS(2.9i)-luc reporter plasmid and cytomegalovirus-β-galactosidase as an internal control for transfection efficiency, along with a combination of FoxH1/Smad2/Smad4, Smad3/Smad4 or empty vector. The transfections were performed using the Superfect reagent for αT3-1 and HepG2 cells and polyethyleneimine for HEK293T cells. Following the 6-h (αT3-1 or HepG2) or the 18-h (HEK293T) incubation period with the mix of DNA and transfection reagent, the cells were supplemented with fresh complete medium and treated for 15 h with vehicle or activin A. The cells were harvested 16–18 h after treatment, and the luciferase activity (arbitrary light units) of each sample was internally normalized to that of β-galactosidase. The data are reported as the ratio of these normalized luciferase values relative (Rel) to that of the empty pGL2 vector under identical conditions. The experiments were performed in triplicate, and the reported mean ± S.E. values are from a representative experiment with each cell line (**, p < 0.001 relative to the corresponding untreated basal; #, p < 0.05; ##, p < 0.001 relative to basal of the group).
FIGURE 2.
A luciferase reporter plasmid, rFS(0.3ex45)-luc, that incorporates the Smad-binding element of intron 1 but not the region upstream of –312 of the rat Fst is activated in response to activin A (1 nm) in αT3-1 but not HEK293T cells. The transfection conditions were the same as those described for Fig. 1. FoxH1 plasmid was included along with Smad2/4. The reported data (arbitrary light units) reflect luciferase activity normalized to β-galactosidase and then calculated relative (Rel) to pGL2 activity obtained from cells treated under the same conditions. The experiments were performed in triplicate, and the reported mean ± S.E. values are from a representative experiment with each cell line (**, p < 0.001 relative to the corresponding untreated basal; #, p < 0.05 relative to basal of the group).
Mass Spectrometric Identification of Smad2/3 and FoxL2 as SBE1-associated Transcription Factors—To evaluate the mechanism underlying SBE1-mediated activation of the follistatin gene in αT3-1 cells and to identify endogenous proteins that, along with Smad3, participate in mediating this effect of activin, we employed biotinylated probes harboring wild type or mutant SBE1 and enriched associated proteins by an oligonucleotide pulldown strategy. We used lysates of αT3-1 cells treated or untreated with activin A and analyzed the proteins associated with the probes under basal and stimulated conditions by subjecting them to ESI-LC/MS/MS analysis. Initially, we focused on proteins migrating on SDS-PAGE in the expected mobility range for Smad2, -3, or -4 (45–65 kDa) and analyzed them following in-gel trypsin digestion. One set of tryptic peptides derived from proteins of ∼50-kDa mobility corresponded to endogenous Smad2/3 of mouse αT3-1 cells. These peptides were significant by Mascot criteria, and the protein identification was consistent across all experimental sets. As shown in Table 2, these Smad2/3-derived peptides were recovered exclusively in activin-treated samples incubated with the wild type but not the corresponding SBE1 mutant oligonucleotide probe, consistent with our previous data on the role of SBE1 in recruiting Smad3 and mediating activin effects (35). The analysis of equivalent samples by MudPIT yielded similar results and led to the same conclusions regarding the pattern of Smad2/3 association with the SBE1 probes (Table 2). These observations confirmed the specificity of the interactions with wild type versus mutant probe and validated our strategy. Further analysis of the samples identified yet another protein migrating with an apparent mobility of ∼45 kDa. This protein was identified as FoxL2 (also known as PFrk) (50). Unlike Smad2/3, mass spectrometric analysis of tryptic peptides from gel slices suggested that FoxL2 interacted with both wild type and mutant probes in a manner largely independent of activin A, whereas MudPIT data suggested that FoxL2 binding was dependent on an intact SBE1 site (Table 2). We resolved this issue through a series of biochemical and functional assays, and as shown in this study, we determined that FoxL2 action on the follistatin gene in αT3-1 cells is indeed dependent on a functionally intact SBE1.
TABLE 2.
Identification by mass spectrometry of Smad2/3 and FoxL2 from αT3-1 lysates following oligonucleotide pulldown with biotinylated probes that incorporate a wild type or a mutant SBE1
|
Gel
slicesa
|
MudPITb
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
Wild type
|
Mutant
|
Wild type
|
Mutant
|
|||||
| Basal | Activin A | Basal | Activin A | Basal | Activin A | Basal | Activin A | |
| Smad2/3 | 79-99c | 99-201 | NDd | ND | ||||
| FoxL2 | 77-109 | 83-97 | 72-87 | 78-84 | 159-205 | 202-253 | ND | ND |
Three independent experiments were performed.
Two independent experiments were performed.
Individual ion scores of peptides >42 indicate identity or extensive homology (p < 0.05) to the identified protein.
ND, not determined.
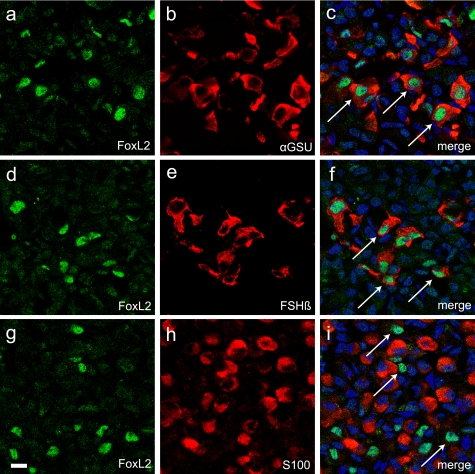
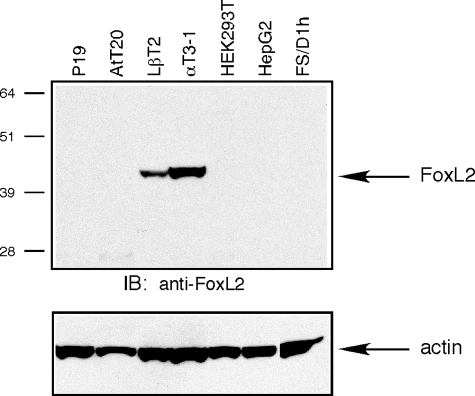
FoxL2 Is Present in αGSU- and FSHβ-positive Pituitary Cells—The identification of FoxL2 from cellular extracts of αT3-1 cells suggested that it participates in the regulation of follistatin expression in anterior pituitary gonadotropes. FoxL2 is a member of the winged-helix forkhead family of transcription factors (51). It is one of the earliest known markers of ovarian differentiation in mammals and is essential for the maintenance of ovarian follicles in the adult (52). Deficiencies of FoxL2 cause the disorder known as blepharophimosis/ptosis/epicanthus inversus syndrome (BPES) associated with eyelid problems and varying degrees of premature ovarian failure (53, 54). Consistent with the BPES phenotype, FoxL2 is expressed in ovarian follicles and the developing eyelid (53). FoxL2 (initially named PFrk) is also present in the developing pituitary around e10–10.5 (55). Moreover, FoxL2 expression persists in the adult pituitary and localizes mostly to cells positive for the glycoprotein α subunit (αGSU) shared by LH, FSH, and thyroid-stimulating hormone (TSH) (56). To further validate that FoxL2 co-localizes with markers of gonadotropes, we performed immunohistochemical studies using pituitary tissue from adult male mice. These analyses confirmed that FoxL2 immunoreactivity is present in a sub-set of pituitary cells (Fig. 3, a, d, and g). The majority of FoxL2 immunoreactivity was restricted to the population of anterior pituitary cells immunopositive for αGSU (Fig. 3, a–c). A sub-set of FoxL2 immunopositive cells also express FSHβ, a marker of gonadotropes (Fig. 3, d–f). These observations confirmed that FoxL2 expression indeed persists in the adult pituitary and localizes primarily to gonadotropes and thyrotropes. Interestingly, FoxL2 immunoreactivity was excluded from the population of S100-positive folliculostellate cells previously established to be a major source of pituitary-derived follistatin (Fig. 3, g–i). A screen of a number of pituitary- and non-pituitary-derived cell lines by Western blot analysis confirmed the restricted pattern of FoxL2 expression. These experiments demonstrated that a protein of ∼45 kDa recognized by anti-FoxL2 IgG was detectable only in the extracts of αT3-1 and LβT2 cells, two gonadotrope-derived cell lines, and not in other cell types that were tested (Fig. 4).
FIGURE 3.
FoxL2 immunofluorescence in pituitary tissue of an adult mouse co-localizes with αGSU and FSHβ. Confocal images of double-immunolabeled pituitary sections (20 μm) from an adult wild type mouse show nuclear FoxL2 staining in a sub-set of cells that are positive for αGSU or FSHβ. FoxL2 expression (a, d, and g) was evaluated using goat anti-FoxL2. For co-localization studies, rabbit anti-αGSU (b), guinea pig anti-FSHβ (e), or rabbit anti-S100 (h) were used. FoxL2 immunoreactivity was detected with donkey anti-goat IgG conjugated to Alexa-488 (green), αGSU, and S100 with donkey anti-rabbit IgG conjugated to Cy3 (red) and FSHβ with donkey anti-guinea pig IgG conjugated to Cy3 (red). Cell nuclei were labeled with 4′,6′-diamidino-2-phenylindole (DAPI)(blue). Merged confocal images of FoxL2 and αGSU (c) or FoxL2 and FSHβ (f) show that FoxL2 co-localizes with both pituitary markers. Merged images of FoxL2 and S100 show lack of co-localization (i). Arrows in c, f, and i denote representative FoxL2-positive cells that also display cytoplasmic αGSU or FSHβ but not S100. Scale bar, 10 μm.
FIGURE 4.
FoxL2 protein is detectable by Western blot in gonadotrope-derived αT3-1 and LβT2 cells. Protein extracts prepared from the indicated cell lines were analyzed by SDS-PAGE (150 μg of protein/lane), and endogenous FoxL2 was detected using goat anti-FoxL2 (upper panel). The blots were probed with a mAb anti-actin to assess differences in loading (lower panel). The cell lines evaluated were as follows: mouse embryonal carcinoma P19, mouse pituitary corticotropic AtT20, mouse pituitary gonadotropic αT3-1 and LβT2, human embryonic kidney HEK293T, human hepatocellular carcinoma HepG2, rat pituitary folliculostellate FS/D1h. Protein bands were visualized by ECL. IB, immunoblot.
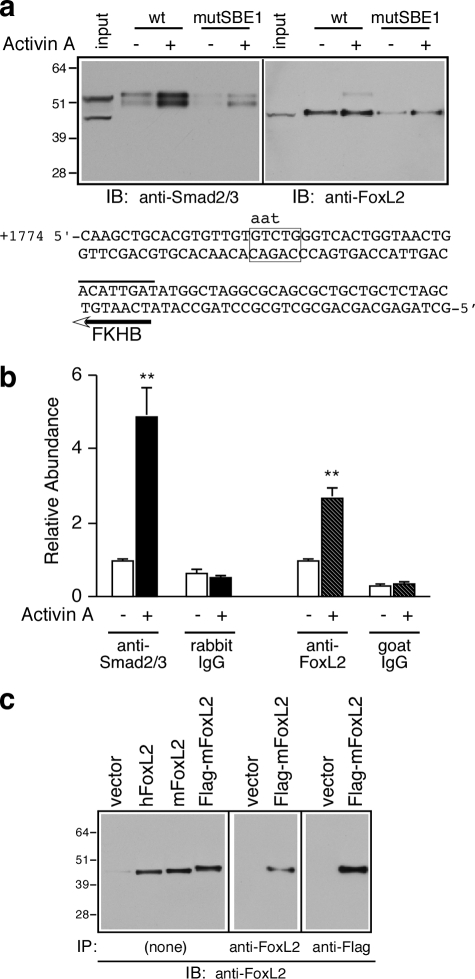
FoxL2 and Smad3 Recruitment to the Follistatin Gene Is Dependent on an Intact SBE1 in Intron 1—To validate the mass spectrometric identification of Smad2/3 and FoxL2 as SBE1-binding proteins expressed in αT3-1 cells, we carried out oligonucleotide precipitation experiments with the same biotinylated oligonucleotides followed by Western analysis of specifically bound proteins. Consistent with the mass spectrometric analyses and our recent work (35), this experiment confirmed that activin A signaling in αT3-1 cells induces the association of endogenous Smad2/3 to the biotinylated SBE1 oligonucleotide probe and that this association is dependent on an intact SBE1 site (Fig. 5a, left panel). In parallel oligonucleotide precipitation experiments, we confirmed that FoxL2 is also recruited to the wild type biotinylated SBE1 oligonucleotide probe (Fig. 5a, right panel). In contrast to Smad3, FoxL2 binding to the wild type probe was largely constitutive and only marginally sensitive to activin A (Fig. 5a, right panel). Similar to Smad3, FoxL2 binding was compromised to the equivalent probe that incorporates a mutant SBE1 site (Fig. 5a, right panel). We also performed ChIP experiments to further evaluate the functional importance of endogenous FoxL2 and determine whether it is indeed recruited to the intronic enhancer of the endogenous Fst in αT3-1 cells. As we reported previously (35), Smad2/3 recruitment to this region was induced 5-fold in cells treated with 1 nm activin A compared with vehicle (Fig. 5b). FoxL2 recruitment to the same intronic enhancer region was also induced, albeit to a lesser extent, in cells treated with activin A (Fig. 5b). The specificity of the goat FoxL2 antibody was validated by testing it against recombinant mouse and human FoxL2 and FLAG-tagged mFoxL2 (Fig. 5c). The results of this series of experiments suggest that follistatin transcription in αT3-1 cells is regulated by the concerted actions of constitutively bound FoxL2 and activin-dependent recruitment of Smad2/3 to SBE1.
FIGURE 5.
Activin A induces the recruitment of Smad2/3 and FoxL2 to the intronic SBE1 of follistatin. a, oligonucleotide precipitation experiments utilized extracts of αT3-1 cells treated with vehicle or 1 nm activin A for 30 min. The cell extracts were precipitated with wild type (wt) or mutant (mutSBE1) SBE1 double-stranded biotinylated probes and subjected to Western blot analysis with either rabbit anti-Smad2/3 (left panel) or goat anti-FoxL2 (right panel) IgGs. The numbers to the left of the immunoblots (IB) correspond to protein size markers (kDa). The nucleotide sequence corresponds to the double-stranded biotinylated probe used for these experiments. The sequence shows the wild type SBE1 site (boxed) and the corresponding mutant form (indicated by lowercase letters) as well as the FKHB (marked by a line above and an arrow below the sequence). b, αT3-1 cells were treated with vehicle or activin A (1 nm for 30 min) and subjected to ChIP analysis using rabbit anti-Smad2/3, goat anti-FoxL2, or the corresponding normal rabbit or goat IgGs. The samples were then analyzed by real time PCR using a primer set surrounding the intronic SBE1 site of the endogenous mouse Fst or a primer set that amplifies a fragment located 5 kb upstream of the follistatin transcription start site. Real time PCR results (mean ± S.E. of triplicate determinations) of a representative experiment were calculated using the ΔΔCt method (**, p < 0.001 relative to the corresponding basal). c, protein extracts prepared from αT3-1 cells transfected with appropriate plasmids to express human FoxL2 (hFoxL2), mouse FoxL2 (mFoxL2), FLAG-tagged mFoxL2, or vector only were directly resolved by SDS-PAGE and subjected to Western blot analysis (15 μg/lane; left panel). Alternatively, FLAG-mFoxL2 was first immunoprecipitated (IP) from αT3-1 extracts (200 μg) by incubation with either goat anti-FoxL2 (middle panel) or agarose-coupled mAb anti-FLAG (right panel) and then subjected to Western blot analysis using goat anti-FoxL2.
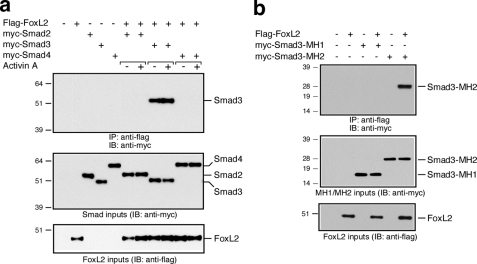
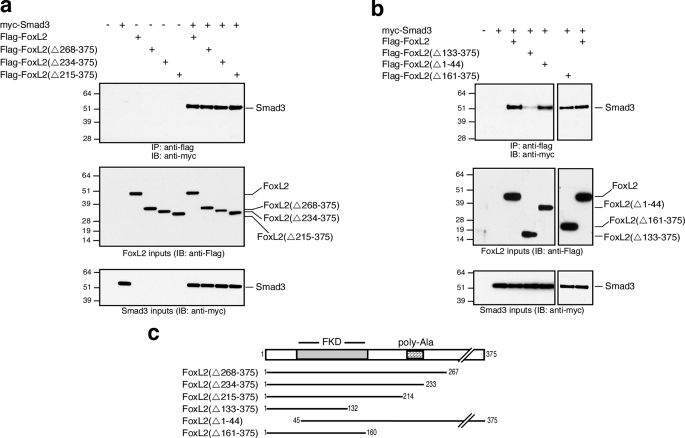
FoxL2 Is a Smad3 Partner—The results thus far raise the possibility that FoxL2 interacts and forms a complex with Smad2/3. We tested this possibility by performing co-immunoprecipitation experiments using Myc-tagged hSmad2, -3, or -4 and FLAG-tagged mFoxL2 transiently expressed in HEK293T cells. These co-immunoprecipitation experiments conclusively demonstrated that FoxL2 associates with only Smad3 but not Smad2 or Smad4 (Fig. 6a). The association of FoxL2 and Smad3 was marginally induced by activin treatment and was largely independent of activin signaling (Fig. 6a). Equivalent results were observed when the duration of activin treatment was varied from 5 min to as long as 1 h (data not shown). Further analysis using Myc-tagged MH1 or MH2 domains of Smad3 confirmed that the MH2 domain of Smad3 mediates complex formation between FoxL2 and Smad3 (Fig. 6b). We also generated N- and C-terminally truncated FLAG-tagged mFoxL2 variants to map the Smad3-interacting domain within FoxL2. Serial C-terminal truncations of mFoxL2 that ultimately remove the entire C terminus following the forkhead domain, amino acids 161–375, did not significantly disrupt Smad3 binding (Fig. 7). A further truncation that removes 28 more residues, including the second wing-like loop region of the forkhead domain, significantly disrupted the ability of this recombinant mutant form of FoxL2 (FoxL2(Δ133–375)) to bind Smad3 (Fig. 7b). These observations suggest that an intact forkhead domain in FoxL2 is necessary for Smad3 binding. Consistently, FoxL2(Δ1–44), which incorporates an intact forkhead domain and the entire C terminus, retains full Smad3 binding (Fig. 7b).
FIGURE 6.
FoxL2 associates with the MH2 domain of Smad3. a, HEK293T cells were transfected with vectors for co-expression of FLAG-mFoxL2 with myc-Smad2, myc-Smad3, or myc-Smad4. After 3 days of protein expression, the cells were treated with vehicle or 1 nm activin A for 30 min. Cell lysates were immunoprecipitated (IP) by incubation with agarose-coupled mAb anti-FLAG, and immunoprecipitates were analyzed by Western blots using mAb anti-Myc. Inputs of Myc-tagged Smads (middle panel) and FLAG-mFoxL2 (bottom panel) were validated by Western blot, as shown. IB, immunoblot. b, HEK293T cells were transfected with FLAG-mFoxL2, myc-Smad3-MH1, or myc-Smad3-MH2. Cell lysates containing mFoxL2 were combined with equivalent amounts of those expressing myc-Smad3-MH1 or myc-Smad3-MH2 and then immunoprecipitated by incubation with agarose-coupled mAb anti-FLAG and subjected to Western blot analysis using mAb anti-Myc. Inputs of myc-Smad3-MH1 or myc-Smad3-MH2 (middle panel) and FLAG-mFoxL2 (bottom panel) were validated by Western blot, as shown.
FIGURE 7.
An intact forkhead binding domain in FoxL2 is necessary for Smad3 binding. a and b, HEK293T cells were transfected with vectors for co-expression of full-length FLAG-mFoxL2, the indicated truncations of N-terminally FLAG-tagged FoxL2, or full-length myc-Smad3. After 2–3 days of protein expression, cell lysates were prepared and used for co-immunoprecipitation studies with agarose-coupled mAb anti-FLAG. Immunoprecipitates (IP) were analyzed by Western blots using mAb anti-Myc. Inputs of FLAG-mFoxL2 or truncated forms (middle panel) and myc-Smad3 (bottom panel) were validated by Western blot, as shown. IB, immunoblot. c, schematic representation of the truncated FoxL2 forms tested in a and b.
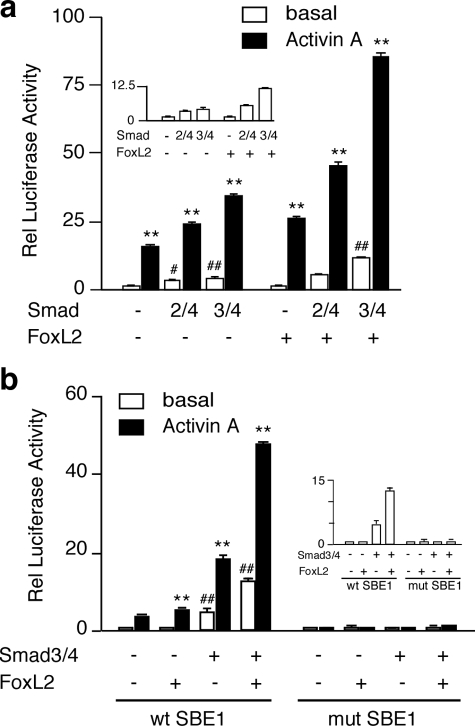
FoxL2 Participates in SBE1-mediated Activation of Follistatin Transcription in αT3-1 Cells—Having demonstrated that FoxL2 and Smad3 form a complex and that the intronic enhancer of the follistatin gene engages both Smad3 and FoxL2, we sought to evaluate FoxL2 effects on Smad3-dependent, SBE1-mediated transcription. We first tested the effects of FoxL2 overexpression in αT3-1 cells even though they express ample levels of endogenous FoxL2. Consistent with our previous findings, the rFS(0.3ex45)-luc reporter was activated by the individual and combined actions of activin A and co-transfected Smad3/4 but not Smad2/4 (Fig. 8a). In cells co-transfected with a FoxL2 expression plasmid, the rFS(0.3ex45)-luc reporter displayed ∼1.5-fold greater basal activity, which was further amplified in cells treated with 1 nm activin A and/or co-transfected with Smad3/4 (Fig. 8a). FoxL2 had a marginal effect on the same reporter when co-transfected with Smad2/4 (Fig. 8a). Smad4 alone or in combination with Smad2, Smad3, FoxL2, or combinations thereof did not have a significant effect (data not shown). Next, we determined if the functional effects of FoxL2/Smad3 on the rFS(0.3ex45)-luc reporter are dependent on an intact SBE1. Consistent with the results shown in Fig. 8a, Smad3/4 and, to a much greater extent Smad3/4+FoxL2, augmented the effects of activin A on the wild type rFS(0.3ex45)-luc reporter (Fig. 8b). The same reporter harboring a mutant SBE1 site, however, was totally unresponsive to activin A in αT3-1 cells transfected with FoxL2, Smad3/4, or both (Fig. 8b). These data suggest that FoxL2-dependent transcriptional activation of follistatin is facilitated by the recruitment of Smad3 to the SBE1 site.
FIGURE 8.
FoxL2 and Smad3 transactivate the rFS(0.3ex45)-luc plasmid that incorporates a wild type SBE1 site. a, to evaluate the effects of FoxL2, αT3-1 cells were co-transfected with the rFS(0.3ex45)-luc plasmid along with the indicated expression plasmids (Smad2/4, Smad3/4, with or without FoxL2). FoxH1 was substituted for FoxL2 to test Smad2/4 effects. b, functional importance of the intronic SBE1 site in transactivation by activin A, FoxL2, and Smad3 was evaluated by comparing the wild type (wt) rFS(0.3ex45)-luc plasmid to the equivalent one in which the SBE1 is mutated. In all experiments, pGL2 luciferase reporter was measured in parallel to assess nonspecific effects. The transfected cells were treated with vehicle or 1 nm activin A and harvested 16–18 h later. Luciferase activity (arbitrary light units) was internally normalized to that of β-galactosidase and then calculated relative (Rel) to the activity of the pGL2 vector under identical conditions. The inset panels show only the basal activity of the reporter. The reported results (mean ± S.E.) are from a representative experiment performed in triplicate (**, p < 0.001 relative to the corresponding untreated basal; #, p < 0.05; ##, p < 0.001 relative to basal of the group).
FoxL2 Effects Are Mediated by a Regulatory Element Located Just Downstream of the SBE1 Site—A putative inverted FoxL2-binding site (lower strand, 5′-ATCAATGT-3′) is located just downstream of SBE1, within the intronic enhancer of the follistatin gene (shown in Fig. 5a). This site is quite similar to the recently characterized FoxL2-binding site (5′-GT(c/g)AAGG(g/t)-3′) (57). We tested the functional importance of this motif by mutating the site within the context of the rFS(0.3ex45)-luc reporter plasmid. All five of the mutants we tested displayed varying degrees of compromised responsiveness to activin A (Fig. 9a). FKHBmut#2 and FKHB#3 did not show a statistically significant response to co-transfected mFoxL2 (Fig. 9a). FKHBmut#4 was fully responsive to mFoxL2, whereas #1 and #5 displayed attenuated responses (Fig. 9a). We further tested the activities of FKHBmut#2 and FKHBmut#5 and determined that although both retained a marginal response to activin A or Smad3 alone, they were essentially unresponsive to mFoxL2 with or without Smad3 (Fig. 9b). These results highlighted the importance of this motif as a mediator of FoxL2 effects.
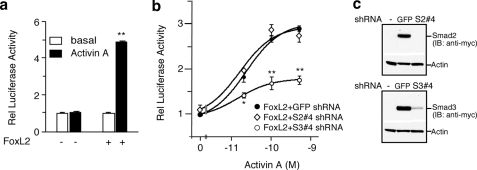
FoxL2 Enables SBE1-mediated Follistatin Transcription in a Heterologous Cell Type—Next we determined if heterologous expression of FoxL2 confers activin responsiveness from the intronic SBE1 in an otherwise FoxL2-deficient cell type. We utilized HEK293T cells because they lack detectable levels of endogenous FoxL2 (as shown in Fig. 4). Consistent with the data shown in Fig. 2, the rFS(0.3ex45)-luc reporter was not responsive to activin A when transiently transfected and evaluated in HEK293T cells (Fig. 10a). However, activin responsiveness of rFS(0.3ex45)-luc was enabled by co-transfecting HEK293T cells with an expression plasmid encoding either FLAG-mFoxL2 (data not shown) or hFoxL2 (Fig. 10a). Given our observation that FoxL2 forms a complex with Smad3 and that SBE1-mediated transcription of follistatin in αT3-1 cells is dependent on the partnership of Smad3/FoxL2, we hypothesized that activin-dependent activation of rFS(0.3ex45)-luc in HEK293T cells expressing FoxL2 is also mediated by Smad3. We tested this possibility by individually knocking down Smad2 or Smad3 with specific lentiviral shRNA vectors (S2#4 shRNA or S3#4 shRNA) under optimized conditions, as shown (Fig. 10b and c) and as reported previously (35). These experiments illustrated that in FoxL2-transfected HEK293T cells, transcriptional activation of the rFS(0.3ex45)-luc reporter is largely dependent on Smad3, although knockdown of Smad2 has no appreciable effect, knockdown of Smad3 blunts the effect at all doses of activin tested (Fig. 10b).
FIGURE 10.
FoxL2 transfected into HEK293T facilitates activin-mediated transactivation of rFS(0.3ex45)-luc through a Smad3-dependent mechanism. a, HEK293T cells were co-transfected with rFS(0.3ex45)-luc along with either FLAG-mFoxL2 or vector and then treated with 1 nm activin A or vehicle for 16–18 h. b, HEK293T cells expressing control GFP-expressing shRNA vector, Smad2 targeted shRNA (S2#4), or Smad3 targeted shRNA (S3#4) were co-transfected with rFS(0.3ex45)-luc and β-galactosidase plasmids and then evaluated for activin-dependent transactivation of this luciferase reporter. c, Western analysis showing the effectiveness of Smad2 (S2#4) or Smad3 (S3#4) shRNA to knock down transfected myc-Smad2 or -Smad3 levels in HEK293T cells expressing the corresponding shRNA or the control GFP-expressing lentiviral vector. The reported luciferase values (mean ± S.E.) are from representative experiments each performed in triplicate (*, p < 0.05; **, p < 0.001 relative to GFP shRNA). IB, immunoblot.
shRNA Knockdown of FoxL2 Attenuates SBE1-mediated Follistatin Transcription in Response to Activin Signaling in αT3-1 Cells—To further confirm the functional importance and participation of endogenous FoxL2 in mediating SBE1-dependent activation of the follistatin gene, we performed knockdown studies using a FoxL2-targeted shRNA selected for its effectiveness. Lentivirus-mediated delivery of this shRNA produced significant knockdown of FLAG-tagged mFoxL2 in transfected HEK293T cells (Fig. 11a). Endogenous mouse FoxL2 levels of FACS-sorted GFP+ αT3-1 cells expressing the FoxL2 shRNA was also significantly lower than FoxL2 levels of cells expressing the GFP or the scrambled control shRNA (Fig. 11b). Importantly, in sorted GFP+ αT3-1 expressing FoxL2 shRNA, the response of rFS(0.3ex45)-luc to all doses of activin A tested was significantly blunted compared with the response in cells expressing either of the control vectors (Fig. 11c). The latter was not because of compromised activin signaling because activin-dependent P-Smad2 accumulation in sorted GFP+ cells expressing FoxL2 shRNA was indistinguishable from those expressing a control vector (Fig. 11d).
FIGURE 11.
shRNA knockdown of endogenous FoxL2 compromises transactivation of the rFS(0.3ex45)-luc plasmid in response to activin A in αT3-1 cells. a, Western analysis of FLAG-mFoxL2 levels in HEK293T cells expressing control GFP or FoxL2 shRNA. Increasing amounts of HEK293T cell lysates were analyzed by SDS-PAGE, and FLAG-mFoxL2 was detected with mAb anti-FLAG. IB, immunoblot. b, Western analysis of endogenous FoxL2 levels in FACS-sorted GFP+ αT3-1 cells infected with lentivirus for delivery of FoxL2 shRNA, control GFP, or a scrambled control shRNA. Varying amounts of αT3-1 lysate were resolved by SDS-PAGE, and endogenous FoxL2 was detected using goat anti-FoxL2 IgG. c, transactivation of rFS(0.3ex45)-luc by activin A in FACS-sorted GFP+ αT3-1, corresponding to those shown in b. The cells were treated with the indicated concentrations of activin A or vehicle for 16–18 h then harvested for evaluation of luciferase activity relative (Rel) to β-galactosidase. Normalized luciferase values (arbitrary light units, mean ± S.E.) from a representative experiment are reported relative to that of pGL2 measured under identical conditions (**, p < 0.001 relative to scramble or GFP shRNA). d, Western analysis of P-Smad2 accumulation in response to the indicated concentrations of activin A in the FACS-sorted GFP+ αT3-1 cells expressing control GFP or FoxL2 shRNA. A rabbit anti-P-Smad2 IgG was used to detect phosphorylated Smad2 (P-Smad2, upper panel), and rabbit anti-Smad2/3 IgG was used to detect total Smad2/3 input.
DISCUSSION
Follistatin plays a critical role in many tissues as a local modulator of activin and several other members of the TGF-β family (6). Using gonadotrope-derived αT3-1 cells, we recently demonstrated that follistatin is a transcriptional target of activin and that this action is primarily mediated by Smad3 via SBE1 within an activin-responsive region of intron 1 (35). Unexpectedly, further evaluation of this conserved SBE1 element in various cell lines suggested that it is not obligatory for activin/TGF-β effects on follistatin expression in all cell types and that it might rather be involved in mediating cell type-specific actions of activin on the follistatin gene. This possibility was revealed by our observations that follistatin-luciferase reporters that incorporate the upstream promoter region of the rat follistatin gene but not SBE1 are responsive to activin A (Fig. 1) or TGF-β (data not shown) in HepG2 and HEK293T cells. Studies of the human FST in HepG2 cells previously led to similar conclusions (42). We sought to understand the mechanism underlying these observations and postulated that they reflect differential utilization of factors and the existence of cell type-specific mechanisms for the control of follistatin expression in response to activin signaling.
Our analysis has identified FoxL2 as an endogenous protein of αT3-1 that participates in Smad3-dependent transcription of follistatin via the intronic SBE1. FoxL2 is a member of the forkhead family of transcription factors that share a conserved DNA-binding domain that, in most cases, recognizes a core consensus sequence (51). Forkhead proteins mostly act as transcriptional activators but are known to also mediate trans-repression (51). Members of this large family have key regulatory roles in developmental processes and are implicated in the control of cellular functions ranging from metabolism, cell proliferation, and survival that often reflect tissue-restricted expression and actions. Indeed, consistent with their restricted expression pattern, mutations of individual forkhead proteins are known to be associated with certain human anomalies and, in some cases, the equivalent mouse phenotype with null mutations of the orthologous genes (51). Loss of function mutations of FoxL2 are associated with a genetic disorder known as BPES characterized by developmental malformation of the eyelids and premature ovarian failure in type I but not type II BPES patients (53, 54). FoxL2 knock-out mice exhibit the equivalent characteristics of craniofacial defects and female infertility because of disruptions of granulosa cell differentiation and failure of oocyte growth (58, 59). These defects are in line with the relatively restricted pattern of FoxL2 expression in the eyelid and ovary.
The pituitary is yet another site of FoxL2 expression (55, 56). The exact role of this forkhead protein in the pituitary, however, has not been extensively explored. FoxL2 (also designated PFrk) was first identified in the embryonic pituitary at e10.5 as a potential player in pituitary organogenesis (55). A more recent report has extended these initial observations by demonstrating that FoxL2 expression from e11.5 becomes restricted largely to a population of quiescent cells that are αGSU-positive, consistent with the possibility that it has a role in regulating the cellular processes or promoting the differentiation of gonadotropes and thyrotropes (56). Indeed, of the very few mammalian FoxL2 targets identified to date, two are expressed in the pituitary (56, 60–62). It has been reported that the glycoprotein α subunit shared by LH and FSH in gonadotropes and TSH in thyrotropes is a transcriptional target of FoxL2 (56). Furthermore, in gonadotrope-derived αT3-1 cells, it has been shown that FoxL2 participates in activin-dependent regulation of the GnRH receptor gene via a composite regulatory element known as GRAS (GnRH Activating Sequence) that also recruits Smad3/4 and AP-1 (60). The identification of follistatin as yet another FoxL2 target of gonadotropes further supports the argument that it has regulatory roles in this cell type.
By employing oligonucleotide pulldown assays coupled with mass spectrometry, our strategy has identified FoxL2 as an endogenous protein of αT3-1 cells and elucidated a novel role for this forkhead protein in mediating Smad3-dependent regulation of follistatin in gonadotropes. We present several lines of evidence to support our conclusion for a key role of FoxL2 in the regulation of the follistatin gene in gonadotropes. Immunohistochemical studies localized FoxL2 to FSHβ-positive gonadotropes of the adult pituitary. A previous study also localized FoxL2 to LHβ-positive cells of the embryonic pituitary (56). These findings highlight the likelihood that FoxL2 and follistatin co-localize, given that gonadotropes also express follistatin (24, 30). The evaluation of several pituitary cell lines further demonstrated the restricted pattern of FoxL2 expression in gonadotrope-derived αT3-1 and LβT2 cells but not in those representative of several other pituitary lineages. A series of functional studies confirmed that endogenous FoxL2 indeed has a role in regulating follistatin expression in αT3-1 gonadotropes. Notably, shRNA-mediated knockdown of FoxL2 expression in αT3-1 cells significantly compromised the transcriptional activation of the rFS(0.3ex45)-luc reporter in response to activin A.
The function of FoxL2 was largely dependent on Smad3 binding to SBE1. As we reported previously (35), compared with the equivalent wild type version, the modified rFS(0.3ex45)-luc luciferase reporter that incorporates a mutant SBE1 site was not responsive to activin and/or Smad3. Similarly, activin/Smad3 failed to activate the mutant rFS(0.3ex45)-luc plasmid even in αT3-1 cells overexpressing FoxL2. Experiments with HEK293T cells devoid of detectable FoxL2 further substantiated the obligatory role of FoxL2 in mediating activin/Smad3 effects via the intronic SBE1 site. FoxL2 overexpression in HEK293T conferred activin/Smad3 sensitivity to the rFS(0.3ex45)-luc plasmid. Similarly, it is likely that ectopic FoxL2 expression in FS/D1h folliculostellate cells, which lack FoxL2 and fail to show activin-dependent regulation of follistatin (26–29), would also enable SBE1-dependent follistatin transcription. These observations suggest that, whereas Smad3 is the essential mediator of activin via SBE1, FoxL2 must somehow be functionally or physically engaged with Smad3 to facilitate the actions of activin/Smad3 within the context of this intronic regulatory region of the follistatin gene. Within this fragment, SBE1 is the primary Smad3-binding site and an FKHB is located just downstream of it. Mutagenesis studies outlined in Fig. 9 demonstrated that a functional FKHB is indeed required for the ability of FoxL2 to partner with Smad3 and activate follistatin transcription in response to activin A. This intronic FKHB site differs from the recently characterized core FoxL2-binding element at only two positions and incorporates the two central amino acids suggested to be critical for FoxL2 binding and function (57).
Through co-immunoprecipitation studies we have clearly established that FoxL2 discriminates between the two downstream mediators of activin, namely Smad2 and Smad3, and specifically associates with Smad3. Consistent with our functional data, these experiments did not provide evidence of a direct association between Smad4 and FoxL2, although the possibility of a trimeric complex involving FoxL2-Smad3-Smad4 cannot be ruled out at this time. Smad4 is a component of the transcriptionally active complex of two analogous partnerships, Smad2-FoxH1 and Smad3-FoxO factors (63–67). In the case of Smad2-FoxH1, receptor-mediated phosphorylation promotes Smad4 binding and facilitates recruitment to promoter elements of targets such as Xenopus Mix2 downstream of activin/nodal signals (64–66). The regulation of TGF-β targets such as p21 Cip1, on the other hand, seems to involve the inducible formation of a transcriptionally active Smad3-Smad4 complex that recruits FoxO proteins for targeting to specific DNA elements (67). Under the conditions we employed involving HEK293T cells, the association of Smad3 with FoxL2 was only marginally induced by activin when evaluated at time points ranging from 5 to 60 min. These experiments were optimized for protein overexpression in HEK293T cells, and it is possible that more robust activin-inducible association indeed does occur under conditions when protein levels are more limiting. Alternatively, activin mainly provides the signal for Smad3 activation/nuclear translocation rather than assembly of the FoxL2-Smad3 complexes.
The N-terminal MH1 and C-terminal MH2 domains of Smad2 and Smad3 have both been shown to mediate protein-protein interactions with their respective partners (2, 9). Our experiments demonstrate that the FoxL2-Smad3 association is mediated by the C-terminal MH2 domain of Smad3. A yeast two-hybrid strategy has previously suggested a similar mode of association (60). The MH2 domain of Smad2 also mediates its interactions with FoxH1, whereas FoxO proteins bind to Smad3 and Smad4 via their MH1 domains (9, 67). Two Smad interaction motifs in FoxH1 mediate binding to Smad2 (68). These studies have suggested that the highly proline-rich Smad interaction motif, similar to that found in SARA, is involved in the recognition of both phosphorylated and unphosphorylated Smad2, whereas the Fast-FoxH1 interaction motif recognizes only active Smad2-Smad 4 complexes (68). To gain further insight into the mode of complex formation between FoxL2 and Smad3, we performed co-immunoprecipitation studies with truncated forms of FoxL2. These experiments indicated that an intact forkhead domain in FoxL2 is necessary for Smad3 binding. Although these studies also suggested that the C- and N-terminal regions flanking the forkhead domain are not required for this interaction, they do not rule out the possibility that residues within these domains contribute to or modulate complex formation between these two partners. For example, it is possible that the proline-rich stretches at the C terminus of both the human and mouse FoxL2 proteins contribute to Smad3 binding (50). Similarly, it would be of interest to determine whether the conserved polyalanine tract found at the C terminus influences Smad3 binding because this region seems to be a “hot spot” of FoxL2 mutations in BPES and is implicated in FoxL2 target selection and sensitivity (57, 69, 70). Within the forkhead domain, a number of point mutations that cause BPES have recently been characterized to also influence target selection and differential transactivating function of hFoxL2 (71). Whether some of the same mutations also disrupt Smad3 binding and thus alter FoxL2 function remains a question for future studies.
Our data on follistatin and those of others on αGSU and GnRH receptor lend support to the possibility that FoxL2 may have a central role in regulating Smad-dependent actions of activin on key targets and for the generation of cell type-specific responses to the activin signal. For example, the differential pattern of FoxL2 expression in αGSU-positive pituitary cells but not in S100-positive folliculostellate cells (26–29), which also express follistatin, may serve to safeguard against excessive follistatin secretion and limit the self-modulating actions of activin. Preliminary data from gonadotrope-derived LβT2 cells reveal that FSHβ is yet another target of FoxL2 in this cell lineage4 and highlight the possibility that FoxL2 has a central role in coordinating the dynamic expression of those targets of gonadotropes with critical roles in the control of the reproductive axis. Whether pituitary level disruptions of FoxL2 contribute to the infertility or the ovarian defects in BPES is not known but is a worthwhile question for future studies. Developmental studies of the pituitary have previously shown that FoxL2 expression precedes that of signals required for the expression of gonadotrope-specific markers and suggested that FoxL2 may be involved in the differentiation of this lineage (55, 56). During this process, FoxL2-dependent activation of the follistatin gene might provide a mechanism for the modulation of Smad signaling and thereby contribute to the developmental program of this lineage. The restricted pattern of FoxL2 expression and the differential partnership it forms with Smad3 collectively point to a role for this forkhead protein in mediating cell-restricted and target-selective actions of activin and possibly other ligands such as TGF-β that also signal via Smad3.
Acknowledgments
We acknowledge the technical expertise of William Low and Jessica Read in the analysis of samples by mass spectrometry. We also thank Camille Huwyler for technical contributions, Dr. Peter Gray for helpful discussions and feedback on the manuscript, and Dave Dalton and Sandra Guerra for assistance with manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R01HD046941-05 (NICHD). This work was also supported in part by the Clayton Medical Research Foundation, Inc., and the Adler Foundation. The mass spectrometry facility at the Salk Institute is supported by the Vincent J. Coates Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TGF-β, transforming growth factor β; SBE1, Smad-binding element; FSH, follicle-stimulating hormone; FKHB, forkhead-binding site; ChIP, chromatin immunoprecipitation; shRNA, short hairpin RNA; hSmad, human Smad; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; BPES, blepharophimosis/ptosis/epicanthus inversus syndrome; mAb, monoclonal antibody; MOPS, 4-morpholinepropanesulfonic acid; FACS, fluorescence-activated cell sorting; GFP, green fluorescent protein; LH, luteinizing hormone; ESI-LC/MS/MS, electrospray ionization-liquid chromatography/tandem mass spectrometry; αGSU, α-glycoprotein subunit; MH2, Mad homology 2; GnRH, gonadotropin-releasing hormone.
A. L. Blount, W. W. Vale, and L. M. Bilezikjian, unpublished observations, and D. J. Bernard, personal communication.
References
- 1.Vale, W., Hsueh, A., Rivier, C., and Yu, J. (1990) Handb. Exp. Pharmacol. 95 211–248 [Google Scholar]
- 2.Shi, Y., and Massague, J. (2003) Cell 113 685–700 [DOI] [PubMed] [Google Scholar]
- 3.Ethier, J. F., and Findlay, J. K. (2001) Reproduction (Camb.) 121 667–675 [DOI] [PubMed] [Google Scholar]
- 4.Welt, C., Sidis, Y., Keutmann, H., and Schneyer, A. (2002) Exp. Biol. Med. (Maywood) 227 724–752 [DOI] [PubMed] [Google Scholar]
- 5.Phillips, D. J., and deKretser, D. M. (1998) Front. Neuroendocrinol. 19 287–322 [DOI] [PubMed] [Google Scholar]
- 6.Harrison, C. A., Gray, P. C., Vale, W. W., and Robertson, D. M. (2005) Trends Endocrinol. Metab. 16 73–78 [DOI] [PubMed] [Google Scholar]
- 7.Wiater, E., and Vale, W. (2008) in The Biology of the TGFβ Family (Derynck, R., and Miyazono, K., eds) pp. 79–120, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 8.Attisano, L., and Wrana, J. L. (2002) Science 296 1646–1647 [DOI] [PubMed] [Google Scholar]
- 9.Feng, X. H., and Derynck, R. (2005) Annu. Rev. Cell Dev. Biol. 21 659–693 [DOI] [PubMed] [Google Scholar]
- 10.Shimonaka, M., Inouye, S., Shimasaki, S., and Ling, N. (1991) Endocrinology 128 3313–3315 [DOI] [PubMed] [Google Scholar]
- 11.Thompson, T. B., Lerch, T. F., Cook, R. W., Woodruff, T. K., and Jardetzky, T. S. (2005) Dev. Cell 9 535–543 [DOI] [PubMed] [Google Scholar]
- 12.Harrington, A. E., Morris-Triggs, S. A., Ruotolo, B. T., Robinson, C. V., Ohnuma, S., and Hyvonen, M. (2006) EMBO J. 25 1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sidis, Y., Mukherjee, A., Keutmann, H., Delbaere, A., Sadatsuki, M., and Schneyer, A. (2006) Endocrinology 147 3586–3597 [DOI] [PubMed] [Google Scholar]
- 14.Esch, F. S., Shimasaki, S., Mercado, M., Cooksey, K., Ling, N., Ying, S., Ueno, N., and Guillemin, R. (1987) Mol. Endocrinol. 1 849–855 [DOI] [PubMed] [Google Scholar]
- 15.Robertson, D. M., Klein, R., deVos, F. L., McLachlan, R. I., Wettenhall, R. E. H., Hearn, M. T. W., Burger, H. G., and de Kretser, D. M. (1987) Biochem. Biophys. Res. Commun. 149 744–749 [DOI] [PubMed] [Google Scholar]
- 16.Shimasaki, S., Koga, M., Buscaglia, M. L., Simons, D. M., Bicsak, T. A., and Ling, N. (1989) Mol. Endocrinol. 3 651–659 [DOI] [PubMed] [Google Scholar]
- 17.Nakamura, T., Takio, K., Eto, Y., Shibai, H., Titani, K., and Sugino, H. (1990) Science 247 836–838 [DOI] [PubMed] [Google Scholar]
- 18.Kogawa, K., Nakamura, T., Sugino, K., Takio, K., Titani, K., and Sugino, H. (1991) Endocrinology 128 1434–1440 [DOI] [PubMed] [Google Scholar]
- 19.Kogawa, K., Ogawa, K., Hayashi, Y., Nakamura, T., Titani, K., and Sugino, H. (1991) Endocrinol. Jpn. 38 383–391 [DOI] [PubMed] [Google Scholar]
- 20.Matzuk, M. M., Lu, N., Vogel, H., Sellheyer, K., Roop, D. R., and Bradley, A. (1995) Nature 374 360–363 [DOI] [PubMed] [Google Scholar]
- 21.Guo, Q., Kumar, T. R., Woodruff, T., Hadsell, L. A., DeMayo, F. J., and Matzuk, M. M. (1998) Mol. Endocrinol. 12 96–106 [DOI] [PubMed] [Google Scholar]
- 22.Shimasaki, S., Koga, M., Esch, F., Cooksey, K., Mercado, M., Koba, A., Ueno, N., Ying, S. Y., Ling, N., and Guillemin, R. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 4218–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugino, K., Kurosawa, N., Nakamura, T., Takio, K., Shimasaki, S., Ling, N., Titani, K., and Sugino, H. (1993) J. Biol. Chem. 268 15579–15587 [PubMed] [Google Scholar]
- 24.Bilezikjian, L. M., Blount, A., Leal, A. M. O., Donaldson, C., Fischer, W., and Vale, W. (2004) Mol. Cell. Endocrinol. 225 29–36 [DOI] [PubMed] [Google Scholar]
- 25.Burger, L. L., Haisenleder, D. J., Dalkin, A. C., and Marshall, J. C. (2004) J. Mol. Endocrinol. 33 559–584 [DOI] [PubMed] [Google Scholar]
- 26.Corrigan, A. Z., Bilezikjian, L. M., Carroll, R. S., Bald, L. N., Schmelzer, C. H., Fendly, B. M., Mason, A. J., Chin, W. W., Schwall, R. H., and Vale, W. W. (1991) Endocrinology 128 1682–1684 [DOI] [PubMed] [Google Scholar]
- 27.DePaolo, L. V., Mercado, M., Guo, Y., and Ling, N. (1993) Endocrinology 132 2221–2228 [DOI] [PubMed] [Google Scholar]
- 28.Besecke, L. M., Guendner, M. J., Schneyer, A. L., Bauer-Dantoin, A. C., Jameson, J. L., and Weiss, J. (1996) Endocrinology 137 3667–3673 [DOI] [PubMed] [Google Scholar]
- 29.Bilezikjian, L. M., Leal, A. M. O., Blount, A., Corrigan, A. Z., Turnbull, A. V., and Vale, W. (2003) Endocrinology 144 732–740 [DOI] [PubMed] [Google Scholar]
- 30.Kaiser, U. B., Lee, B. L., Carroll, R. S., Unabia, G., Chin, W. W., and Childs, G. V. (1992) Endocrinology 130 3048–3056 [DOI] [PubMed] [Google Scholar]
- 31.Michel, U., McMaster, J. W., and Findlay, J. K. (1992) J. Mol. Endocrinol. 9 147–156 [DOI] [PubMed] [Google Scholar]
- 32.Bilezikjian, L. M., Corrigan, A. Z., Vaughan, J. M., and Vale, W. W. (1993) Endocrinology 133 2554–2560 [DOI] [PubMed] [Google Scholar]
- 33.Dalkin, A. C., Haisenleder, D. J., Yasin, M., Gilrain, J. T., and Marshall, J. C. (1996) Endocrinology 137 548–554 [DOI] [PubMed] [Google Scholar]
- 34.Burger, L. L., Dalkin, A. C., Aylor, K. W., Haisenleder, D. J., and Marshall, J. C. (2002) Endocrinology 143 3243–3249 [DOI] [PubMed] [Google Scholar]
- 35.Blount, A., Vaughan, J., Vale, W., and Bilezikjian, L. M. (2008) J. Biol. Chem. 283 7016–7026 [DOI] [PubMed] [Google Scholar]
- 36.Thomas, T. Z., Wang, H., Niclasen, P., O'Bryan, M. K., Evans, L. W., Groome, N. P., Pedersen, J., and Risbridger, G. P. (1997) J. Clin. Endocrinol. Metab. 82 3851–3858 [DOI] [PubMed] [Google Scholar]
- 37.Danila, D. C., Inder, W. J., Zhang, X., Alexander, J. M., Swearingen, B., Hedley-Whyte, E. T., and Klibanski, A. (2000) J. Clin. Endocrinol. Metab. 85 1009–1015 [DOI] [PubMed] [Google Scholar]
- 38.Ogino, H., Yano, S., Kakiuchi, S., Muguruma, H., Ikuta, K., Hanibuchi, M., Uehara, H., Tsuchida, K., Sugino, H., and Sone, S. (2008) Clin. Cancer Res. 14 660–667 [DOI] [PubMed] [Google Scholar]
- 39.Sardana, G., Jung, K., Stephan, C., and Diamandis, E. P. (2008) J. Proteome Res. 7 3329–3338 [DOI] [PubMed] [Google Scholar]
- 40.Lebrun, J.-J., Chen, Y., and Vale, W. W. (1997) in Inhibin, Activin and Follistatin, Recent Advances and Future Views (Aono, T., ed) pp. 1–20, Springer-Verlag, Tokushima, Japan
- 41.Winters, S. J., Dalkin, A. C., and Tsujii, T. (1997) Endocrinology 138 4324–4329 [DOI] [PubMed] [Google Scholar]
- 42.Bartholin, L., Maguer-Satta, V., Hayette, S., Martel, S., Gadoux, M., Corbo, L., Magaud, J. P., and Rimokh, R. (2002) Oncogene 21 2227–2235 [DOI] [PubMed] [Google Scholar]
- 43.Horn, F., Bilezikjian, L. M., Perrin, M. H., Bosma, M. M., Windle, J. J., Hille, B., Vale, W. W., and Mellon, P. L. (1990) Mol. Endocrinol. 5 347–355 [DOI] [PubMed] [Google Scholar]
- 44.Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. (2002) Anal. Chem. 74 5383–5392 [DOI] [PubMed] [Google Scholar]
- 45.Qian, W. J., Liu, T., Monroe, M. E., Strittmatter, E. F., Jacobs, J. M., Kangas, L. J., Petritis, K., Camp, D. G., II, and Smith, R. D. (2005) J. Proteome Res. 4 53–62 [DOI] [PubMed] [Google Scholar]
- 46.Washburn, M. P., Wolters, D., and Yates, J. R., III (2001) Nat. Biotechnol. 19 242–247 [DOI] [PubMed] [Google Scholar]
- 47.Ding, Y., Chan, C. Y., and Lawrence, C. E. (2005) RNA (N. Y.) 11 1157–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding, Y., Chan, C. Y., and Lawrence, C. E. (2004) Nucleic Acids Res. 32 W135–W141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tiscornia, G., Singer, O., and Verma, I. M. (2006) Nat. Protoc. 1 234–240 [DOI] [PubMed] [Google Scholar]
- 50.Cocquet, J., Pailhoux, E., Jaubert, F., Servel, N., Xia, X., Pannetier, M., De Baere, E., Messiaen, L., Cotinot, C., Fellous, M., and Veitia, R. A. (2002) J. Med. Genet. 39 916–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlsson, P., and Mahlapuu, M. (2002) Dev. Biol. 250 1–23 [DOI] [PubMed] [Google Scholar]
- 52.Uhlenhaut, N. H., and Treier, M. (2006) Mol. Genet. Metab. 88 225–234 [DOI] [PubMed] [Google Scholar]
- 53.Crisponi, L., Deiana, M., Loi, A., Chiappe, F., Uda, M., Amati, P., Bisceglia, L., Zelante, L., Nagaraja, R., Porcu, S., Ristaldi, M. S., Marzella, R., Rocchi, M., Nicolino, M., Lienhardt-Roussie, A., Nivelon, A., Verloes, A., Schlessinger, D., Gasparini, P., Bonneau, D., Cao, A., and Pilia, G. (2001) Nat. Genet. 27 159–166 [DOI] [PubMed] [Google Scholar]
- 54.De Baere, E., Dixon, M. J., Small, K. W., Jabs, E. W., Leroy, B. P., Devriendt, K., Gillerot, Y., Mortier, G., Meire, F., Van Maldergem, L., Courtens, W., Hjalgrim, H., Huang, S., Liebaers, I., Van Regemorter, N., Touraine, P., Praphanphoj, V., Verloes, A., Udar, N., Yellore, V., Chalukya, M., Yelchits, S., De Paepe, A., Kuttenn, F., Fellous, M., Veitia, R., and Messiaen, L. (2001) Hum. Mol. Genet. 10 1591–1600 [DOI] [PubMed] [Google Scholar]
- 55.Treier, M., Gleiberman, A. S., O'Connell, S. M., Szeto, D. P., McMahon, J. A., McMahon, A. P., and Rosenfeld, M. G. (1998) Genes Dev. 12 1691–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellsworth, B. S., Egashira, N., Haller, J. L., Butts, D. L., Cocquet, J., Clay, C. M., Osamura, R. Y., and Camper, S. A. (2006) Mol. Endocrinol. 20 2796–2805 [DOI] [PubMed] [Google Scholar]
- 57.Benayoun, B. A., Caburet, S., Dipietromaria, A., Bailly-Bechet, M., Fellous, M., Vaiman, D., and Veitia, R. A. (2009) Hum. Mol. Genet., in press [DOI] [PubMed]
- 58.Schmidt, D., Ovitt, C. E., Anlag, K., Fehsenfeld, S., Gredsted, L., Treier, A. C., and Treier, M. (2004) Development (Camb.) 131 933–942 [DOI] [PubMed] [Google Scholar]
- 59.Uda, M., Ottolenghi, C., Crisponi, L., Garcia, J. E., Deiana, M., Kimber, W., Forabosco, A., Cao, A., Schlessinger, D., and Pilia, G. (2004) Hum. Mol. Genet. 13 1171–1181 [DOI] [PubMed] [Google Scholar]
- 60.Ellsworth, B. S., Burns, A. T., Escudero, K. W., Duval, D. L., Nelson, S. E., and Clay, C. M. (2003) Mol. Cell. Endocrinol. 206 93–111 [DOI] [PubMed] [Google Scholar]
- 61.Pisarska, M. D., Bae, J., Klein, C., and Hsueh, A. J. (2004) Endocrinology 145 3424–3433 [DOI] [PubMed] [Google Scholar]
- 62.Pannetier, M., Fabre, S., Batista, F., Kocer, A., Renault, L., Jolivet, G., Mandon-Pepin, B., Cotinot, C., Veitia, R., and Pailhoux, E. (2006) J. Mol. Endocrinol. 36 399–413 [DOI] [PubMed] [Google Scholar]
- 63.Yeo, C. Y., Chen, X., and Whitman, M. (1999) J. Biol. Chem. 274 26584–26590 [DOI] [PubMed] [Google Scholar]
- 64.Labbe, E., Silvestri, C., Hoodless, P. A., Wrana, J. L., and Attisano, L. (1998) Mol. Cell 2 109–120 [DOI] [PubMed] [Google Scholar]
- 65.Chen, X., Weisberg, E., Fridmacher, V., Watanabe, M., Naco, G., and Whitman, M. (1997) Nature 389 85–89 [DOI] [PubMed] [Google Scholar]
- 66.Liu, F., Pouponnot, C., and Massague, J. (1997) Genes Dev. 11 3157–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seoane, J., Le, H. V., Shen, L., Anderson, S. A., and Massague, J. (2004) Cell 117 211–223 [DOI] [PubMed] [Google Scholar]
- 68.Randall, R. A., Howell, M., Page, C. S., Daly, A., Bates, P. A., and Hill, C. S. (2004) Mol. Cell. Biol. 24 1106–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nallathambi, J., Laissue, P., Batista, F., Benayoun, B. A., Lesaffre, C., Moumne, L., Pandaranayaka, P. E., Usha, K., Krishnaswamy, S., Sundaresan, P., and Veitia, R. A. (2008) Hum. Mutat. 29 E123–E131 [DOI] [PubMed] [Google Scholar]
- 70.Beysen, D., De Jaegere, S., Amor, D., Bouchard, P., Christin-Maitre, S., Fellous, M., Touraine, P., Grix, A. W., Hennekam, R., Meire, F., Oyen, N., Wilson, L. C., Barel, D., Clayton-Smith, J., de Ravel, T., Decock, C., Delbeke, P., Ensenauer, R., Ebinger, F., Gillessen-Kaesbach, G., Hendriks, Y., Kimonis, V., Laframboise, R., Laissue, P., Leppig, K., Leroy, B. P., Miller, D. T., Mowat, D., Neumann, L., Plomp, A., Van Regemorter, N., Wieczorek, D., Veitia, R. A., De Paepe, A., and De Baere, E. (2008) Hum. Mutat. 29 E205–E219 [DOI] [PubMed] [Google Scholar]
- 71.Benayoun, B. A., Batista, F., Auer, J., Dipietromaria, A., L'Hote, D., De Baere, E., and Veitia, R. A. (2009) Hum. Mol. Genet., in press [DOI] [PubMed]