Abstract
Although leukemogenic tyrosine kinases (LTKs) activate a common set of downstream molecules, the phenotypes of leukemia caused by LTKs are rather distinct. Here we report the molecular mechanism underlying the development of hypereosinophilic syndrome/chronic eosinophilic leukemia by FIP1L1-PDGFRα. When introduced into c-KithighSca-1+Lineage- cells, FIP1L1-PDGFRα conferred cytokine-independent growth on these cells and enhanced their self-renewal, whereas it did not immortalize common myeloid progenitors in in vitro replating assays and transplantation assays. Importantly, FIP1L1-PDGFRα but not TEL-PDGFRβ enhanced the development of Gr-1+IL-5Rα+ eosinophil progenitors from c-KithighSca-1+Lineage- cells. FIP1L1-PDGFRα also promoted eosinophil development from common myeloid progenitors. Furthermore, when expressed in megakaryocyte/erythrocyte progenitors and common lymphoid progenitors, FIP1L1-PDGFRα not only inhibited differentiation toward erythroid cells, megakaryocytes, and B-lymphocytes but aberrantly developed eosinophil progenitors from megakaryocyte/erythrocyte progenitors and common lymphoid progenitors. As for the mechanism of FIP1L1-PDGFRα-induced eosinophil development, FIP1L1-PDGFRα was found to more intensely activate MEK1/2 and p38MAPK than TEL-PDGFRβ. In addition, a MEK1/2 inhibitor and a p38MAPK inhibitor suppressed FIP1L1-PDGFRα-promoted eosinophil development. Also, reverse transcription-PCR analysis revealed that FIP1L1-PDGFRα augmented the expression of C/EBPα, GATA-1, and GATA-2, whereas it hardly affected PU.1 expression. In addition, short hairpin RNAs against C/EBPα and GATA-2 and GATA-3KRR, which can act as a dominant-negative form over all GATA members, inhibited FIP1L1-PDGFRα-induced eosinophil development. Furthermore, FIP1L1-PDGFRα and its downstream Ras inhibited PU.1 activity in luciferase assays. Together, these results indicate that FIP1L1-PDGFRα enhances eosinophil development by modifying the expression and activity of lineage-specific transcription factors through Ras/MEK and p38MAPK cascades.
During the last decade, it has become clear that hematopoietic growth factors regulate only growth and survival of hematopoietic cells, whereas lineage-specific transcription factors, such as GATA-1, GATA-3, PU.1, Pax-5, C/EBPα, and C/EBPε, crucially control the lineage commitment and lineage-specific differentiation. For example, granulocyte colony-stimulating factor signaling induced megakaryopoiesis in granulocyte colony-stimulating factor receptor-transgenic mice (1). Also, erythropoietin (EPO)2 was found to promote terminal granulocytic differentiation in EPO receptor-transgenic mice. From these data, we speculated that signal transduction molecules activated by hematopoietic growth factors would not influence the lineage commitment of hematopoietic stem cells/progenitor cells (HSCs/HPCs) or subsequent lineage-specific differentiation (2). However, it has very recently been shown that the MEK/ERK pathway is involved in myeloid lineage commitment (3). Also, PKB (c-Akt) was shown to be involved in lineage decision during myelopoiesis (4). In addition, FLT3-activating mutations were proved to inhibit C/EBPα activity through ERK1/2-mediated phosphorylation (3, 5). These results suggest that signal transduction molecules activated by hematopoietic growth factors or their genetic mutations would not only promote growth and survival but also influence lineage commitment and subsequent differentiation of hematopoietic cells.
Activating mutations of the tyrosine kinases (TKs), such as c-Kit, platelet-derived growth factor receptor (PDGFR), FLT3, and c-ABL, are provoked by several mechanisms, including chromosomal translocations and various mutations involving their self-regulatory regions. These mutations are often involved in the pathogenesis of various types of hematologic malignancies. BCR-ABL is known to cause chronic myelogenous leukemia and acute lymphoblastic leukemia. Most patients with PDGFRβ rearrangement reveal common clinical features resembling chronic myelogenous leukemia or choronic myelomonocytic leukemia. In contrast, FLT3 mutations (ITD and point mutations in the TK domain) are primarily detectable in acute myeloid leukemia (AML) or myelodysplastic syndrome (6–8). Also, c-KIT mutations in the TK domain (Asp816 → Val, Tyr, Phe, or His) are found in patients with aggressive mastocytosis, myelodysplastic syndrome, and AML (9–15). Although these leukemogenic TKs (LTKs) activate a common set of downstream signaling molecules, such as Ras/MAPK, PI3-K/Akt/mTOR, and STATs, the mechanisms by which LTKs cause different disease phenotypes remain to be clarified.
FIP1L1-PDGFRα is a fusion gene, which was originally identified in the patients with hypereosinophilic syndrome (HES)/chronic eosinophilic leukemia (CEL) (16, 17). FIP1L1-PDGFRα fusion protein supports cytokine-independent growth and survival of hematopoietic cells as a constitutively active TK (16, 18–21). As for the downstream signaling molecules, FIP1L1-PDGFRα was shown to activate STAT5, phosphatidylinositol 3-kinase, and Ras/ERK pathways like other LTKs, such as BCR-ABL, TEL-ABL, TEL-JAK2, and TEL-PDGFRβ (18). In addition, Buitenhuis et al. (22) recently reported that activation of phosphatidylinositol 3-kinase, ERK1/2, and STAT5 is pivotal for FIP1L1/PDGFRα-induced myeloproliferation.
The concept of “cancer stem cell” has widely been recognized and validated in various types of cancers, including breast cancer, brain tumors, colon cancer, lung cancer, and malignant melanoma. This concept was originally established in AML as a “leukemic stem cell (LSC)” (23, 24). In this concept, LSCs are defined as specific leukemic cells that can cause leukemia when transplanted into NOD/SCID mice. In AML, although leukemic blasts often display relatively homogenous features, they are organized in a hierarchy. Among them, LSCs reveal the most immature CD34+CD38- phenotype similar to normal HSCs, whereas several antigen expressions are different. LSCs, which account for only 0.2–1.0% of AML cells in the bone marrow (BM), have both abilities to self-renew and to produce restrictedly differentiated leukemia cells, thereby maintaining themselves and yielding leukemia cells composing the majority (23, 25, 26). It is still unclear whether LSCs originate solely from HSCs or are generated from nonstem immature cells that have acquired de novo self-renewal ability. It has been shown that, although common myeloid progenitors (CMPs) and granulocyte/monocyte progenitors (GMPs) have very limited life spans, several leukemogenic oncogenes, such as MLL-ENL, MOZ-TIF2, and MLL-AF9, have an ability to immortalize these cells, thereby enabling them to act as LSCs (27, 28). On the other hand, although LSCs in a chronic phase of chronic myelogenous leukemia are at an HSC level, chronic myelogenous leukemia cells at a CMP/GMP level can act as LSCs in an accelerated phase, suggesting that additional gene mutations can change the main LSC population during disease progression. From these findings, it is now speculated that the leukemia phenotype is determined by the biologic property of the mutated gene and/or the lineage and the differentiation state of LSCs.
In an attempt to analyze the molecular mechanisms by which each LTK causes leukemia with the specific phenotype, we introduced FIP1L1-PDGFRα, which plays a causal role in HES/CEL, into murine HSCs and various types of HPCs. As a result, we found that FIP1L1-PDGFRα specifically enhanced eosinophil development from HSCs/HPCs and imposed the lineage conversion to eosinophil lineage on megakaryocyte/erythroid progenitors (MEPs) and common lymphoid progenitors (CLPs) through Ras/MEK and p38MAPK cascades by modifying the expression and activity of lineage-specific transcription factors.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—Recombinant human TPO was provided by Kirin Brewery (Tokyo, Japan). Recombinant human FLT3L, human IL-6, murine SCF, murine IL-5, murine IL-7, murine granulocyte-macrophage colony-stimulating factor, and human EPO were purchased from Peprotech (Hamburg, Germany).
Antibodies, Cell Staining, and Sorting—To isolate KSLs and CLPs, murine BM cells were stained with phycoerythrin-conjugated anti-IL-7Rα chain (SB/199) (eBioscience, San Diego, CA), fluorescein isothiocyanate-conjugated anti-Sca-1 (E13-161-7), and APC-conjugated anti-c-Kit (2B8) monoclonal antibodies, and biotinylated rat antibodies specific for the lineage markers Ter119, CD3ε (145-2C11), B220 (RA3–6B2), and Gr-1 (RB6-8C5), followed by staining with streptavidin-PerCP/Cy5.5 (BD Biosciences). Then KSLs and CLPs were sorted as IL-7Rα-Lin-Sca-1hic-Kithi and IL-7Rα+Lin-Sca-1loc-Kitlo populations, respectively. For myeloid progenitor sorting, murine BM cells were stained with phycoerythrin-conjugated anti-FcγRII/III (2.4G2), fluorescein isothiocyanate-conjugated anti-CD34 (RAM34) (BD Biosciences), APC-conjugated anti-c-Kit, biotinylated anti-Sca-1, and anti-IL-7Rα (SB/199) (Serotec, Raleigh, NC) monoclonal antibodies, and the above described lineage mixture of monoclonal antibodies (BD Biosciences), followed by staining with avidin-APC/Cy7 (BD Biosciences). After the staining, IL-7Rα-Lin-Sca-1-c-Kit+CD34+FcγRII/IIIlo were sorted as CMPs, IL-7Rα-Lin-Sca-1-c-Kit+CD34+FcγRII/IIIhi as GMPs, and IL-7Rα-Lin-Sca-1-c-Kit+CD34-FcγRII/IIIlo as MEPs, as described previously (29). All of these HSCs and HPCs were isolated using a FACS Aria (BD Bioscience, San Jose, CA). In all analyses and sorting, dead cells were excluded by staining with 7-amino-actinomycin D (Calbiochem). Cells were stained with phycoerythrin-conjugated CD125 (IL-5 receptor α-subunit, T21) and APC-conjugated Gr-1(RB6–8C5) (BD Biosciences) for detection of eosinophil lineage.
Plasmids—Expression vectors for FIP1L1-PDGFRα and TEL-PDGFRβ were kindly provided by Dr. D. Gary Gilliland (Harvard Medical School, Boston, MA). Expression vectors for PDGFRα V561D and D842V were kindly provided by Dr. S. Hirota (Hyogo Medical School, Hyogo, Japan). FIP1L1-PDGFRα, TEL-PDGFRβ, PDGFRαV561D, and PDGFRαD842V were cloned into the murine stem cell virus-internal ribosome entry site-EGFP (pMie) vector. Also, we constructed FIP1L1-PDGFRβ and TEL-PDGFRα by the PCR method and subcloned them into pMie.
Short hairpin RNA (shRNA) interference oligonucleotides against GATA-2 and C/EBPα described previously (30, 31) were cloned into an shRNA expression vector, pCS-RfA-CG, which was kindly provided by Dr. Miyoshi H (RIKEN Bio-Resource center, Tsukuba, Japan). The retrovirus expression vector for dominant negative GATA was constructed by cloning human GATA-3KRR cDNA that can inhibit GATA-1, GATA-2, and GATA-3 (32) into pMie.
Cell Culture and Preparation—Murine BM cells were obtained from 6–8-week-old C57BL/6J mice, which were purchased from CLEA (Tokyo, Japan). After sedimentation of the red blood cells with 6% hydroxyethyl starch, mononuclear cells were separated by density gradient centrifugation, using HIS-TOPAQUE 1083 (Sigma). KSLs, CMPs, GMPs, and MEPs were purified from mononuclear cells and cultured in RPMI1641 medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal calf serum (EQUITECH-BIO, Kerrville, TX) in the presence of murine SCF (50 ng/ml), human FLT3L (10 ng/ml), human IL-6 (50 ng/ml), and human TPO (50 ng/ml) for 48 h, and the cells were subjected to the retrovirus infection.
Retrovirus Transduction—The conditioned media containing high titer retrovirus particles were prepared as described previously (33). Briefly, an ecotropic packaging cell line, 293gp, kindly provided by Dr. H. Miyoshi (RIKEN BioResource Center, Tsukuba, Japan), was transfected with each retrovirus vector by the calcium phosphate coprecipitation method. After 12 h, the cells were washed and cultured for 48 h. To produce lentivirus, 293T cells were transfected with each shRNA expression vector together with a packaging vector (pCAG-HIVgp) and a lentivirus envelope and Rev construct (pCMV-VSV-G-RSV-Rev), both of which were provided by Dr. Miyoshi. Then the supernatant containing virus particles was collected, centrifuged, and concentrated 50-fold in volume. The precultured murine BM cells were infected with each retrovirus in the RPMI1641 medium supplemented with the same medium containing protamine sulfate for 48 h in 6-well dishes coated with RetroNectin (Takara Bio Inc., Shiga, Japan).
Colony Assays—Cells were seeded into methylcellulose medium (MethoCult GF M3434; Stem Cell Technologies, Vancouver, Canada) at a density 2.5 × 102 cells/35-mm dish and were cultured with 5% CO2 at 37 °C. All cultures were performed in triplicate, and the numbers of colonies were counted after 10 days.
In Vitro Immortalization Assays for HPCs—Immortalization assays of HPCs in vitro were performed as previously described (34). In brief, 104 cells were plated in 1.1 ml of methylcellulose medium (Methocult M3434). After the 1 week of culture, colony numbers were counted, and single-cell suspensions of colonies (104 cells) were subsequently replated under identical conditions. Replating was repeated every week in the same way.
Luciferase Assays—Luciferase assays were performed with a dual luciferase reporter system (Promega, Madison, WI), as previously described (35). Briefly, 293T or NIH3T3 cells (2 × 105 cells) were seeded in a 60-mm dish and cultured for 24 h. Using the calcium phosphate coprecipitation method, cells were transfected with 2 μg of reporter gene (pGL3-3×MαP-luciferase, 3×MHC-luciferase, or 1×MPO-luciferase) in combination with 2 μg of pcDNA3-GATA1, pcDNA3-PU.1 (36), or pcDNA3-C/EBPα together with 6 μg of an empty vector or an effector vector for FIP1L1-PDGFRα, H-RasG12V, 1*6-STAT5A (37), or CAAX-p110 (38) and 10 ng of pRL-CMV, a Renilla luciferase expression vector. After 12 h, cells were washed, serum-starved for 24 h, and subjected to luciferase assays. After 36 h, the cells were lysed and subjected to a measurement for luciferase activity. The relative firefly luciferase activity was calculated by normalizing transfection efficiency according to the Renilla luciferase activity.
Semiquantitative RT-PCR Analysis—Total RNA was isolated from 5 × 104 FACS-sorted GFP-positive cells using TRIzol reagent (Invitrogen). RT-PCR was performed using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The cDNA product (1 μl) was resuspended in 20 μl of the PCR buffer containing 0.5 units of TaqGold DNA polymerase (PerkinElmer Life Sciences), 2 mm MgCl2, and 15 pmol of forward and reverse primers. The sequences of forward/reverse primer sets were as follows: C/EBPα, 5′-GCC TGG CCT TGA CCA AGG AG-3′ and 5′-CAC AGG ACT AGA ACA CCT GC-3′; GATA-1, 5′-GGA ATT CGG GCC CCT TGT GAG GCC AGA GAG-3′ and 5′-CGG GGT ACC TCA CGC TCC AGC CAG ATT CGA CCC-3′; GATA-2, 5′-CGG AAT TCG ACA CAC CAC CCG ATA CCC ACC TAT-3′ and 5′-CGG AAT TCG CCT ACG CCA TGG CAG TCA CCA TGC T-3′; IL5-Rα, 5′-GCC CTT TGA TCA GCT GTT CAG TCC AC-3′ and 5′-CGG AAC CGG TGG AAA CAA CCT GGT C-3′; MBP, 5′-ACC TGT CGC TAC CTC CTA-3′ and 5′-GTG GTG GCA GAT GTG TGA-3′; PU.1, 5′-GAT GGA GAA AGC CAT AGC GA-3′ and 5′-TTG TGC TTG GAC GAG AAC TG-3′; HPRT, 5′-CAC AGG ACT AGA ACA CCT GC-3′ and 5′-GCT GGT GAA AAG GAC CTC T-3′.
The PCR products were electrophoresed in agarose gels containing ethidium bromide, and their amounts were analyzed with a Fluor Imager595 and ImageQuant software (Amersham Biosciences).
Transplantation Assays—Transplantation assays were performed according to procedures described previously (39). Briefly, 8–12-week-old CD45.2 mice were lethally irradiated (900 rads) 24 h before the transplantation. BM cells isolated from congenic C57BL/6 (B6-Ly5.1) mice were transduced with FIP1L1-PDGFRα, and 10,000 GFP-positive cells were injected intravenously in combination with 2 × 105 normal BM cells with CD45.2 phenotype. Chimeric analyses were performed at 4 weeks and 8 weeks, and mice were sacrificed 16 weeks after transplantation. Animal care was performed according to institutional guidelines.
Measurement of Phosphorylation of Intracellular Signaling Molecules—Phosphorylation of intracellular molecules was assessed using Phosflow technology (BD Biosciences) according to the manufacturer's recommendation. Briefly, cells were fixed with Phosflow Fix Buffer and incubated at 37 °C for 10–15 min. After permeabilization at room temperature for 10 min, cells were washed twice with Phosflow Perm/Wash Buffer and incubated at room temperature for 10 min. After the binding reaction to each antibody, cells were washed once with Phosflow Perm/Wash buffer, resuspended in 500 μl of BD Pharmingen stain buffer (BD Bioscience), and then subjected to flow cytometric analysis. All experiments were repeated independently at least three times, and reproducibility was confirmed.
Statistical Analyses—Statistical analyses were performed using Student's t test.
RESULTS
Effects of FIP1L1-PDGFRα on the Growth and Survival of Murine KSL Cells—To investigate the effects of LTKs on the growth, differentiation, and survival of HSCs/HPCs, we constructed bicistronic retrovirus vectors for FIP1L1-PDGFRα and TEL-PDGFRβ, which express these cDNAs together with EGFP through the internal ribosome entry site in the infected cells. At first, we introduced these retrovirus vectors into KSL cells. After a 48-h infection, 55–65% of KSLs were found to be GFP-positive in all of transfectants by flow cytometric analysis (data not shown). Next, we isolated retrovirus-infected cells as GFP-positive cells and cultured them in the medium with or without SCF, TPO, FLT3L, and IL-6. As shown in Fig. 1A (left), neither FIP1L1-PDGFRα nor TEL-PDGFRβ further augmented cytokine-dependent growth of KSLs. However, these LTKs enabled KSLs to survive and proliferate under cytokine-deprived conditions at least for 96 h, whereas mock (an empty retrovirus)-infected KSLs rapidly led to apoptosis in this condition (Fig. 1A, right).
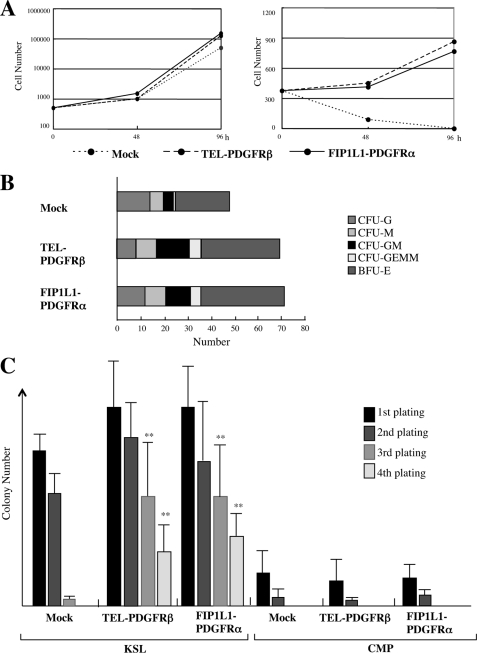
FIGURE 1.
The effects of leukemogenic tyrosine kinases on proliferation and survival of hematopoietic stem/progenitor cells. A, KSLs were isolated from murine bone marrow mononuclear cells. After the retrovirus (mock, FIP1L1-PDGFRα, or TEL-PDGFRβ) infection, retrovirus-infected KSLs were sorted as GFP+ cells and cultured with (left) or without (right) SCF, TPO, FLT3L, and IL-6 for 96 h. During these cultures, total viable cell numbers were counted at the time indicated. B, KSLs infected with each retrovirus were sorted and seeded into the methylcellulose medium containing EPO, TPO, SCF, granulocyte colony-stimulating factor, and IL-3. Colony numbers were counted on day 12. C, immortalization assays for retrovirus-infected KSLs and CMPs. Retrovirus-infected KSLs and CMPs (103 cells) were plated into methylcellulose medium, and colony numbers were counted after 1 week. Then single-cell suspensions of colonies (103 cells) were serially replated every week in the same way. Bars, number of colonies obtained after each round of replating in methylcellulose as means ± S.D. (n = 3). **, p < 0.01 compared with the value of mock-transduced cells.
Next, we performed colony assays using these retrovirus-infected KSLs. After 2-day retrovirus infection, GFP-positive cells were sorted and plated into methylcellulose medium containing the cytokine mixture (EPO, TPO, SCF, granulocyte colony-stimulating factor, and IL-3), and numbers of colonies were counted after 10 days. As shown in Fig. 1B, the total number of colonies that developed from FIP1L1-PDGFRα- or TEL-PDGFRβ-infected KSLs was increased by 40–50% as compared with that from mock-infected KSLs. Also, these colonies were larger than those yielded from mock-infected KSLs (data not shown). However, the proportion of CFU-GEMM, CFU-GM, CFU-G, CFU-M, and BFU-E was roughly the same among three transfectants, indicating that these LTKs scarcely influence the lineage commitment and differentiation of KSLs in colony assays performed in this cytokine combination (Fig. 1B).
We next performed an in vitro immortalization assay using FIP1L1-PDGFRα-, TEL-PDGFRβ-, or mock-transduced KSLs and CMPs. After the first and second plating, both FIP1L1-PDGFRα- and TEL-PDGFRβ-transduced KSLs yielded a slightly increased number of colonies relative to mock-transduced KSLs, whereas these differences were not significant (Fig. 1C, left). Also, in contrast to mock-transduced KSLs, FIP1L1-PDGFRα- or TEL-PDGFRβ-transduced KSLs still kept colony-forming activities even after the third and fourth plating (FIP1L1-PDGFRα versus mock at the third plating, p < 0.01; at the fourth plating, p < 0.01), although these activities were rather reduced (Fig. 1C, left). On the other hand, even if FIP1L1-PDGFRα or TEL-PDGFRβ was introduced, CMPs could not form any colony at the third plating, as was the case with mock-infected CMPs (Fig. 1C, right). To evaluate leukemogenic potential of FIP1L1-PDGFRα-transduced KSLs in vivo, we transplanted these cells into lethally irradiated mice in combination with freshly prepared competitor KSLs. As a result, although none of the mice transplanted with mock-transduced KSLs developed leukemia or MPD, FIP1L1-PDGFRα-transduced KSLs developed MPD in three mice and acute leukemia in one mouse of five recipient mice within 15 weeks after transplantation (Table 1). However, in agreement with the previous report (16), none of the five recipient mice developed eosinophilic disorders. In addition, none of the 10 mice transplanted with FIP1L1-PDGFRα-transduced CMPs developed MPD or leukemia (data not shown). Together, these results indicate that FIP1L1-PDGFRα can confer the ability of cytokine-independent growth/survival on KSLs and enhance their self-renewal, whereas it cannot immortalize CMPs in vitro or in vivo.
TABLE 1.
Peripheral blood examinations 16 weeks after transplantation
| Mouse | White blood cells × 109/liter | Eosinophil |
|---|---|---|
| % | ||
| Mock-1 | 88.3 | 1.2 |
| Mock-2 | 96.2 | 2.4 |
| Mock-3 | 83.5 | 2.6 |
| Mock-4 | 102.2 | 0.8 |
| Mock-5 | 88.2 | 2 |
| FIP1L1-PDGFRα-1 | 563.2 | 3.6 |
| FIP1L1-PDGFRα-2 | 121.1 | 1.2 |
| FIP1L1-PDGFRα-3 | 492.3 | 4.8 |
| FIP1L1-PDGFRα-4 | 140.1 | 3.6 |
| FIP1L1-PDGFRα-5 | 662.3a | 0.2 |
CD3(+)CD8(+) cells: 96%
Effects of FIP1L1-PDGFRα and TEL-PDGFRβ on Differentiation from KSLs—We next investigated whether FIP1L1-PDGFRα or TEL-PDGFRβ influences the lineage commitment and subsequent differentiation of KSLs. For this purpose, we infected retrovirus harboring FIP1L1-PDGFRα or TEL-PDGFRβ into KSLs; cultured them with SCF, TPO, FLT3L, and IL-6; and examined the expression of a granulocyte marker (Gr-1) and an eosinophil marker (IL-5 receptor α, CD125) in GFP-positive cells by flow cytometry. After 4-day cultures, there was not an apparent difference in the expression pattern of these markers among FIP1L1-PDGFRα-, TEL-PDGFRβ-, and mock-transduced KSLs (Fig. 2A, top). However, after 6-day cultures, TEL-PDGFRβ- or FIP1L1-PDGFRα-transduced KSLs yielded significantly increased Gr-1+ fraction (66.8 and 77.5%, respectively) compared with mock-transduced KSLs (49.6%). In addition, it was of particular interest that 51.8% of FIP1L1-PDGFRα-transduced KSLs grew to express CD125 and Gr-1 simultaneously, whereas only 6.0% of mock-transduced and 14.0% of TEL-PDGFRβ-transduced KSLs revealed this phenotype (FIP1L1-PDGFRα versus mock, p < 0.01; Fig. 2A, bottom). These results imply that FIP1L1-PDGFRα but not TEL-PDGFRβ preferentially imposes the commitment and differentiation to the eosinophilic lineage.
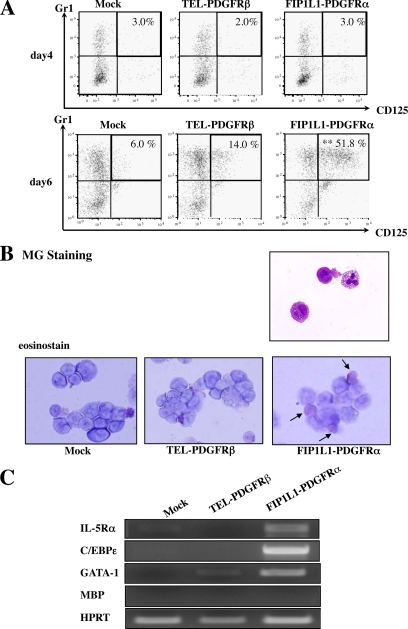
FIGURE 2.
Eosinophil development from KSLs. A, after retrovirus transduction, KSLs were cultured with SCF, TPO, IL-6, and FLT3L, and FACS analysis was performed after 4 days (top) and 6 days (bottom). GFP+ cells were gated, and the expression of Gr-1 and CD125 was analyzed. **, p < 0.01 compared with the value of mock-transduced cells (n = 3). B, after 6-day cultures with SCF, TPO, IL-6, and FLT3L, retrovirus-infected KSLs were further cultured with a cytokine mixture containing IL-5 for 5 days. Transduced cells were subjected to May-Giemsa staining (top) and eosinostain (bottom). C, after 10-day cultures with TPO, IL-6, FLT3L, and SCF, GFP-positive cells were sorted, and the expression of eosinophil-related genes was analyzed by RT-PCR analysis.
To examine whether Gr-1+CD125+ cells that developed from FIP1L1-PDGFRα-transduced KSLs are actually eosinophil precursors, we further cultured these KSLs with a cytokine mixture containing IL-5 for an additional 5 days. As a result, most of FIP1L1-PDGFRα-transduced but not mock- or TEL-PDGFRβ-transduced KSLs came to possess large granule characteristics of mature eosinophil in the MG staining, which were positive for the eosinostain (Fig. 2B). Furthermore, after 10-day cultures, we examined the mRNA expression of eosinophil-related genes, GATA-1, IL-5Rα, and C/EBPε, by RT-PCR analysis using sorted GFP-positive cells. As shown in Fig. 2C, IL-5Rα and C/EBPε mRNAs were detected only in FIP1L1-PDGFRα-transduced KSLs. Also, GATA-1 mRNA was more intensively expressed in FIP1L1-PDGFRα-transduced KSLs than in mock- or TEL-PDGFRβ-transduced KSLs. These data indicate that Gr-1+CD125+ cells that developed from FIP1L1-PDGFRα-transduced KSLs can indeed differentiate into mature eosinophils.
Effects of FIP1L1-PDGFRα on Differentiation of CMPs, MEPs, and CLPs—It was previously shown that eosinophil precursors stochastically develop from HSCs through MMP, CMP, and GMP (40, 41). Therefore, at first, we examined whether FIP1L1-PDGFRα can enhance the development of eosinophils from CMPs. For this purpose, we isolated CMPs from murine BM mononuclear cells by FACS using several markers (Fig. 3A). Then we introduced FIP1L1-PDGFRα into these cells and cultured them with SCF, IL-6, FLT3L, and TPO for 6 days. As was the case with KSLs, FIP1L1-PDGFRα remarkably enhanced the development of Gr-1+CD125+ cells from CMPs compared with mock cultures (57% versus 6%, p < 0.01; Fig. 3B).
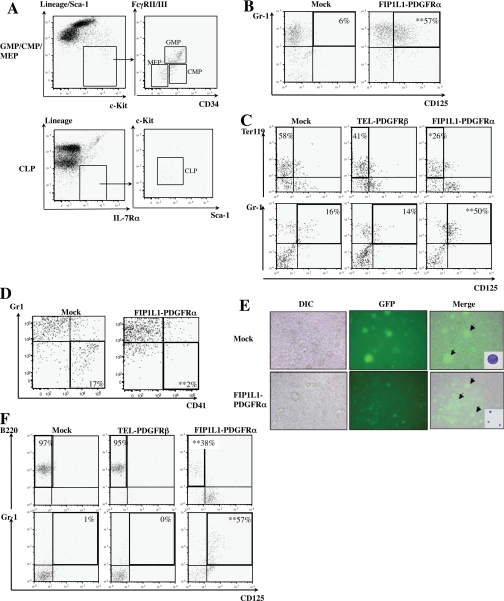
FIGURE 3.
FIP1L1-PDGFRα-induced eosinophil development from CMPs, MEPs, and CLPs. A, isolation of GMPs/CMPs/MEPs and CLPs from murine mononuclear cells by a FACS using several markers. B, mock-, FIP1L1-PDGFRα-, or TEL-PDGFRβ-transduced CMPs were cultured with SCF, IL-6, FLT3L, and TPO for 6 days. Then the expression of CD125 and Gr-1 was analyzed by flow cytometry. C, mock-, FIP1L1-PDGFRα-, or TEL-PDGFRβ-transduced MEPs were cocultured with OP-9 cells in the presence of EPO and SCF for 8 days and then subjected to FACS analysis. D and E, mock- or FIP1L1-PDGFRα-transduced MEPs were cultured in serum-free medium supplemented with TPO and IL-11 for 9 days and subjected to FACS analysis. Transduced cells were observed with differential interference contrast (DIC) and fluorescence microscopy. Mock- and FIP1L1-PDGFRα-transduced GFP-positive cells (arrows) were sorted and subjected to May-Giemsa staining. F, retrovirus-transduced CLPs were cocultured with OP-9 cells in the presence of SCF, IL-7, and FLT3L for 2 days. Then granulocyte-macrophage colony-stimulating factorwas added into the medium, and cells were cultured for an additional 8 days. **, p < 0.01 compared with the value of mock-transduced cells (n = 3).
Our next question was whether FIP1L1-PDGFRα could convert the lineages of MEPs and CLPs, which were already committed to the other lineages, into the eosinophil lineage. To address this issue, we introduced FIP1L1-PDGFRα or TEL-PDGFRβ into MEPs. When cocultured with a stroma cell line OP-9 in the presence of SCF and EPO for 9 days, 58% of mock-infected and 41% of TEL-PDGFRβ-infected MEPs came to reveal the Ter119+CD125- erythroid phenotype. In contrast, only 26% of FIP1L1-PDGFRα-infected MEPs revealed this phenotype (FIP1L1-PDGFRα versus mock, p < 0.05; Fig. 3C, top). Moreover, 50% of FIP1L1-PDGFRα-transduced MEPs differentiated into CD125+Gr-1+ cells, whereas only 16% of mock-infected and 14% of TEL-PDGFRβ-infected MEPs revealed this phenotype (FIP1L1-PDGFRα versus mock, p < 0.01; Fig. 3C, bottom). Similarly, after 9-day cultures in serum-free medium supplemented with TPO and IL-11, although mock-transduced MEPs effectively gave rise to CD41+Gr-1- cells (17%), only 2% of FIP1L1-PDGFRα-infected MEPs revealed this phenotype (FIP1L1-PDGFRα versus mock, p < 0.01; Fig. 3D). Also, mock-transduced MEPs were found to become large polyploid megakaryocytes in morphological analysis, whereas most of the FIP1L1-PDGFRα-transduced MEPs remained small and mononuclear (Fig. 3E). Together, these results indicate that FIPIL1-PDGFRα inhibits erythroid and megakaryocytic differentiation from MEPs and imposes lineage conversion to the eosinophil lineage.
Next, we introduced FIP1L1-PDGFRα into CLPs and cocultured them with OP-9 cells in the presence of SCF, IL-7, and FLT3L. After 10-day cultures, 97% of mock- and 95% of TEL-PDGFRβ-transduced CLPs came to have the B220+CD125- B-lymphoid phenotype, whereas only 38% of FIP1L1-PDGFRα-transduced CLPs had this phenotype (FIP1L1-PDGFRα versus mock, p < 0.01). Furthermore, a considerable proportion of FIP1L1-PDGFRα-transduced CLPs but not mock- or TEL-PDGFRβ-transduced CLPs aberrantly differentiate into Gr-1+CD125+ cells (percentage of Gr-1+CD125+ cells as follows: FIP1L1-PDGFRα, 57% versus mock (1%) (p < 0.01); TEL-PDGFRβ, 0% (Fig. 3F). We further cultured Gr-1+CD125+ cells that developed from FIP1L1-PDGFRα-transduced CLPs with a cytokine mixture containing IL-5 and confirmed that these cells became positive for eosinostain (data not shown). These results indicate that FIP1L1-PDGFRα inhibits B-lymphoid differentiation from CLPs and instructs them to differentiate into the eosinophil lineage.
Function of FIP1L1 and PDGFRα in the Fusion Protein—It was previously shown that the FIP1L1 moiety is dispensable for kinase activation and for transforming properties of FIP1L1-PDGFRα (42). To determine the role of FIP1L1 in FIP1L1-PDGFRα-enhanced eosinophil development, we generated two artificial chimeric constructs, FIP1L1-PDGFRβ and TEL-PDGFRα, in which FIP1L1 in FIP1L1-PDGFRα and TEL in TEL-PDGFRβ were completely replaced (Fig. 4A). In addition, we generated retrovirus vectors for constitutively active PDGFRα (PDGFRαV561D and PDGFRαD842V), which are considered to be causative mutations of gastrointestinal stromal tumors (43) (Fig. 4A). When expressed in a murine IL-3-dependent cell line, Ba/F3, all of the four PDGFR mutants conferred IL-3-independent growth on these cells (data not shown). Also, Western blot analysis demonstrated that these PDGFR mutants phosphorylated various cellular proteins, including themselves (data not shown), indicating that these proteins act as constitutively active tyrosine kinases.
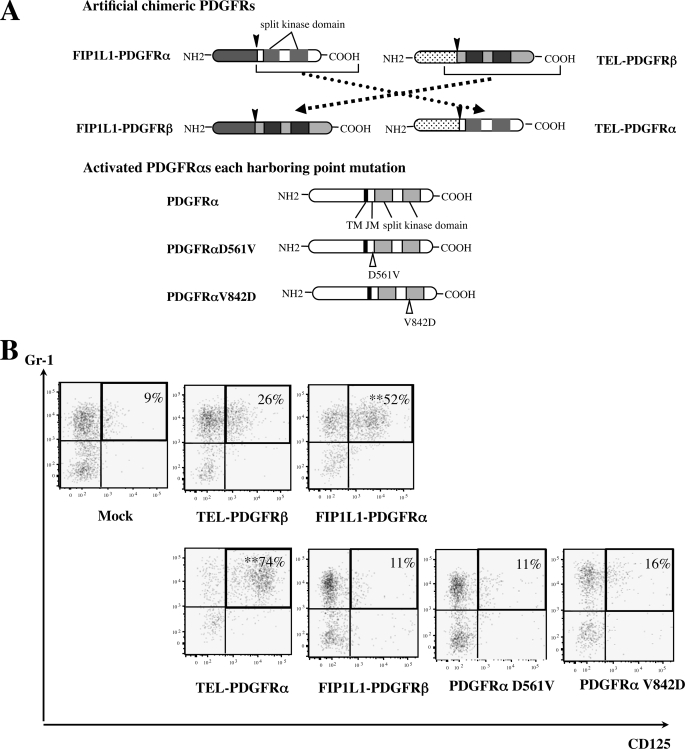
FIGURE 4.
Function of FIP1L1 and PDGFRα in FIP1L1-PDGFRα-induced eosinophil development. A, schematic representation of FIP1L1-PDGFRβ and TEL-PDGFRα, PDGFRαD561V, and PDGFRαD842V. In FIP1L1-PDGFRβ and TEL-PDGFRα, FIP1L1 in FIP1L1-PDGFRα and TEL in TEL-PDGFRβ were completely exchanged one another. Splicing sites are indicated with black arrows, and point mutation sites are indicated with vacant arrows. TM, transmembrane domain; JM, juxtamembrane domain. B, murine KSLs were infected with the retrovirus, as indicated, and cultured with SCF, TPO, IL-6, and FLT3L for 6 days. Then expression of CD125 and Gr-1 was analyzed by flow cytometry. **, p < 0.01 compared with the value of mock-transduced cells (n = 3).
We transduced these retrovirus expression vectors into KSLs and cultured them with SCF, TPO, FLT3L, and IL-6 for 6 days. As shown in Fig. 4B, only TEL-PDGFRα and not FIP1L1-PDGFRβ, PDGFRαV561D, or PDGFRαD842V promoted eosinophil development from KSLs (percentage of Gr-1+CD125+ fraction as follows: TEL-PDGFRα, 74%; FIP1L1-PDGFRβ, 11%; PDGFRαV561D, 11%; PDGFRαD842V, 16%) (TEL-PDGFRα versus mock, p < 0.01) (Fig. 4B), indicating that FIP1L1 is dispensable for FIP1L1-PDGFRα-mediated eosinophil development and that PDGFRα-mediated signaling but not PDGFRβ-mediated signaling is required for inducing eosinophil development. However, because neither PDGFRαV561D nor PDGFRαD842V promoted eosinophil development, specific kinase activity transmitted from chimeric PDGFRα was supposed to be necessary to enhance eosinophil development.
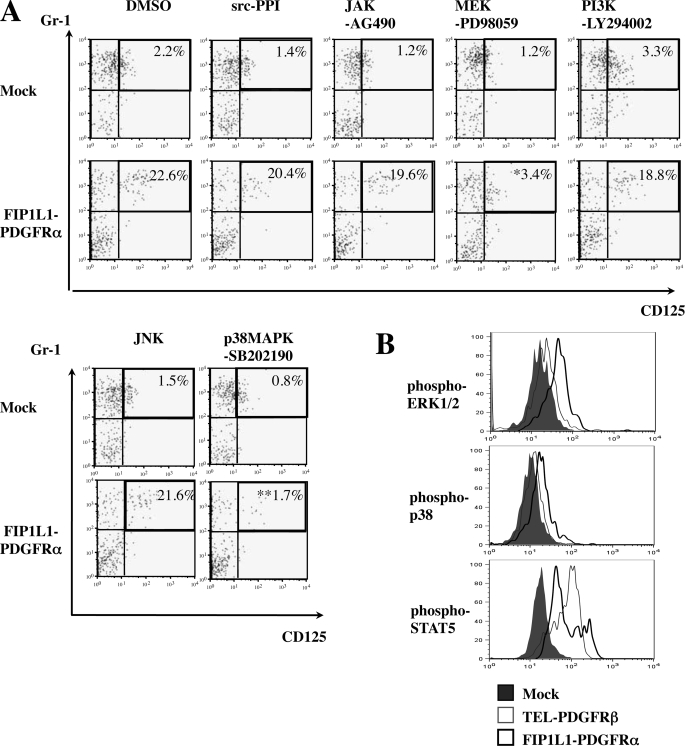
Both a MEK1/2 Inhibitor and a p38MAPK Inhibitor Blocked FIP1L1/PDGFRα-induced Eosinophil Development from KSLs— PDGFRα activates various downstream cascades, thereby exerting its biologic activity (44). To seek out the mechanism underlying instructive eosinophil differentiation induced by FIP1L1-PDGFRα, FIP1L1-PDGFRα- or mock-transduced KSLs were cultured with or without several kinase inhibitors as indicated (Fig. 5A).
FIGURE 5.
Roles of signal cascades in FIP1L1-PDGFRα-induced eosinophil development. A, murine KSLs were infected with the retrovirus indicated and cultured with SCF, TPO, IL-6, and FLT3L with or without kinase inhibitors as indicated and then subjected to FACS analysis. *, p < 0.05; **, p < 0.01 compared with the value of DMSO-treated cells (n = 4). B, FIP1L1-PDGFRα-, TEL-PDGFRβ-, or mock-transduced KSLs were cultured for 2 days, and the phosphorylation status of ERK1/2, p38MAPK, and STAT5 was analyzed using Phosflow technology.
As shown in Fig. 5A (top), neither a c-Jun N-terminal kinase inhibitor, a phosphatidylinositol 3-kinase inhibitor (LY294002), an Src inhibitor (PPI), nor a JAK2/STAT inhibitor (AG490) influenced FIP1L1-PDGFRα-enhanced eosinophil development, since about 20% of cells came to be CD125+Gr1+ after 5-day cultures as was seen after the culture without an inhibitor (Fig. 2A). In contrast, a MEK inhibitor (PD98059) and a p38MAPK inhibitor (SB202190) reduced the CD125+Gr1+ fraction to 3.4% (p < 0.05) and 1.7% (p < 0.01), respectively (Fig. 5A, bottom). We also analyzed the phosphorylation states of ERK, STAT5, and p38MAPK in FIP1L1-PDGFRα- or TEL-PDGFRβ-transduced KSLs by flow cytometry. As shown in Fig. 5B, ERK1/2 and p38MAPK but not STAT5 were phosphorylated more intensely in FIP1L1-PDGFRα-transduced KSLs than in mock- or TEL-PDGFRβ-transduced KSLs. These data suggest that FIP1L1-PDGFRα instructs HSCs/HPCs to differentiate into eosinophil progenitors through the activation of MEK1/2-ERK1/2 and p38 pathways.
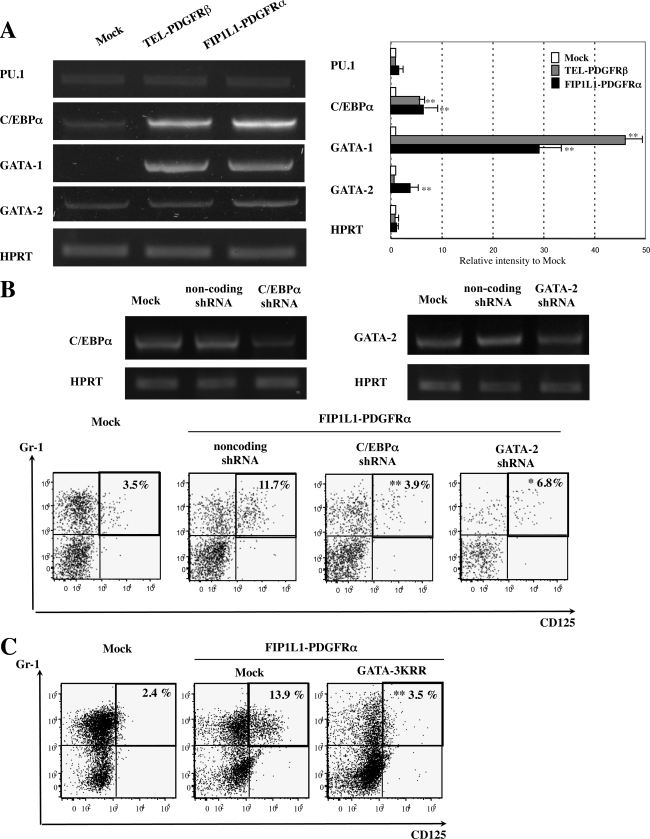
Effects of FIP1L1-PDGFRα on the Expression and Activity of Lineage-specific Transcription Factors in KSLs—To further clarify the mechanism through which FIP1L1-PDGFRα enhanced eosinophil development, we analyzed the effects of FIP1L1-PDGFRα on the expression of GATA-1, GATA-2, C/EBPα, and PU.1, all of which have been reported to be key transcription factors for eosinophil development (45–47). To detect the changes in the expression of these factors that precede the phenotypic change, we isolated mRNA from sorted GFP-positive KSLs after 48-h retrovirus infection and performed semiquantitative RT-PCR analysis, since an apparent phenotypic change was not observed until 4 days (Fig. 2A, top). As shown in Fig. 6A, although the expression of PU.1 was not so different among three transfectants, FIP1L1-PDGFRα augmented the expression of C/EBPα (p < 0.01) and GATA-1 (p < 0.01) compared with mock-transduced KSLs. Furthermore, the expression of GATA-2 was significantly higher in FIP1L1-PDGFRα-transduced KSLs than in mock- or TEL-PDGFRβ-transduced KSLs (FIP1L1-PDGFRα versus mock, p < 0.01).
FIGURE 6.
Effects of FIP1L1-PDGFRα and its downstream molecules on the expressions of eosinophil-related transcription factors and effects of inhibition of these molecules. A, the expressions of eosinophil-related transcription factors in KSLs were analyzed by RT-PCR analysis 48 h after retrovirus transfection. PCR products were electrophoresed and visualized by ethidium bromide staining (left), and their intensities were quantified using a Fluor Imager595 and ImageQuant software. Relative intensities to the products from mock-transduced cells are indicated (right). *, p < 0.05; **, p < 0.01 as compared with the value in mock-transduced cells. Data represent means ± S.D. (n = 3). B, murine KSLs were infected with lentivirus-expressing noncoding or encoding shRNA against C/EBPα or GATA-2 to evaluate the suppression efficacy of each shRNA. After a 48-h culture, cells were subjected to RT-PCR analyses (top). Next, FIP1L1-PDGFRα-transduced murine KSLs were further infected with these shRNAs and cultured with SCF, TPO, IL-6, and FLT3L, which were subjected to FACS analyses upon the expression of CD125 and Gr-1 (bottom). *, p < 0.05; **, p < 0.01 as compared with the value in the cells coexpressing FIP1L1-PDGFRα and noncoding shRNA (n = 3). C, FIP1L1-PDGFRα-transduced murine KSLs were further infected with retrovirus encoding mock or a dominant negative form of GATAs (GATA-3KRR). **, p < 0.01 as compared with the value in FIP1L1-PDGFRα- and mock-cotransduced cells (n = 3).
To evaluate the roles for these transcription factors in FIP1L1-PDGFRα-induced eosinophil development, we inhibited the expression or function of these transcription factors using shRNAs or a dominant negative mutant. At first, we confirmed that these shRNAs suppressed the expression of C/EBPα and GATA-2 considerably (Fig. 6B, top). When coexpressed with FIP1L1-PDGFRα in this condition, shRNA against C/EBPα reduced the FIP1L1-PDGFRα-induced CD125+Gr1+ fraction from 11.7 to 3.9% (p < 0.01) (Fig. 6B, bottom). Similarly, shRNA against GATA-2 suppressed this fraction to 6.8% (p < 0.05). Also, GATA-3KRR, which can inhibit both GATA-1 and GATA-2, reduced FIP1L1-PDGFRα-induced CD125+Gr1+ fraction from 13.9 to 3.5% (p < 0.01) (Fig. 6C). These results indicate that both GATA-2 and C/EBPα are required for FIP1L1-PDGFRα-induced eosinophil development.
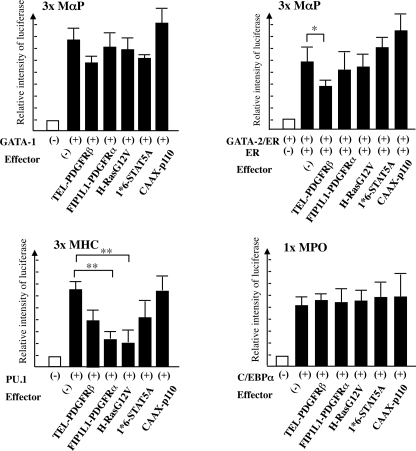
We also examined the effects of FIP1L1-PDGFRα and its downstream signaling molecules (i.e. Ras, STAT5, and PI3-K) on transcription activities of these factors with luciferase assays using reporter genes and effector genes in combinations, as indicated in Fig. 7. In NIH3T3 cells, transiently transduced reporter genes for GATAs (3×MαP-luciferase), PU.1 (3×MHC-luciferase), and C/EBPα (1×MPO-luciferase) were activated by cotransfected GATA-1, PU.1, and C/EBPα by 7-fold, 7-fold, and 5-fold, respectively (Fig. 7). Also, the estradiol treatment activated 3×MαP-luciferase in GATA-2/ER-transfected cells. When FIP1L1-PDGFRα or a constitutively active form of H-Ras (H-RasG12V), STAT5 (1*6-STAT5A), or phosphatidylinositol 3-kinase (CAAX-p110) was further cotransfected, 1*6-STAT5A and CAAX-p110 scarcely affected transcription activities of GATA-1, GATA-2, PU.1, and C/EBPα. In contrast, both FIP1L1-PDGFRα and H-RasG12V reduced PU.1 activities to 30–40% (p < 0.01). Similar results were also obtained from 293T cells (data not shown). These results indicate that FIP1L1-PDGFRα regulates the expression and activities of various transcription factors, thereby promoting eosinophil development, and suggest that Ras may be a pivotal downstream mediator of FIP1L1-PDGFRα in this process.
FIGURE 7.
Effects of FIP1L1-PDGFRα and its downstream molecules on the activities of eosinophil-related transcription factors. The activities of GATA-1, GATA-2, PU.1, and C/EBPα were analyzed by luciferase assays. After transfection of several effector genes and the appropriate reporter genes, as indicated, NIH3T3 cells were cultured for 48 h and subjected to luciferase assays. 3×MαP-luciferase, 3×MHC-luciferase, and 1×MPO-luciferase contain biding sites for GATA, PU.1, and C/EBPα, respectively. *, p < 0.05; **, p < 0.01. Data represent means ± S.D. (n = 3).
DISCUSSION
In this study, we found that TEL-PDGFRα, but not FIP1L1-PDGFRβ, PDGFRαD562V, or PDGFRαD842V, promoted eosinophil development from KSLs as efficiently as FIP1L1-PDGFRα. This result indicates that constitutive TK activity transmitted from chimeric structure of PDGFRα is necessary to augment eosinophil development. In agreement with our finding, novel mutations identified in CEL were restricted to the chimeric form of PDGFRα (i.e. KIF5B-PDGFRα formed by t(4; 10)(q12;p11), STRN-PDGFRα by t(2;4)(p24;q12), and ETV6-PDGFRα by t(4;12)(q2?3;p1?2)). As for the roles for downstream signaling molecules, the current results indicate that Ras/MEK and p38MAPK play essential roles in FIP1L1-PDGFRα-induced eosinophil development. However, this finding seems to be inconsistent with the fact that Ras/MEK is activated by various LTKs and normal hematopoietic growth factors. As for this reason, because FIP1L1-PDGFRα more intensely activated MEK/ERK and p38MAPK than TEL-PDGFRβ, we speculated that leukemogenic signals transmitted from chimeric PDGFRα would be quantitatively and qualitatively different from those from wild type TKs or other LTKs, thereby specifically promoting eosinophil development. In addition to the regulation of neoplastic cell proliferation, ERK has also been implicated in the control of signaling cascades associated with eosinophilia in asthma. Duan et al. (48) reported that an MEK inhibitor dramatically inhibited OVA-induced lung tissue eosinophilia and airway hyperresponsiveness. Also, p38MAPK is important for the induction of eosinophilia and function of terminal differentiated eosinophils in allergic airway inflammation (49, 50). In addition, our data suggest that p38MAPK would regulate eosinophil development at the early stage of hematopoiesis. Further studies to elucidate the crucial signal transduction mechanisms that control eosinophil development will provide a better rationale for the design of drug therapy not only for FIP1L1-PDGFRα-associated HES/CEL but also for allergic inflammation.
Our in vitro studies showed that FIP1L1-PDGFRα confers cytokine independence on KSLs and enhances their self-renewal activity, whereas it did not immortalize CMPs. In addition, although FIP1L1-PDGFRα-transduced KSLs caused MPD in recipient mice, FIP1L1-PDGFRα-transduced CMPs did not. These results indicate that FIP1L1-PDGFRα cannot confer self-renewal activity on CMPs and that the genetic alternation of FIP1L1-PDGFRα that causes CEL/HES occurs at an HSC level but not at a CMP level. In addition, we confirmed that mature eosinophils were generated from FIP1L1-PDGFRα-transduced KSLs in the presence of IL-5, indicating that FIP1L1-PDGFRα does not impair terminal differentiation of eosinophils. Also, when expressed in MEPs or CLPs, FIP1L1-PDGFRα brought about lineage conversion to eosinophil lineage. Together, these results suggest that, although LSCs harboring FIP1L1-PDGFRα derived from HSCs would continuously produce an excess number of mature eosinophils, a part of the eosinophils might be derived from FIP1L1-PDGFRα-harboring MEPs or CLPs.
In a previous report, FIP1L1-PDGFRα-transduced HSCs/HPCs caused myeloproliferative disorder in the recipient mice like BCR-ABL- or TEL-PDGFRβ-transduced KSLs (16, 51, 52), which was rather different from simple eosinophilia observed in human HES/CEL. Also in our transplantation experiment, none of the five mice transplanted with FIP1L1-PDGFRα-expressing KSLs developed eosinophilic disorders. However, we also observed that, whereas FIP1L1-PDGFRα-introduced KSLs differentiated up to IL-5Rα+ eosinophil precursors under the cultures without IL-5, supplement of IL-5 let these IL-5Rα+ cells undergo eosinophilic terminal differentiation. In accord with this hypothesis, Yamada et al. (52) reported that transplantation of FIP1L1-PDGFRα-transduced HSCs/HPCs obtained from IL-5 transgenic mice resulted in marked eosinophilia resembling HES/CEL in the recipient mice. Since p210BCR-ABL-transduced HSCs/HPCs did not cause eosinophilia even in the presence of IL-5 overexpression in the recipient mice, the induction of eosinophilia was attributable to FIP1L1-PDGFRα, Together with our results, these lines of evidence suggest that, although FIP1L1-PDGFRα is a major etiologic factor causing eosinophilia, it is not sufficient to induce HES/CEL but requires additional events, such as IL-5 overexpression. In fact, some patients with FIP1L1-PDGFRα-associated HES were complicated with T-cell lymphoma (53–55). The frequency of FIP1L1-PDGFRα-induced HES/CEL was not as high (about 10%) as initially reported. However, similar LTK is supposed to be involved in the pathogenesis of HES/CEL, because imatinib is effective in some patients who do not have a FIP1L1-PDGFRα mutation (56). Also, a significant proportion of patients with HES/CEL have abnormal T-lymphocyte populations, such as CD3+CD4-CD8- and CD3-CD8+ T cells, which secret high levels of IL-5 (57). Currently, HES is categorized into two groups, “myeloproliferative variant” and “T-cell-mediated HES,” and these groups are thought to be independent of each other (58, 59). However, because T-cell differentiation might be perturbed by FIP1L1-PDGFRα, it may be meaningful for the better understanding of the pathogenesis of HES/CEL to clarify the relationship between these two groups.
Iwasaki et al. (60) isolated eosinophil progenitors from murine BM, and they concluded that eosinophil developmental pathway would diverge from neutrophils and monocytes at the GMP stage. The lineage commitment of HSCs/HPCs and subsequent lineage-specific differentiation are crucially regulated by lineage-specific transcription factors, such as GATA-1, GATA-3, PU.1, C/EBPα, and C/EBPε. Among them, GATA-1 and PU.1 are known to antagonize each other and induce differentiation to erythroid/megakalyocyte or myeloid lineage, respectively (61–63). The CEBP family (CEBPα and CEBPε) is essential for the differentiation to myeloid lineage (64–66). FOG (Friend of GATA) and C/EBPβ regulate the eosinophil lineage induction antagonistically (67). Furthermore, enforced expression of C/EBPα converts MEPs to eosinophils (68), and expression of PU.1 converts them to GMPs (61, 67). Also, forced expression of GATA-1 in myeloid cells induces the formation of either MEPs or eosinophils, depending on the concentration of the factor (69). In addition, it was recently reported that C/EBPα expression followed by GATA-2 expression in GMPs is critical for eosinophil lineage specification (46). However, it is plausible that the mechanism of lineage commitment in leukemic cells is somewhat different from that in normal hematopoietic cells. In this study, we found that FIP1L1-PDGFRα enhanced the expression of GATA-1, GATA-2, and C/EBPα and suppressed PU.1 expression. Also, FIP1L1-PDGFRα suppressed transcription activities of PU.1. These results suggest that LTKs can influence the lineage commitment of HSCs/HPCs and subsequent differentiation by modifying the expression and activity of lineage-specific transcription factors.
In conclusion, we here found that FIP1L1-PDGFRα can enhance eosinophil development from HSCs/HPCs through the MEK/ERK and p38MAPK cascades by controlling the expression and activity of lineage-specific transcription factors. Furthermore, as far as we explored, this is the first report providing evidence that LTK has an ability to convert the lineage of committed progenitor cells. Further studies based on these findings would undoubtedly provide more useful information to understand the pathophysiology of various hematologic malignancies caused by LTKs.
Acknowledgments
We thank Dr. Gary Gilliland for provision of plasmids encoding FIP1L1-PDGFRα and TEL-PDGFRβ, Dr. Seiichi Hirota for provision of the plasmids encoding mutated PDGFRαs, and Dr. Hiroyuki Miyoshi for provision of 293gp cells.
This work was supported by grants from the Ministry of Education, Science, Sports, and Culture and Technology of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: EPO, erythropoietin; LTK, leukemogenic tyrosine kinase; PDGFRα, platelet-derived factor receptor α; PDGFRβ, platelet-derived factor receptor β; KSL, Lin-Sca-1hic-Kithi cell; CMP, common myeloid progenitor; CLP, common lymphoid progenitor; GMP, common granulocyte/monocyte progenitor; HSC, hematopoietic stem cell; HPC, hematopoietic progenitor cell; C/EBP, CCAAT/enhancer-binding protein; MPD, myeloproliferative disorder; TK, tyrosine kinase; AML, acute myeloid leukemia; MAPK, mitogen-activated protein kinase; STAT, signal transducers and activators of transcription; HES, hypereosinophilic syndrome; CEL, chronic eosinophilic leukemia; ERK, extracellular signal-regulated kinase; LSC, leukemic stem cell; MEP, megakaryocyte/erythroid progenitor; FACS, fluorescence-activated cell sorter; shRNA, short hairpin RNA; RT, reverse transcription; GFP, green fluorescent protein; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase.
References
- 1.Semerad, C. L., Poursine-Laurent, J., Liu, F., and Link, D. C. (1999) Immunity 11 153-161 [DOI] [PubMed] [Google Scholar]
- 2.Arcasoy, M. O., Maun, N. A., Perez, L., Forget, B. G., and Berliner, N. (2001) Eur. J. Haematol. 67 77-87 [PubMed] [Google Scholar]
- 3.Hsu, C. L., Kikuchi, K., and Kondo, M. (2007) Blood 110 1420-1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buitenhuis, M., Verhagen, L. P., van Deutekom, H. W., Castor, A., Verploegen, S., Koenderman, L., Jacobsen, S. E., and Coffer, P. J. (2008) Blood 111 112-121 [DOI] [PubMed] [Google Scholar]
- 5.Radomska, H. S., Basseres, D. S., Zheng, R., Zhang, P., Dayaram, T., Yamamoto, Y., Sternberg, D. W., Lokker, N., Giese, N. A., Bohlander, S. K., Schnittger, S., Delmotte, M. H., Davis, R. J., Small, D., Hiddemann, W., Gilliland, D. G., and Tenen, D. G. (2006) J. Exp. Med. 203 371-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto, Y., Kiyoi, H., Nakano, Y., Suzuki, R., Kodera, Y., Miyawaki, S., Asou, N., Kuriyama, K., Yagasaki, F., Shimazaki, C., Akiyama, H., Saito, K., Nishimura, M., Motoji, T., Shinagawa, K., Takeshita, A., Saito, H., Ueda, R., Ohno, R., and Naoe, T. (2001) Blood 97 2434-2439 [DOI] [PubMed] [Google Scholar]
- 7.Levis, M., Tse, K. F., Smith, B. D., Garrett, E., and Small, D. (2001) Blood 98 885-887 [DOI] [PubMed] [Google Scholar]
- 8.Griffith, J., Black, J., Faerman, C., Swenson, L., Wynn, M., Lu, F., Lippke, J., and Saxena, K. (2004) Mol. Cell. 13 169-178 [DOI] [PubMed] [Google Scholar]
- 9.Bene, M. C., Bernier, M., Casasnovas, R. O., Castoldi, G., Knapp, W., Lanza, F., Ludwig, W. D., Matutes, E., Orfao, A., Sperling, C., and van't Veer, M. B. (1998) Blood 92 596-599 [PubMed] [Google Scholar]
- 10.Nagata, H., Worobec, A. S., Oh, C. K., Chowdhury, B. A., Tannenbaum, S., Suzuki, Y., and Metcalfe, D. D. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 10560-10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Longley, B. J., Tyrrell, L., Lu, S. Z., Ma, Y. S., Langley, K., Ding, T. G., Duffy, T., Jacobs, P., Tang, L. H., and Modlin, I. (1996) Nat. Genet. 12 312-314 [DOI] [PubMed] [Google Scholar]
- 12.Ikeda, H., Kanakura, Y., Tamaki, T., Kuriu, A., Kitayama, H., Ishikawa, J., Kanayama, Y., Yonezawa, T., Tarui, S., and Griffin, J. D. (1991) Blood 78 2962-2968 [PubMed] [Google Scholar]
- 13.Longley, B. J., Jr., Metcalfe, D. D., Tharp, M., Wang, X., Tyrrell, L., Lu, S. Z., Heitjan, D., and Ma, Y. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 1609-1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritsche-Polanz, R., Jordan, J. H., Feix, A., Sperr, W. R., Sunder-Plassmann, G., Valent, P., and Fodinger, M. (2001) Br. J. Haematol. 113 357-364 [DOI] [PubMed] [Google Scholar]
- 15.Furitsu, T., Tsujimura, T., Tono, T., Ikeda, H., Kitayama, H., Koshimizu, U., Sugahara, H., Butterfield, J. H., Ashman, L. K., Kanayama, Y., Matsuzawa, Y., Kitamura, Y., and Kanakura, Y. (1993) J. Clin. Invest. 92 1736-1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cools, J., Stover, E. H., Boulton, C. L., Gotlib, J., Legare, R. D., Amaral, S. M., Curley, D. P., Duclos, N., Rowan, R., Kutok, J. L., Lee, B. H., Williams, I. R., Coutre, S. E., Stone, R. M., DeAngelo, D. J., Marynen, P., Manley, P. W., Meyer, T., Fabbro, D., Neuberg, D., Weisberg, E., Griffin, J. D., and Gilliland, D. G. (2003) Cancer Cell 3 459-469 [DOI] [PubMed] [Google Scholar]
- 17.Klion, A. D., Noel, P., Akin, C., Law, M. A., Gilliland, D. G., Cools, J., Metcalfe, D. D., and Nutman, T. B. (2003) Blood 101 4660-4666 [DOI] [PubMed] [Google Scholar]
- 18.Stover, E. H., Chen, J., Lee, B. H., Cools, J., McDowell, E., Adelsperger, J., Cullen, D., Coburn, A., Moore, S. A., Okabe, R., Fabbro, D., Manley, P. W., Griffin, J. D., and Gilliland, D. G. (2005) Blood 106 3206-3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cools, J., DeAngelo, D. J., Gotlib, J., Stover, E. H., Legare, R. D., Cortes, J., Kutok, J., Clark, J., Galinsky, I., Griffin, J. D., Cross, N. C., Tefferi, A., Malone, J., Alam, R., Schrier, S. L., Schmid, J., Rose, M., Vandenberghe, P., Verhoef, G., Boogaerts, M., Wlodarska, I., Kantarjian, H., Marynen, P., Coutre, S. E., Stone, R., and Gilliland, D. G. (2003) N. Engl. J. Med. 348 1201-1214 [DOI] [PubMed] [Google Scholar]
- 20.Griffin, J. H., Leung, J., Bruner, R. J., Caligiuri, M. A., and Briesewitz, R. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7830-7835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Starza, R., Specchia, G., Cuneo, A., Beacci, D., Nozzoli, C., Luciano, L., Aventin, A., Sambani, C., Testoni, N., Foppoli, M., Invernizzi, R., Marynen, P., Martelli, M. F., and Mecucci, C. (2005) Haematologica 90 596-601 [PubMed] [Google Scholar]
- 22.Buitenhuis, M., Verhagen, L. P., Cools, J., and Coffer, P. J. (2007) Cancer Res. 67 3759-3766 [DOI] [PubMed] [Google Scholar]
- 23.Bonnet, D., and Dick, J. E. (1997) Nat. Med. 3 730-737 [DOI] [PubMed] [Google Scholar]
- 24.Hope, K. J., Jin, L., and Dick, J. E. (2004) Nat. Immunol. 5 738-743 [DOI] [PubMed] [Google Scholar]
- 25.Sutherland, H. J., Blair, A., and Zapf, R. W. (1996) Blood 87 4754-4761 [PubMed] [Google Scholar]
- 26.Blair, A., Hogge, D. E., and Sutherland, H. J. (1998) Blood 92 4325-4335 [PubMed] [Google Scholar]
- 27.Cozzio, A., Passegue, E., Ayton, P. M., Karsunky, H., Cleary, M. L., and Weissman, I. L. (2003) Genes Dev. 17 3029-3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huntly, B. J., Shigematsu, H., Deguchi, K., Lee, B. H., Mizuno, S., Duclos, N., Rowan, R., Amaral, S., Curley, D., Williams, I. R., Akashi, K., and Gilliland, D. G. (2004) Cancer Cell 6 587-596 [DOI] [PubMed] [Google Scholar]
- 29.Akashi, K., Traver, D., Miyamoto, T., and Weissman, I. L. (2000) Nature 404 193-197 [DOI] [PubMed] [Google Scholar]
- 30.Segawa, K., Matsuda, M., Fukuhara, A., Morita, K., Okuno, Y., Komuro, R., and Shimomura, I. (2009) J. Endocrinology 200 107-116 [DOI] [PubMed] [Google Scholar]
- 31.Okitsu, Y., Takahashi, S., Minegishi, N., Kameoka, J., Kaku, M., Yamamoto, M., Sasaki, T., and Harigae, H. (2007) Biochem. Biophys. Res. Commun. 364 383-387 [DOI] [PubMed] [Google Scholar]
- 32.Smith, V. M., Lee, P. P., Szychowski, S., and Winoto, A. (1995) The J. Biol. Chem. 270 1515-1520 [DOI] [PubMed] [Google Scholar]
- 33.Ezoe, S., Matsumura, I., Nakata, S., Gale, K., Ishihara, K., Minegishi, N., Machii, T., Kitamura, T., Yamamoto, M., Enver, T., and Kanakura, Y. (2002) Blood 100 3512-3520 [DOI] [PubMed] [Google Scholar]
- 34.Ono, R., Ihara, M., Nakajima, H., Ozaki, K., Kataoka-Fujiwara, Y., Taki, T., Nagata, K., Inagaki, M., Yoshida, N., Kitamura, T., Hayashi, Y., Kinoshita, M., and Nosaka, T. (2005) Mol. Cell. Biol. 25 10965-10978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ezoe, S., Matsumura, I., Gale, K., Satoh, Y., Ishikawa, J., Mizuki, M., Takahashi, S., Minegishi, N., Nakajima, K., Yamamoto, M., Enver, T., and Kanakura, Y. (2005) J. Biol. Chem. 280 13163-13170 [DOI] [PubMed] [Google Scholar]
- 36.Matsumura, I., Kawasaki, A., Tanaka, H., Sonoyama, J., Ezoe, S., Minegishi, N., Nakajima, K., Yamamoto, M., and Kanakura, Y. (2000) Blood 96 2440-2450 [PubMed] [Google Scholar]
- 37.Matsumura, I., Kitamura, T., Wakao, H., Tanaka, H., Hashimoto, K., Albanese, C., Downward, J., Pestell, R. G., and Kanakura, Y. (1999) EMBO J. 18 1367-1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doornbos, R. P., Theelen, M., van der Hoeven, P. C., van Blitterswijk, W. J., Verkleij, A. J., and van Bergen en Henegouwen, P. M. (1999) J. Biol. Chem. 274 8589-8596 [DOI] [PubMed] [Google Scholar]
- 39.Tanaka, H., Matsumura, I., Itoh, K., Hatsuyama, A., Shikamura, M., Satoh, Y., Heike, T., Nakahata, T., and Kanakura, Y. (2006) Stem Cells 24 2592-2602 [DOI] [PubMed] [Google Scholar]
- 40.Abkowitz, J. L., Golinelli, D., Harrison, D. E., and Guttorp, P. (2000) Blood 96 3399-3405 [PubMed] [Google Scholar]
- 41.Huang, S., Law, P., Francis, K., Palsson, B. O., and Ho, A. D. (1999) Blood 94 2595-2604 [PubMed] [Google Scholar]
- 42.Stover, E. H., Chen, J., Folens, C., Lee, B. H., Mentens, N., Marynen, P., Williams, I. R., Gilliland, D. G., and Cools, J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8078-8083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinrich, M. C., Corless, C. L., Duensing, A., McGreevey, L., Chen, C. J., Joseph, N., Singer, S., Griffith, D. J., Haley, A., Town, A., Demetri, G. D., Fletcher, C. D., and Fletcher, J. A. (2003) Science 299 708-710 [DOI] [PubMed] [Google Scholar]
- 44.Heldin, C. H., and Westermark, B. (1999) Physiol. Rev. 79 1283-1316 [DOI] [PubMed] [Google Scholar]
- 45.McNagny, K., and Graf, T. (2002) J. Exp. Med. 195 F43-F47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwasaki, H., Mizuno, S., Arinobu, Y., Ozawa, H., Mori, Y., Shigematsu, H., Takatsu, K., Tenen, D. G., and Akashi, K. (2006) Genes Dev. 20 3010-3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du, J., Stankiewicz, M. J., Liu, Y., Xi, Q., Schmitz, J. E., Lekstrom-Himes, J. A., and Ackerman, S. J. (2002) J. Biol. Chem. 277 43481-43494 [DOI] [PubMed] [Google Scholar]
- 48.Duan, W., Chan, J. H., Wong, C. H., Leung, B. P., and Wong, W. S. (2004) J. Immunol. 172 7053-7059 [DOI] [PubMed] [Google Scholar]
- 49.Kampen, G. T., Stafford, S., Adachi, T., Jinquan, T., Quan, S., Grant, J. A., Skov, P. S., Poulsen, L. K., and Alam, R. (2000) Blood 95 1911-1917 [PubMed] [Google Scholar]
- 50.Wong, C. K., Zhang, J. P., Ip, W. K., and Lam, C. W. (2002) Clin. Exp. Immunol. 128 483-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daley, G. Q., Van Etten, R. A., and Baltimore, D. (1990) Science 247 824-830 [DOI] [PubMed] [Google Scholar]
- 52.Yamada, Y., Rothenberg, M. E., Lee, A. W., Akei, H. S., Brandt, E. B., Williams, D. A., and Cancelas, J. A. (2006) Blood 107 4071-4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McPherson, T., Cowen, E. W., McBurney, E., and Klion, A. D. (2006) Br. J. Dermatol. 155 824-826 [DOI] [PubMed] [Google Scholar]
- 54.Robyn, J., Lemery, S., McCoy, J. P., Kubofcik, J., Kim, Y. J., Pack, S., Nutman, T. B., Dunbar, C., and Klion, A. D. (2006) Br. J. Haematol. 132 286-292 [DOI] [PubMed] [Google Scholar]
- 55.Capovilla, M., Cayuela, J. M., Bilhou-Nabera, C., Gardin, C., Letestu, R., Baran-Marzak, F., Fenaux, P., and Martin, A. (2008) Eur. J. Haematol. 80 81-86 [DOI] [PubMed] [Google Scholar]
- 56.Jovanovic, J. V., Score, J., Waghorn, K., Cilloni, D., Gottardi, E., Metzgeroth, G., Erben, P., Popp, H., Walz, C., Hochhaus, A., Roche-Lestienne, C., Preudhomme, C., Solomon, E., Apperley, J., Rondoni, M., Ottaviani, E., Martinelli, G., Brito-Babapulle, F., Saglio, G., Hehlmann, R., Cross, N. C., Reiter, A., and Grimwade, D. (2007) Blood 109 4635-4640 [DOI] [PubMed] [Google Scholar]
- 57.Simon, H. U., Plotz, S. G., Dummer, R., and Blaser, K. (1999) N. Engl. J. Med. 341 1112-1120 [DOI] [PubMed] [Google Scholar]
- 58.Roufosse, F., Cogan, E., and Goldman, M. (2003) Annu. Rev. Med. 54 169-184 [DOI] [PubMed] [Google Scholar]
- 59.Roufosse, F. E., Goldman, M., and Cogan, E. (2003) N. Engl. J. Med. 348 2687; Author Reply 2687 [DOI] [PubMed] [Google Scholar]
- 60.Iwasaki, H., Mizuno, S., Mayfield, R., Shigematsu, H., Arinobu, Y., Seed, B., Gurish, M. F., Takatsu, K., and Akashi, K. (2005) J. Exp. Med. 201 1891-1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rekhtman, N., Radparvar, F., Evans, T., and Skoultchi, A. I. (1999) Genes Dev. 13 1398-1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, P., Behre, G., Pan, J., Iwama, A., Wara-Aswapati, N., Radomska, H. S., Auron, P. E., Tenen, D. G., and Sun, Z. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 8705-8710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nerlov, C., Querfurth, E., Kulessa, H., and Graf, T. (2000) Blood 95 2543-2551 [PubMed] [Google Scholar]
- 64.Smith, L. T., Hohaus, S., Gonzalez, D. A., Dziennis, S. E., and Tenen, D. G. (1996) Blood 88 1234-1247 [PubMed] [Google Scholar]
- 65.Hohaus, S., Petrovick, M. S., Voso, M. T., Sun, Z., Zhang, D. E., and Tenen, D. G. (1995) Mol. Cell. Biol. 15 5830-5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, N. D., Finegold, M. J., Bradley, A., Ou, C. N., Abdelsayed, S. V., Wilde, M. D., Taylor, L. R., Wilson, D. R., and Darlington, G. J. (1995) Science 269 1108-1112 [DOI] [PubMed] [Google Scholar]
- 67.Querfurth, E., Schuster, M., Kulessa, H., Crispino, J. D., Doderlein, G., Orkin, S. H., Graf, T., and Nerlov, C. (2000) Genes Dev. 14 2515-2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McNagny, K. M., and Graf, T. (2003) Blood 101 1103-1110 [DOI] [PubMed] [Google Scholar]
- 69.Kulessa, H., Frampton, J., and Graf, T. (1995) Genes Dev. 9 1250-1262 [DOI] [PubMed] [Google Scholar]