Abstract
Cisplatin is a widely used chemotherapeutic agent for treatment of ovarian, testicular, lung, and stomach cancers. The initial response to the drug is robust; however, tumor cells commonly develop resistance to cisplatin, which complicates treatment. Recently, overexpression of the Cu-ATPase ATP7B in ovary cells was linked to the increased cellular resistance to cisplatin; and the role for Cu-ATPases in the export of cisplatin from cells was proposed. Our results support functional interactions between cisplatin and ATP7B but argue against the active transport through the copper translocation pathway as a mechanism of drug resistance. In hepatocytes, we observed no correlation between the levels of endogenous ATP7B and the resistance of cells to cisplatin. Unlike copper, cisplatin does not induce trafficking of ATP7B in hepatoma cells, neither does it compete with copper in a transport assay. However, cisplatin binds to ATP7B and stimulates catalytic phosphorylation with EC50 similar to that of copper. Mutations of the first five N-terminal copper-binding sites of ATP7B do not inhibit the cisplatin-induced phosphorylation of ATP7B. In contrast, the deletion of the first four copper-binding sites abolishes the effect of cisplatin on the ATP7B activity. Thus, cisplatin binding to ATP7B and/or general changes in cellular copper homeostasis are likely contributors to the increased resistance to the drug. The link between changes in copper homeostasis and cisplatin resistance was confirmed by treating the Huh7 cells with copper chelator and increasing their resistance to cisplatin.cisplatin
Cisplatin, cis-diamminedichloroplatinum (DDP),3 is a common anti-tumor agent that is used to treat many types of cancer. It is especially prescribed for testicular, ovarian, bladder, liver, lung, and stomach cancers (1–5). DDP mediates its cytotoxic effects by binding to DNA, forming the intrastrand cross-links and thus causing an inhibition of DNA synthesis and repair with eventual cell death (6, 7). The initial tumor response to the treatment with DDP is robust; however, the efficacy of treatment decreases with longer and repetitive therapy cycles. The resistance arises rapidly and is sufficient to cause a failure of DDP therapy (8). The mechanisms by which cells develop resistance to DDP are not fully understood. Detoxification of DDP, enhanced repair and tolerance of DNA adducts, inhibition of apoptosis, impaired uptake and increased efflux of the drug may contribute to the acquired resistance of cells (9–13).
Recently, an unexpected connection was discovered between the resistance of cells to DDP and cellular copper metabolism (14–16). Either down-regulation of CTR1 (49), a transporter responsible for the uptake of copper, and/or up-regulation of the copper-transporting ATPases (Cu-ATPases) responsible for copper efflux were found to increase cells resistance to DDP, although correlation between the levels of CTR1 and resistance were not always observed (17). The DDP-resistant cells were shown to have a lower copper content (18), and a cross-resistance was detected between cells selected for resistance to either copper or DDP (19, 20). The link between the Cu-ATPase ATP7B and cell response to DDP has been particularly well documented. ATP7B was found to be overexpressed in several solid tumors and in cultured cells; the increase in the ATP7B levels correlated with an increased resistance to DDP (18, 21–24). In ovarian carcinoma cells, ATP7B was detected in vesicles resembling exosomes, prompting the suggestion that ATP7B facilitates DDP efflux (25). The evidence was also provided for colocalization of the enhanced cyan fluorescent protein-tagged ATP7B and fluorescent DDP analogue in vesicles (26).
These observations have raised significant interest and many questions. ATP7B is a P1-type-ATPase with high selectivity to Cu(I) (27). Cu(I) stimulates catalytic activity of ATP7B, inducing the hydrolysis of ATP via formation of an acyl-phosphate intermediate, a step necessary for subsequent transport of copper across membranes (28–30). Neither Cu(II) nor other divalent metals such as Zn2+ or Cd2+ stimulate the formation of phospho-intermediate (29). Therefore, it is not apparent how ATP7B may coordinate and transport DDP or even platinum, a metal with electronic structure different from Cu(I). Recent studies provided the in vitro evidence for the DDP-dependent ATP-hydrolysis by ATP7B and the ATP7B-dependent transport of DDP at acidic pH (pH 4.6) (31). Because these conditions do not match the physiological conditions at which ATP7B operates, it remains uncertain whether the transport of DDP to vesicles represents the primary mechanism through which ATP7B mediates resistance of cells to DDP.
ATP7B is highly expressed in hepatocytes. Given the poor response of hepatic tumors to chemotherapy and the poor prognosis of advanced hepatomas (32, 33), it was interesting and important to investigate the role of ATP7B in the sensitivity of hepatocytes and hepatoma cells to DDP. Hepatocytes, unlike ovary cells, express only one Cu-ATPase, ATP7B; this simplifies evaluation of the ATP7B contribution to DDP resistance. Contrary to expectation, in hepatoma cells and in primary hepatocytes, we did not observe a correlation between the levels of ATP7B and cell sensitivity to DDP, nor did we see the effect of DDP on ATP7B transport activity or trafficking. Instead, we demonstrate that DDP binds to ATP7B and induces formation of a phosphorylated catalytic intermediate. We found that the mode of DDP binding to ATP7B differs from that of copper and requires the presence of the N-terminal metal-binding domain. We propose a model of how the overexpression of ATP7B increases cell resistance to the drug.
EXPERIMENTAL PROCEDURES
Reagents—The DDP solution (GRY-Pharma, Kirchzarten, Germany or Novaplus purchased through Ben Venue Laboratories, Bedford, OH) was obtained via the pharmacies of the University of Leipzig or the Oregon Health & Science University, respectively. It was kept as a 3.33 mm solution in 0.9% NaCl in the dark at room temperature. Other chemicals, unless otherwise specified, were provided by Sigma.
Cell Lines—Sf9 cells (Invitrogen) were maintained at 27 °C in suspension cultures in SF900 II (Invitrogen). The human hepatoma cell lines HepG2 and Huh7 were cultured at 37 °C in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 2 mm l-glutamine, and 100 units/ml penicillin and streptomycin (Invitrogen).
Mouse Strains—The generation of Atp7b-/- mice has been described previously (34). The Atp7b-/- mice were housed at the Medizinisch Experimentelles Zentrum (MEZ) of the University of Leipzig according to the university guidelines on the use of laboratory and experimental animals. The animals were euthanized by phenobarbital injection (intraperitoneal). Eight-week-old mice of either sex were used for the experiments.
Primary Liver Cells—Hepatocytes were isolated according to Seglen et al. (35) with modifications by Gebhardt et al. (36). The isolated cells were centrifuged at room temperature at 37.5 × g for 2 min. The pellets were resuspended in culture medium Williams E (Biochrom AG, Berlin, Germany). The viability was determined by staining with trypan blue. The hepatocytes were plated on collagen-coated tissue culture dishes, prepared 1 day before hepatocyte isolation, and maintained in Williams E. For 24-well plates, hepatocytes were plated at a density 2.5 × 105 cells/dish. All of the cells were then grown in 5% CO2 at 37 °C for 2 h prior to treatment with DDP.
Cytotoxicity Experiments—HepG2 and Huh7 cells were plated in 25-cm2 tissue culture flasks at a density of 7 × 105 cells (HepG2) or 3 × 105 cells (Huh7) in 5 ml of standard growth medium and left for 72 h in a CO2 incubator at 37 °C to form a monolayer. The cells were then treated with 0.1–100 μm DDP for 24 h. Copper deficiency was produced by incubating cells with 200 μm bathocuproine disulfonate (BCS) 48 h prior to and during treatment with DDP. The number of viable cells was determined by Casy TT® (Schärfe System, Reutlingen, Germany).
The primary hepatocytes were cultured in a 24-well plate and then incubated with different concentration of DDP for 24 h. Drug-containing medium was then aspirated, and the cells were washed with PBS twice before the addition of MTT and then incubated with 200 μl of MTT solution (2 mg/ml PBS) for 2 h. MTT solution was removed, and 200 μl of Me2SO was added to each well to lyse cells. The percentage of viable cells after DDP treatment was determined photometrically by measuring the absorbance at 490 nm compared with untreated controls.
ATP7B Expression in Hepatoma Cells—Huh7 and HepG2 cells were grown to >90% confluence and trypsinized in 0.05% trypsin/PBS. Then cells were centrifuged at 500 × g for 10 min at 4 °C. The cell pellets were resuspended in 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 0.1% β-mercaptoethanol/PBS. The cells were homogenized and centrifuged for 10 min at 500 × g. The supernatant was subjected centrifugation for 30 min at 20,000 × g to sediment cell membranes. The membranes were resuspended in buffer containing 333 mm Tris, pH 6.8, 2.6 m urea, 3.3% SDS, and 0.1% β-mercaptoethanol. Protein concentration was determined (37), and Western blot analysis was carried out using antibody against the nucleotide-binding domain; staining with the anti-β-actin antibody was used as a protein loading control.
Immunofluorescence Microscopy—HepG2 cells were grown in 10-cm2 cell culture dishes to >90% confluence before being trypsinized in 1 ml of trypsin solution (0.05% trypsin) and adjusted to 10 ml with the medium. In a 12-well tray, 16 μl of the 10 ml of cell suspension was seeded onto flamed glass coverslips and cultured in the appropriate growth media until the cells reached 80% confluence. The cells were then treated with either 200 μm BCS to decrease free copper levels in the medium, with 10–200 μm CuCl2 or with 10–200 μm DDP for 3 h. The cells were then fixed by immersion in acetone for 30 s at -20 °C. The cells were blocked overnight in 1% gelatin, 1% bovine serum albumin, 0.01% sodium azide in PBS at 4 °C and then incubated with the anti-ATP7B antibodies as in Ref. 38 and trans-Golgi marker Syntaxin 6 (BD Sciences, San Jose, CA) in blocking solution for 1 h at room temperature (each antibody at 1:500 dilution). After washing four times with PBS for 1 h, the cells were treated with fluorescently labeled secondary antibodies (1:2000; Alexa Fluor 488 donkey anti-rat for ATP7B, Alexa Fluor 555 goat anti-mouse (Molecular Probes, Eugene, OR) for Syntaxin 6. The cells were then washed four times in PBS for 1 h, and then coverslips were mounted onto glass slides using mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). The images were analyzed using a Zeiss Confocal Scanning microscope (Carl Zeiss, Göttingen, Germany).
ATP7B Expression in Sf9 Insect Cells and Preparation of Membrane Fractions—The cells were infected with the recombinant virus encoding the wild-type ATP7B, the D1027A mutant (this mutation inactivates phosphorylation of invariant residue D1027), the ATP7B variant with the deletion of metal-binding sites 1–4 (ΔMBS1–4), and the ATP7B with CXXCto AXXA mutation of metal-binding sites 1–5, (mMBS1–5)) as previously described (29, 39). To generate the CPC>SPS mutant, site-directed mutagenesis was performed using primers (forward) 5′-TGT GCA TTG CCA GCC CCA GCT CCC TGG GG-3′ and (reverse) 5′-CCC CAG GGA GCT GGG GCT GGC AAT GCA CA-3′ and using the wild-type ATP7B cDNA as a template; the baculovirus virus was generated as previously described (29). To obtain membrane preparations, the cells from 50 ml of culture were centrifuged at 500 × g for 10 min at 4 °C and resuspended in 5 ml of homogenizing buffer (25 mm imidazole, pH 7.4, 0.25 m sucrose, 1 mm dithiothreitol (DTT), and 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride). One tablet of complete protease inhibitor mixture without EDTA (Roche) was added per 50 ml of buffer solution. The cells were homogenized 20 times in a manual glass homogenizer and then centrifuged for 10 min at 500 × g. The supernatant was subjected to an additional centrifugation for 30 min at 20,000 × g to sediment cell membranes. The pelleted cell membranes were resuspended in the homogenizing buffer. The membrane protein solution was stored frozen at -80 °C. Protein concentration was determined by the method of Lowry et al. (40).
Preparation of Vesicles for Copper Transport—The Sf9 insect cells were infected with recombinant virus or empty vector (used as a control) were harvested after 3 days. Cells from a 50-ml culture were pelleted, resuspended in 6 ml of ice-cold homogenizing buffer for vesicles: 50 mm Tris, pH 7.0, 50 mm mannitol, 2 mm EGTA, antipain, and leupeptin (each diluted 1:1000), phenylmethylsulfonyl fluoride (diluted 1:200). The cells were homogenized 20 times in a semi-automatic homogenizer (Schütt homgen plus, Göttingen, Germany) at 3000 rpm and then centrifuged for 10 min at 500 g at 4 °C. Subsequently, the supernatant was centrifuged for 1 h at 100,000 × g. The pelleted membranes were then resuspended in sterile filtered buffer (SMS; 50 mm sucrose, 100 mm potassium nitrate, 10 mm Hepes/Tris, pH 7.4). Vesicle formation was facilitated by vortexing and passing several times through a 25-gauge ⅝-inch needle. The vesicle solution was stored in liquid nitrogen. Protein concentration was determined by the method of Bradford (37). To confirm the ATP7B expression, the samples were analyzed by Western analysis using antibody against the nucleotide-binding domain of ATP7B (the region that is unaltered in all mutants).
Cu64 Uptake in Vesicles—The transport assay has been carried as described by Gmaj et al. (41). The vesicle solution was thawed, diluted to 5 μg of protein/μl with sterile filtered SMS buffer and kept on ice. For the transport assay 20 μl of vesicle solution was preincubated at 37 °C for 1 min. Subsequently, the reaction was started at 37 °C by the addition of 80 μl of sterile filtered incubation solution (50 mm sucrose, 100 mm potassium nitrate, 10 mm Hepes/Tris pH 7.4, 12.5 mm magnesium nitrate, 12.5 mm DTT, 6.25 mm ATP, 2.5 μm copper chloride (final copper concentration, 2 μm), 6 μCi/ml Cu64 (Rotop Pharmaka, Radeberg, Germany), and 0–10 μm DDP). The reaction was stopped at different time intervals by the addition of 3 ml of ice-cold sterile filtered stopping solution (50 mm sucrose, 100 mm potassium chloride, 10 mm Hepes/Tris, pH 7.4, 0.5 mm EDTA) and filtered immediately through a 0.45-μm nitrocellulose vacuum filter system. The filter was washed with 3 ml of stopping solution before and after filtration, and its radioactivity was measured by the γ-Counter Cobra Quantum (PerkinElmer Life Sciences).
The Effect of Copper or DDP on Phosphorylation of ATP7B with [γ-32P]ATP—75 μg of membrane protein was resuspended in 200 μl of phosphorylation buffer (20 mm bis-Tris propane, pH 6, 200 mm KCl, 5 mm MgCl2). 250 μm BCS was added and incubated on ice for 30 min; BCS was removed by centrifugation (5 min at 20,000 × g at 4 °C). The pellets were washed with phosphorylation buffer, resuspended in the same buffer containing 100 μm ascorbate and 100 μm tris(2-carboxyethyl)phosphine hydrochloride, and then incubated with of copper or DDP (0.5–10 μm) on ice for 10 min. In some cases, incubation with 2 μm DDP for 15 min was done prior to incubation with copper. To test the effect of BCS on phosphorylation, the metals were first removed by incubation with BCS as described above, then ATP7B was incubated for 10 min with either 10 μm copper (control) or DDP, then increasing concentrations of BCS (10–100 μm) were added for 30 min, and phosphorylation was measured.
Radioactive [γ-32P]ATP (5 μCi; specific activity, 20 mCi/μmol; PerkinElmer Life Sciences) was added to a final concentration of 1 μm, and the reaction was incubated on ice for 4 min. In several experiments, 0–500 μm ADP was added on ice for 4 min prior to termination of the reaction. All of the additional treatments were done as described in the figure legends. The reaction was stopped by the addition of 50 μl of ice-cold stop solution (1 mm NaH2PO4 in 50% trichloroacetic acid) and then centrifuged for 10 min at 20,000 × g. The pellet was washed once with 1 ml of ice-cold water and centrifuged again for 5 min. The pellet was dissolved in 40 μl of sample buffer (5 mm Tris-PO4, pH 5.8, 6.7 m urea, 0.4 m DTT, 5% SDS, and bromphenol blue), and 30 μl were loaded on a 7% acidic gel (stacking gel: 5.5% acrylamide, 41.3 mm Tris, pH 5.8, 1% SDS, 5% ammonium persulfate, 5% N,N,N′,N′-tetramethylethylenediamine; separating gel: 7% acrylamide, 64.5 mm Tris-HCl, pH 6.8, 1% SDS, 6.25% ammonium persulfate, 6.25% N,N,N′,N′-tetramethylethylenediamine). After electrophoresis the gels were fixed in 10% acetic acid for 10 min and dried on blotting paper. The dried gels were exposed overnight either to screen for the Fuji BAS reader (Fuji, Düsseldorf, Germany) or Kodak BioMax MR film. The photon-stimulated-luminescence intensity of the bands was quantified using Aida Image Analyzer (Raytest, Straubenhardt, Germany).
Papain Digestion and Western Blot Analysis of ATP7B—The membrane protein was solubilized in 1 mm MgCl2, 0.1% n-decyl-β-d-maltoside, 0.5 mm DTT, 10 mm cysteine, 20% glycerol, 50 mm MOPS·NaOH, pH 7 and centrifuged 5 min at 20,000 × g at 4 °C. The pellet was then resuspended and incubated in the buffer containing 500 μm BCS, 10 μm CuCl2, or 10 μm DDP at 30 °C for 1 h. Papain was activated in papain buffer (5 mm cysteine, 1 mm DTT, 50 mm MOPS·NaOH, pH 7) at room temperature for 30 min. ATP7B and papain were mixed 1:100–1:1000 at 30 °C for 2–200 min. The reaction was stopped by adding 1-trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane (E-64) up to a concentration of 2 mm. trichloroacetic acid was added to 2.5% immediately, and samples were centrifuged 5 min at 20,000 × g at 4 °C. Then the pellet was resuspended in 34 μl of loading buffer, and 30 μl was loaded onto 8–16% gradient gel (Pierce). The samples were transferred to polyvinylidene difluoride or nitrocellulose membrane (Millipore, Bedford, MA). The membranes were then incubated in 3% bovine serum albumin in PBS overnight at 4 °C. For the detection of ATP7B, anti-C-terminal domain antibody (generated against the recombinant protein corresponding to the last 92 residues of human ATP7B) was diluted in 2% bovine serum albumin in 0.1% PBS-T (1:3000) and incubated for 1.5 h at room temperature. Following extensive washing, the membranes were incubated with secondary antibody for 1 h at room temperature. The bands were visualized using Super Signal Chemiluminescent reagent (Pierce) and detected using x-ray films (Biomax O-Mat; Kodak).
Statistical Analysis—The results are presented as the means ± S.D. Statistical differences were analyzed by Mann-Whitney-U test using SPSS software (SPSS Inc., Chicago, IL). The test of significance was two-tailed, and a p value of less than 0.05 was considered statistically significant.
RESULTS
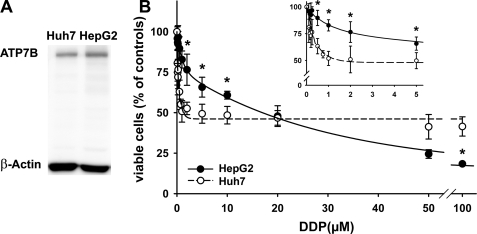
Hepatoma Cells HepG2 and Huh7 Show Different Sensitivity to DDP—Increased cellular ATP7B levels have been linked to changes in cell resistance to DDP and were interpreted as evidence for a major role of ATP7B in DDP efflux. To investigate the contribution of ATP7B to hepatoma cell response to DDP, the effect of DDP on cell survival was investigated using the HepG2 and Huh7 cells. These cells are both of human hepatic origin and have comparable levels of ATP7B (Fig. 1A). Therefore, if ATP7B plays a major role in DDP efflux, it was expected that both cell lines may have comparable sensitivity to the drug. However, the effect of DDP on cell viability was very different. The Huh7 cells showed a biphasic response with a sharp 50% decrease in cell survival and the half-maximum effect at 1.54 μm DDP (Fig. 1B). A further increase of the DDP concentration up to 100 μm induces a much smaller (additional 10%) decline in cell viability. In contrast, the HepG2 cells show monophasic decrease in cell viability with the EC50 for DDP equal to 16.15 μm. Higher DDP concentration in the medium induces larger decline in cell survival down to ∼20% at 100 μm (Fig. 1B).
FIGURE 1.
The effects of DDP on viability of HepG2 and Huh7 cells does not correlate with the level of ATP7B expression. A, analysis of ATP7B expression in Huh7 and HepG2 cells by Western blot. B, Huh7 (○) and HepG2 cells (•) show a different response to increasing concentration of DDP. The data show the means ± S.D. of six separate experiments performed in duplicate and presented as percentages of untreated control. For determination of EC50 values, the curves were fitted with two exponentials. *, p < 0.001.
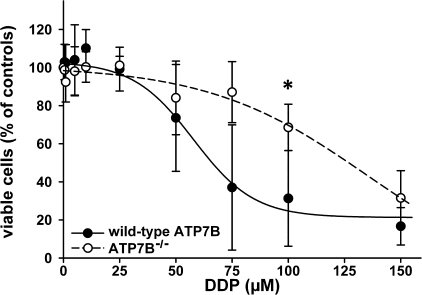
Primary Hepatocytes Lacking ATP7B Show Higher Resistance to DDP—The liver is the major detoxification organ, which mediates its function through multiple pathways. The individual contribution of these pathways may vary in cultured cell lines, thus producing distinct responses to DDP, as we observed for Huh7 and HepG2. Therefore, to examine the role of ATP7B more directly, we determined whether the elimination of endogenous ATP7B would render hepatocytes more sensitive to cisplatin. The primary hepatocytes were isolated from the control and Atp7b-/- mice, and their viability was compared in the presence of increasing concentration of DDP. Although in the absence of DDP the Atp7b-/- hepatocytes are generally less viable (as evidenced by the lower yield of cells during primary culture preparation), they showed higher resistance to cisplatin (EC50 = 135.2 μm) compared with control (EC50 = 57.8 μm) (Fig. 2). These results suggested that, in hepatocytes, ATP7B-mediated efflux did not play a major role in determining cell sensitivity to DDP.
FIGURE 2.
Viability of primary hepatocytes from wild-type (•) and Atp7b-/- (○) livers after treatment with DDP. Cell survival was measured by MTT assay. Control cells were cultured without DDP treatment. The data show the means ± S.D. of four separate experiments performed in triplicate and presented as percentages of untreated control. The difference at a concentration of 100 μm DDP was significant (*, p < 0.05). The curves were fitted using a sigmoidal decay model.
Trafficking of ATP7B in HepG2 Cells Is Not Regulated by DDP—One may argue that the inactivation of ATP7B in the Atp7b-/- liver induces significant remodeling of cell metabolism; this may involve up-regulation of other detoxification pathways and compensate for the loss of ATP7B function. Consequently, we decided to use an additional approach to characterize the DDP-ATP7B interactions. In hepatocytes, the primary function of ATP7B is to deliver cytosolic copper to the secretory pathway and to export excess copper out of the cell (27). Elevated copper facilitates transport activity of ATP7B and induces trafficking of hepatic ATP7B from the trans-Golgi network (TGN) to subapical vesicles (38). The trafficking process is associated with the excretion of excess copper and the restoration of cellular copper balance (42).
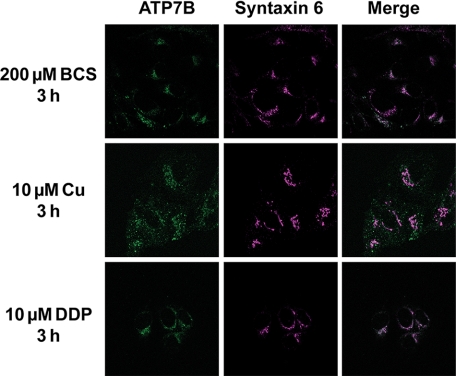
In ovarian cancer cells, DDP was shown to induce redistribution of enhanced cyan fluorescent protein-ATP7B (26), presumably mimicking the response to copper. Consequently, we tested whether DDP can induce trafficking of endogenous ATP7B in hepatocytes. Treatment with copper was used as a positive control. As expected, in cells treated with the copper chelator BCS, ATP7B was located in the perinuclear compartment and colocalized with the TGN marker, Syntaxin 6. In response to 10 μm copper added to the medium, ATP7B trafficked to vesicles, as indicated by the decrease in colocalization with Syntaxin 6 (Fig. 3). This trafficking response was observed at different incubation times (30 min to 24 h; data not shown), and different copper concentrations (supplemental Fig. S1). In contrast, incubation with 10 μm DDP showed no effect on ATP7B localization (Fig. 3). Similarly, no DDP effect was observed with higher DDP concentrations (supplemental Fig. S1) as well as longer treatment times (data not shown).
FIGURE 3.
ATP7B localization and trafficking in response to copper and DDP. HepG2 cells were treated with copper chelator BCS, copper, or DDP, respectively, for 3 h. ATP7B (green) was visualized with the anti N-ATP7B antibody. TGN localization is visualized by staining of Syntaxin 6 (purple). Colocalization was obtained by merging images using the microscope software and is indicated by white.
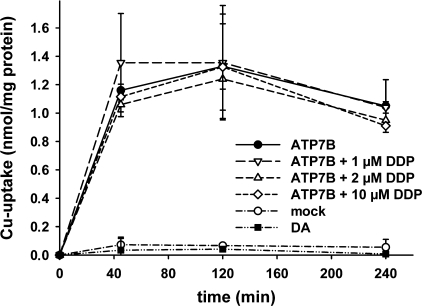
DDP Does Not Inhibit ATP7B-mediated Transport of Copper into Vesicles—Altogether, the above experiments provided little support to the current hypothesis that ATP7B mediates resistance through the transport of DDP out of the cell. Therefore, we tested the ability of ATP7B to transport DDP to vesicles in vitro. ATP7B was expressed in insect cells, and the membrane vesicles were prepared as described under “Experimental Procedures.” In the presence of ATP, the addition of Cu64 results in the increased accumulation of copper in the vesicles (Fig. 4). To confirm that the observed accumulation is due to transport and not simply due to copper binding, we utilized the catalytically inactive D1027A mutant of ATP7B (29). Although this mutant is expressed at levels comparable with those of wild-type ATP7B (Western blot data not shown), it shows no ATP-dependent copper accumulation in vesicles, excluding accumulation caused by binding (Fig. 4).
FIGURE 4.
DDP does not alter ATP7B-mediated copper uptake into vesicles. The kinetic of 64Cu uptake into membrane vesicles containing the wild-type ATP7B (•), the catalytically inactive ATP7B-D1027A variant (▪), and empty vesicles derived from uninfected Sf9 cells (mock, ○). For the competition experiments, vesicles expressing ATP7B were incubated with 64Cu in the presence of increasing DDP concentrations (1 μm, ▿; 2 μm, ▵; and 10 μm, ⋄). The data points show the means ± S.D. of three separate experiments.
To determine whether DDP is a transport substrate for ATP7B, we performed competition experiments. Increasing concentrations of cisplatin up to 10 μm (5-fold excess over copper) did not inhibit the ATP7B-mediated copper uptake into vesicles (Fig. 4). The lack of competition argues against DDP transport through the copper-translocation pathway of ATP7B, although some unusual transport mechanisms cannot be formally excluded.
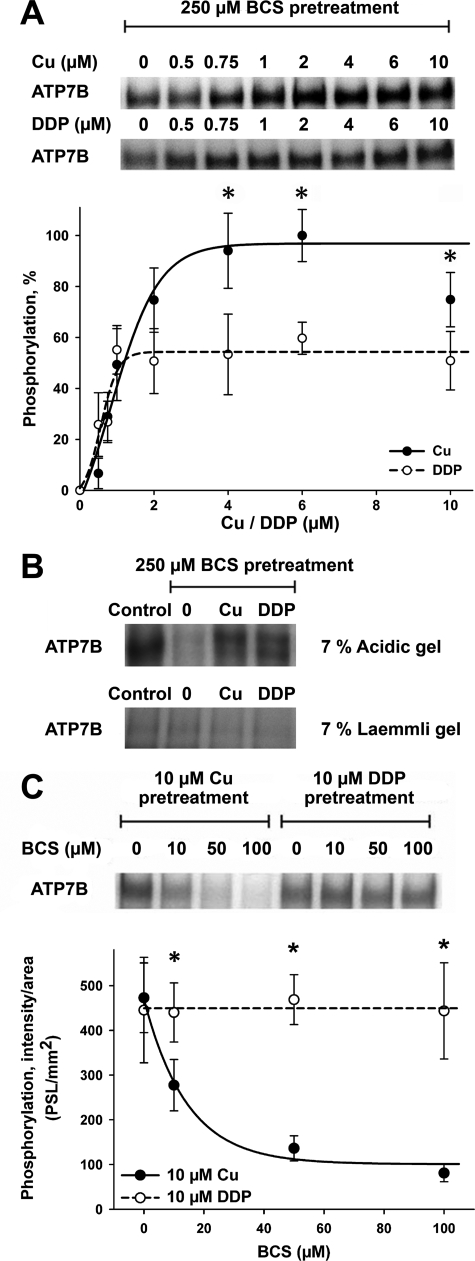
DDP Stimulates Catalytic Phosphorylation of ATP7B—To reconcile our results with the previous data demonstrating a connection between ATP7B levels and cell resistance to DDP, we hypothesized that DDP and ATP7B interact, but this interaction has consequences other than efflux of DDP. To test this hypothesis, we examined whether DDP can bind to ATP7B and regulate its catalytic activity. ATP7B was first inhibited by treatment with the copper chelator BCS, and then the ability of DDP to reactivate ATP7B was characterized. Fig. 5A illustrates that DDP stimulated phosphorylation of ATP7B by ATP with EC50 (0.57 μm) comparable with that of copper (0.84 μm), although the level of reactivation was lower than that for copper.
FIGURE 5.
The effects of copper and DDP on catalytic phosphorylation of ATP7B. A, catalytic activity of ATP7B was first inhibited by incubation with 250 μm BCS. Copper (•) or DDP (○) was then added to ATP7B, and reactivation of catalytic phosphorylation was measured as described under “Experimental Procedures.” The upper panel shows 32P autoradiograms of a representative gel, and the lower panel shows quantification of phosphorylation by densitometry. The phosphorylation level of ATP7B prior to BCS pretreatment was set to 100%. The quantification data show the means ± S.D. of five independent experiments. The curves were fitted with a sigmoidal model. *, p < 0.05. B, ATP7B was phosphorylated as in A; the phosphorylation of ATP7B without any treatments represents the control. The upper panel shows phosphorylation visualized at acidic pH (acidic gel). Phosphorylation is largely absent on a regular Laemmli gel (lower panel). The faint phosphorylation band represents a kinase-induced phosphorylation on serine residues of ATP7B normally found in Sf9 cells. C, ATP7B was phosphorylated as in A using 10 μm copper (•) or DDP (○) in the presence of increasing concentrations of BCS (10–100 μm). The upper panel shows an autoradiogram of a typical gel. The lower panel shows quantification data (means ± S.D.) of four separate experiments. The curves were fitted with a single exponential decay model (*, p < 0.05; *, p < 0.01 when comparing copper and DDP treatments).
ATP7B is known to undergo two types of phosphorylation: the catalytic phosphorylation with the formation of a transient acyl-phosphate intermediate (which is unstable at basic pH, but stable at acidic conditions) (29) and a stable kinase-mediated phosphorylation at a Ser residue(s) (43). To determine whether DDP induces catalytic or kinase-mediated phosphorylation, we phosphorylated ATP7B in the presence of copper (as a control) or DDP and compared the amount of phosphorylated products on acidic and regular Laemmli gels. The bands phosphorylated in the presence of either copper or DDP were clearly visible on the acidic gel but not under the basic conditions (Fig. 5B). We concluded that DDP stimulates catalytic phosphorylation of ATP7B.
To ensure that copper is not introduced with the DDP solution and/or DDP does not simply increase the sensitivity of ATP7B to trace amounts of copper, we repeated the phosphorylation experiments in the presence of increasing concentration of copper chelator BCS. Fig. 5C illustrates that the addition of BCS abolishes copper-dependent phosphorylation of ATP7B but has no effect on stimulation of phosphorylation by DDP.
The N-terminal Domain Has an Important Role in the DDP-dependent Regulation of ATP7B—Chemical dissimilarity of DDP and Cu(I) raise questions about the nature and the location of the DDP-binding sites. ATP7B has six copper-binding sites in the N-terminal domain and presumably one or two sites in the membrane. The binding of copper to the intramembrane sites of ATP7B is necessary to stimulate catalytic phosphorylation, whereas the N-terminal sites play a regulatory role (28, 29, 39). The addition of 2 μm DDP prior to copper had no significant effect on the copper dependence of ATP7B phosphorylation (tested in the 0.1–2 μm range of copper concentration; data not shown) arguing against DDP and copper being bound by the same set of coordinating residues in the membrane.
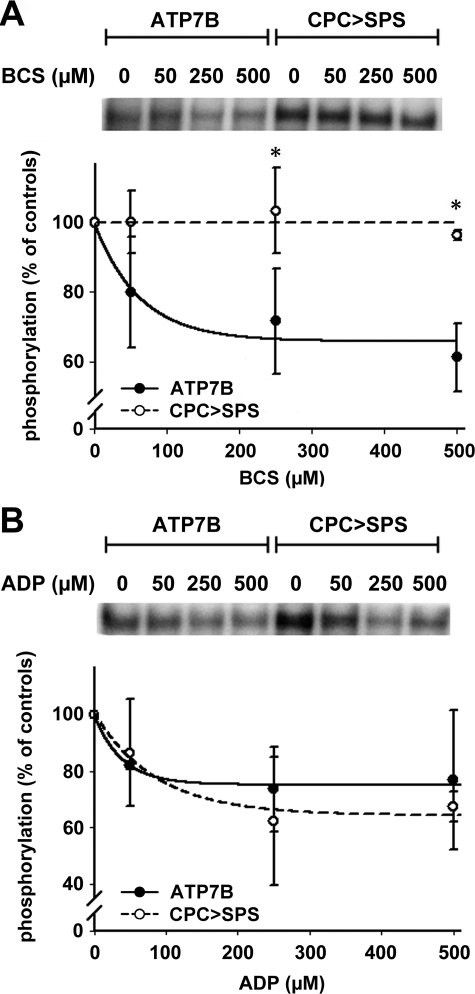
To verify this conclusion, we eliminated the intramembrane copper-binding motif CPC by mutating it to SPS. Phosphorylation of the ATP7B-CPC>SPS mutant yielded very interesting results (Fig. 6). We expected to see no catalytic phosphorylation of this mutant. Instead, the CPC>SPS mutant showed phosphorylation similar to the control ATP7B (the apparently higher level of signal on the gel is due to a higher level of protein expression for the mutant). This phosphorylation is not due to the ability of the mutant to bind copper through the remaining ligands within the membrane portion of ATP7B, because the subsequent addition of BCS (that stimulates dephosphorylation of the wild-type protein) had very little, if any, effect on the dephosphorylation of the mutant (Fig. 6A).
FIGURE 6.
The effects of BCS and ADP on catalytic phosphorylation of ATP7B and the CPC>SPS mutant. A, copper was removed from ATP7B (•) and the CPC>SPS mutant (○) using increasing concentrations of BCS for 30 min at pH 7. 0. In the upper panel a typical autoradiogram is shown. The lower panel shows quantification data (average of four separate experiments; means ± S.D.). The curves were fitted with a single exponential decay model (*, p < 0.05 when comparing ATP7B and CPC>SPS mutant). B, ATP7B and CPC>SPS mutant were treated with radioactive [γ-32P]ATP for 4 min on ice, and then increasing concentrations of ADP were added, and the reaction was stopped after 4 min (upper panel shows a autoradiogram, and the lower panel shows averages of three separate experiments ± S.D.).
The CPC>SPS mutant of ATP7B does not transport copper according to the published reports (30, 44, 45). However, it has a normal sensitivity to ADP, a characteristic enzymatic feature of the P-type ATPases (Fig. 6B). This result indicates that the CPC>SPS mutation stabilizes the conformational state of ATP7B, which is suitable for catalytic phosphorylation even in the absence of metal bound within the membrane. (Another recently reported example of the Cu-ATPase being phosphorylated in the absence of transported ion is CopA from Thermotoga maritima (46)).
We speculated that cisplatin binds to the extramembrane regions of ATP7B and stabilizes such conformation that is suitable for phosphorylation but is not identical to a copper-bound form. To test this hypothesis we first examined whether the N-terminal metal-binding domain may play a role in DDP binding.
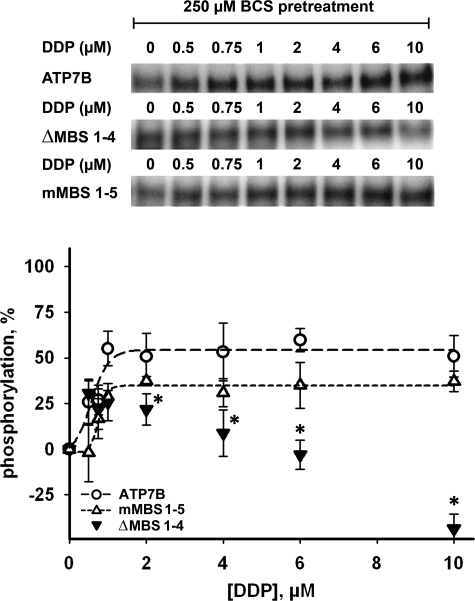
We have previously shown that the deletion of the first four N-terminal metal-binding subdomains (ΔMBS1–4) does not affect the copper dependence of ATP7B phosphorylation (39). In contrast, this deletion completely abolishes the ability of DDP to stimulate phosphorylation of ATP7B (Fig. 7). This result pointed to the important role of the N-terminal domain (in particular, the MBS1–4 region) in mediating the DDP effect. As we discussed above, the coordination environment of Cu(I) and DDP/platinum are likely to be very different; consequently it was interesting to determine whether the copper-binding motifs CXXC play a role in the binding of DDP. To do that we characterized the ATP7B variant in which the first five of six N-terminal CXXC sites (including all in the ΔMBS1–4 region) were substituted with AXXA (mMBS1–5). Remarkably, this mutation, which eliminated most of the copper-binding sites but preserved the N-terminal domain structure (and/or the interactions with other domains), had no significant effect on the DDP potency in the phosphorylation assay (EC50 = 0.78 μm) (Fig. 7). Thus, DDP is unlikely to bind to the N-terminal domain using copper-coordinating Cys.
FIGURE 7.
The effects of DDP on catalytic phosphorylation of ATP7B, ΔMBS1–4 and mMBS1–5. Wild-type ATP7B (○), ΔMBS1–4 (▾), and mMBS1–5 (▵) were inactivated by pretreatment with 250 μm BCS, incubated with DDP (0.5–10 μm) for 10 min and then phosphorylated with [γ-32P]ATP. The phosphorylation levels of ATP7B in the presence of BCS were subtracted, and the maximum phosphorylation level induced by copper was set to 100%. In the upper panel a typical autoradiogram is shown. The lower panel shows quantification (means ± S.D. of three separate experiments). The curves for ATP7B and the mMBS1–5 mutant were fitted with a sigmoidal model. The data for ΔMBS1–4 could not be fitted using the sigmoidal model and are shown without fitting. *, p < 0.05, when comparing ΔMBS1–4 with wild-type ATP7B under DDP treatment.
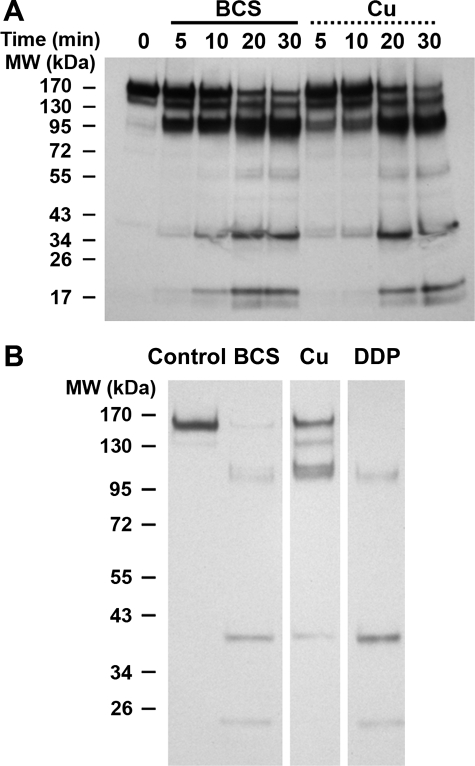
Distinct structural consequences of DDP and copper binding to ATP7B were further illustrated using an independent approach. To monitor the conformational state of ATP7B before and after ligand binding, we utilized a limited proteolysis with papain, as described for the archea Cu-ATPase CopA (47). We first determined whether copper binding to ATP7B can be detected by proteolysis (under used experimental conditions copper does not affect the papain activity (47)). Comparison of the kinetic of ATP7B digestion in the presence of copper and BCS revealed a difference in the rate of generation of the 112-kDa fragment. This fragment was rapidly produced in the absence of copper but was generated less readily when copper was bound to the protein (Fig. 8A). The 112-kDa band is detectable with the antibody against the C terminus of ATP7B, which means that this band is generated by cleaving the 45–50-kDa N-terminal fragment off the full-length ATP7B (165 kDa). The slower appearance of the 112-kDa band indicated a change in the accessibility of the N-terminal portion of ATP7B to papain when copper was bound. Such a protective effect on the 112-kDa band was not observed in the presence of DDP (Fig. 8B). Thus, under conditions used for proteolysis, DDP either does not bind or does not protect, further illustrating different modes of copper and DDP binding to ATP7B.
FIGURE 8.
Limited digestion of ATP7B with papain demonstrates different effect of copper and DDP on the conformation of the protein. A, ATP7B was incubated with 500 μm BCS or 10 μm copper at 30 °C for 1 h. Then protein samples were digested with papain at 1:1000 dilution over a period of 5–30 min at 30 °C. The ATP7B fragments were visualized by Western blotting using the C-terminal antibody for ATP7B. B, ATP7B was incubated with 500 μm BCS, 10 μm copper, and 10 μm DDP as described above and then digested with papain at 1:100 dilution for 5 min at 30 °C. The ATP7B fragments were visualized by Western blotting using the C-terminal antibody for ATP7B (samples were run on the same gel).
Changes in Copper Metabolism Alter Resistance to Cisplatin of the DDP-sensitive Cells—Altogether, our results suggested that the binding of DDP by ATP7B rather than active efflux may contribute to drug resistance in cells with the increased levels of ATP7B. However, ATP7B in many cells is a minor membrane protein, and even a 5–10-fold increase in protein levels (observed in resistant cells) may not be sufficient to sequester DDP. We hypothesized that up-regulation of Cu-ATPases that decreases the cytosolic copper concentration (18, 20) may activate other detoxification pathways (for example, copper deficiency was shown to up-regulate glutathione biosynthesis (48)) and thus indirectly increase resistance to DDP.
To test the prediction that copper deficiency rather than the ATP7B levels per se may affect cells resistance to cisplatin, we incubated hepatic cells with or without BCS and then compared cells resistance to DDP. Fig. 9 illustrates that copper depletion altered the resistance of Huh7 cells to DDP and eliminated the biphasic nature of the DDP effect on cell viability. Furthermore, compared with nontreated cells (EC50 for DDP 1.31 μm), the copper-depleted Huh7 cells showed higher resistance to DDP (EC50 of 13.62 μm). Interestingly, more resistant HepG2 cells were not significantly affected by the pretreatment with BCS (supplemental Fig. S2).
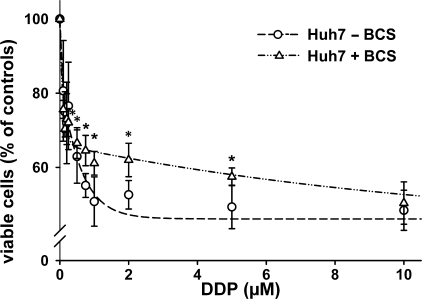
FIGURE 9.
Copper deficiency alters the viability of Huh7 cells exposed to DDP. Huh7 cells were grown under normal conditions (○) and under copper deficiency conditions (200 μm BCS 48 h prior and during DDP treatment ▵) and then exposed to increasing concentrations of DDP (0.1–10 μm) for 24 h. The cells were then harvested, and cytotoxicity was determined by Casy TT®. The data show the means ± S.D. of four to six separate experiments performed in duplicate and presented as percent of untreated control. The curves were fitted with two exponentials (*, p < 0.05; *, p < 0.01 when comparing BCS-treated and nontreated Huh7 cells).
DISCUSSION
In this study we examined the potential mechanism through which Cu-ATPase ATP7B regulates the resistance of human cells to cisplatin. Our results revealed a more complex relationship between the resistance of cells to DDP and ATP7B than is currently thought. It is clear that DDP binds directly to ATP7B and affects its enzymatic activity by stimulating the formation of a phosphorylated intermediate; the N-terminal region of ATP7B is critical for this interaction. Thus, ATP7B can interact with DDP despite distinct chemistry of cisplatin and copper.
However, DDP binding to ATP7B does not result in the competition with copper in the transport assay. An earlier study in ovary cells reported colocalization of enhanced cyan fluorescent protein-ATP7B with the fluorescent DDP analogue as well as a 2-fold increase in the amount of DDP in the vesicles with elevated ATP7B (26). Our results suggest that this modest increase in the amount of platinum associated with the vesicles is likely due to the binding of DDP to the overexpressed ATP7B rather than transport.
DDP does not induce trafficking of endogenous ATP7B in HepG2 cells (Fig. 3). In contrast, the DDP-dependent change in ATP7B localization in ovary cells was reported (26). There could be several reasons for the apparent discrepancy. It is possible that hepatocytes efficiently prevent interaction of DDP with ATP7B using other resistance mechanisms, and therefore in hepatocytes there is simply not enough DDP to induce the ATP7B trafficking. This explanation is less likely because we used very different concentrations of DDP and did not see trafficking of ATP7B in any of those conditions. Previous studies also utilized the overexpressed green fluorescent protein-tagged protein in cells that normally do not express ATP7B in high quantities, which may lead to protein mistargeting to vesicles rather than TGN. Because the localization of the green fluorescent protein-tagged ATP7B in the TGN (or the loss of colocalization in response to copper or DDP) has not been confirmed by colocalization with the TGN marker (as we did for the endogenous ATP7B in the present study), the possibility remains that the distribution/staining of vesicles (rather than the localization of ATP7B) differed in ± DDP conditions.
Copper Metabolism and Resistance to DDP—Cultured and primary hepatocytes show significant resistance to DDP and retain high cell viability (50% in the case of Huh7cells) even at high drug concentrations. This property is likely due to the presence of multiple detoxification pathways and is an in vitro reflection of a poor susceptibility of hepatic tumors to chemotherapeutic treatment. We have shown that in hepatocytes ATP7B does not play a major role in the resistance of cells to DDP and that inactivation of ATP7B does not increase cells susceptibility to the drug. This negative result has important practical implications.
We also found that changes in cellular copper balance (caused by either the loss of ATP7B or copper depletion produced by BCS) do alter cell resistance to DDP. These observations may help to reconcile previous literature reports and our own findings and shed light on additional important mechanisms operating in resistant cells. Indeed, it has been shown that the DDP-resistant ovary cells are copper-deficient (18), i.e. the overexpression of ATP7B, as expected, is accompanied by the loss of copper. Given our data with Huh7 cells, we speculate that copper deficiency (independently of how it was induced) could be a factor in cell resistance to DDP. Although we have not investigated the mechanism behind increased resistance in response to copper deprivation, increased synthesis and export of glutathione have been reported in copper-deficient cells and may increase resistance to DDP (52, 53).
How could the inactivation of Cu-ATPase increase the hepatoma cell resistance to DDP? Hepatic cells perform many detoxification functions and have multiple resistance mechanisms. In the Atp7b-/- mice, in response to copper accumulation, several protective mechanisms are up-regulated that may potentially affect cell sensitivity to DDP. Specifically, inactivation of ATP7B greatly up-regulates expression of metallothionein and increases levels of P-glycoprotein (50). Both events may lead to increased sequestration and/or export of DDP (51), explaining the observed increased resistance of Atp7b-/- cells to the drug.
Interaction with DDP Revealed Conformational Plasticity of ATP7B—Our studies confirmed the effect of DDP on ATP7B catalytic activity (31) and revealed an unexpected conformational plasticity of ATP7B with respect to the formation of phosphorylated intermediate. It is thought that the binding of transported ions to the intramembrane sites is required for the formation of phosphorylated intermediate. We found that DDP induces catalytic phosphorylation while binding at the sites that are distinct from the binding sites for copper. This conclusion is supported by (i) a very different effect of the ΔMBS1–4 deletion on copper and DDP stimulation phosphorylation, (ii) by distinct consequences of the DDP and copper on ATP7B proteolysis, (iii) by the inability of DDP to stimulate trafficking of ATP7B, and (iv) by the ability of CPC>SPS mutant to become phosphorylated in the absence of copper. While this manuscript was in preparation, the studies on CopA from Thermotoga meritima provided further evidence that Cu-ATPases (46), unlike the P2-type ATPases, can undergo phosphorylation in the absence of transported ion.
So far, our results suggest that the N-terminal domain of ATP7B (N-ATP7B) plays an important role in interactions between the transporter and the drug. A deletion of the first four MBS of the N-terminal domain, but not inactivation of the first five MBS via substitution of cysteine by alanine, completely abolished the ability of DDP to stimulate phosphorylation of ATP7B (Fig. 7). We conclude that DDP binds to the N-terminal domain using ligands other than copper-coordinating cysteines. This result is reminiscent of earlier findings by Sarkar and co-workers (54), who investigated interactions between N-ATP7B and Zn2+ and found that zinc and copper bind differently to N-ATP7B. A recent meeting report (55) provided further evidence for the direct interaction of DDP with the recombinant N-ATP7B.
In conclusion, our results show that DDP binds directly to ATP7B and affects its activity. Sequestration of DDP by ATP7B could be one of the components of the cellular DDP detoxification mechanism, particularly in cells in which ATP7B is highly up-regulated. Another possible mechanism of copper-dependent resistance to DDP involves change in cellular copper homeostasis, which may in turn activate secondary drug resistance pathways.
Supplementary Material
Acknowledgments
We thank D. Mahn, I. Sommerer, L. Thiel, and F. Struck for invaluable technical help and R. Linz for generating the antibodies against the C-terminal domain of ATP7B. The assistance of Dr. D. G. Huster for mathematical models and helpful discussions of our results is gratefully acknowledged.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK071865 (to S. L.). This work was also supported by German Research Foundation Grant HU 932/3-1 (to D. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: DDP, cis-diamminedichloroplatinum; BCS, bathocuproine disulfonate; DTT, dithiothreitol; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; TGN, trans-Golgi network; Cu-ATPase, copper-transporting ATPase; PBS, phosphate-buffered saline; MOPS, 3-morpholinopropanesulfonic acid; MBS, metal-binding site.
References
- 1.Barocas, D. A., and Clark, P. E. (2008) Curr. Opin. Oncol. 20 307-314 [DOI] [PubMed] [Google Scholar]
- 2.Ferraldeschi, R., Baka, S., Jyoti, B., Faivre-Finn, C., Thatcher, N., and Lorigan, P. (2007) Drugs 67 2135-2152 [DOI] [PubMed] [Google Scholar]
- 3.Kelland, L. (2007) Nat. Rev. Cancer 7 573-584 [DOI] [PubMed] [Google Scholar]
- 4.Moehler, M., Galle, P. R., Gockel, I., Junginger, T., and Schmidberger, H. (2007) Best Pract. Res. Clin. Gastroenterol. 21 965-981 [DOI] [PubMed] [Google Scholar]
- 5.Yuan, J. N., Chao, Y., Lee, W. P., Li, C. P., Lee, R. C., Chang, F. Y., Yen, S. H., Lee, S. D., and Whang-Peng, J. (2008) Med. Oncol. 25 201-206 [DOI] [PubMed] [Google Scholar]
- 6.Jamieson, E. R., and Lippard, S. J. (1999) Chem. Rev. 99 2467-2498 [DOI] [PubMed] [Google Scholar]
- 7.Wong, E., and Giandomenico, C. M. (1999) Chem. Rev. 99 2451-2466 [DOI] [PubMed] [Google Scholar]
- 8.Andrews, P. A., Jones, J. A., Varki, N. M., and Howell, S. B. (1990) Cancer Commun. 2 93-100 [DOI] [PubMed] [Google Scholar]
- 9.Akiyama, S., Chen, Z. S., Sumizawa, T., and Furukawa, T. (1999) Anticancer Drug Des. 14 143-151 [PubMed] [Google Scholar]
- 10.Mukai, M., Kanzaki, A., Chen, Z. S., Miyashita, H., Sumizawa, T., Furukawa, T., Haraguchi, M., Takebayashi, Y., Takamatsu, H., and Akiyama, S. (2002) Oncol. Rep. 9 839-844 [PubMed] [Google Scholar]
- 11.Jordan, P., and Carmo-Fonseca, M. (2000) Cell Mol. Life Sci. 57 1229-1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siddik, Z. H. (2003) Oncogene 22 7265-7279 [DOI] [PubMed] [Google Scholar]
- 13.Brozovic, A., Fritz, G., Christmann, M., Zisowsky, J., Jaehde, U., Osmak, M., and Kaina, B. (2004) Int. J. Cancer 112 974-985 [DOI] [PubMed] [Google Scholar]
- 14.Safaei, R. (2006) Cancer Lett. 234 34-39 [DOI] [PubMed] [Google Scholar]
- 15.Samimi, G., Safaei, R., Katano, K., Holzer, A. K., Rochdi, M., Tomioka, M., Goodman, M., and Howell, S. B. (2004) Clin. Cancer Res. 10 4661-4669 [DOI] [PubMed] [Google Scholar]
- 16.Kuo, M. T., Chen, H. H., Song, I. S., Savaraj, N., and Ishikawa, T. (2007) Cancer Metastasis Rev. 26 71-83 [DOI] [PubMed] [Google Scholar]
- 17.Yoshizawa, K., Nozaki, S., Kitahara, H., Ohara, T., Kato, K., Kawashiri, S., and Yamamoto, E. (2007) Oncol. Rep. 18 987-991 [PubMed] [Google Scholar]
- 18.Katano, K., Kondo, A., Safaei, R., Holzer, A., Samimi, G., Mishima, M., Kuo, Y. M., Rochdi, M., and Howell, S. B. (2002) Cancer Res. 62 6559-6565 [PubMed] [Google Scholar]
- 19.Safaei, R., and Howell, S. B. (2005) Crit. Rev. Oncol. Hematol. 53 13-23 [DOI] [PubMed] [Google Scholar]
- 20.Safaei, R., Katano, K., Samimi, G., Naerdemann, W., Stevenson, J. L., Rochdi, M., and Howell, S. B. (2004) Cancer Chemother. Pharmacol. 53 239-246 [DOI] [PubMed] [Google Scholar]
- 21.Higashimoto, M., Kanzaki, A., Shimakawa, T., Konno, S., Naritaka, Y., Nitta, Y., Mori, S., Shirata, S., Yoshida, A., Terada, K., Sugiyama, T., Ogawa, K., and Takebayashi, Y. (2003) Int. J. Mol. Med. 11 337-341 [PubMed] [Google Scholar]
- 22.Kanzaki, A., Toi, M., Neamati, N., Miyashita, H., Oubu, M., Nakayama, K., Bando, H., Ogawa, K., Mutoh, M., Mori, S., Terada, K., Sugiyama, T., Fukumoto, M., and Takebayashi, Y. (2002) Jpn. J. Cancer Res. 93 70-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohbu, M., Ogawa, K., Konno, S., Kanzaki, A., Terada, K., Sugiyama, T., and Takebayashi, Y. (2003) Cancer Lett. 189 33-38 [DOI] [PubMed] [Google Scholar]
- 24.Komatsu, M., Sumizawa, T., Mutoh, M., Chen, Z. S., Terada, K., Furukawa, T., Yang, X. L., Gao, H., Miura, N., Sugiyama, T., and Akiyama, S. (2000) Cancer Res. 60 1312-1316 [PubMed] [Google Scholar]
- 25.Safaei, R., Larson, B. J., Cheng, T. C., Gibson, M. A., Otani, S., Naerdemann, W., and Howell, S. B. (2005) Mol. Cancer Ther. 4 1595-1604 [DOI] [PubMed] [Google Scholar]
- 26.Katano, K., Safaei, R., Samimi, G., Holzer, A., Tomioka, M., Goodman, M., and Howell, S. B. (2004) Clin. Cancer Res. 10 4578-4588 [DOI] [PubMed] [Google Scholar]
- 27.Lutsenko, S., Barnes, N. L., Bartee, M. Y., and Dmitriev, O. Y. (2007) Physiol. Rev. 87 1011-1046 [DOI] [PubMed] [Google Scholar]
- 28.Petris, M. J., Voskoboinik, I., Cater, M., Smith, K., Kim, B. E., Llanos, R. M., Strausak, D., Camakaris, J., and Mercer, J. F. (2002) J. Biol. Chem. 277 46736-46742 [DOI] [PubMed] [Google Scholar]
- 29.Tsivkovskii, R., Eisses, J. F., Kaplan, J. H., and Lutsenko, S. (2002) J. Biol. Chem. 277 976-983 [DOI] [PubMed] [Google Scholar]
- 30.Cater, M. A., La Fontaine, S., and Mercer, J. F. (2007) Biochem. J. 401 143-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Safaei, R., Otani, S., Larson, B. J., Rasmussen, M. L., and Howell, S. B. (2008) Mol. Pharmacol. 73 461-468 [DOI] [PubMed] [Google Scholar]
- 32.Wakamatsu, T., Nakahashi, Y., Hachimine, D., Seki, T., and Okazaki, K. (2007) Int. J. Oncol. 31 1465-1472 [PubMed] [Google Scholar]
- 33.Mathurin, P., Rixe, O., Carbonell, N., Bernard, B., Cluzel, P., Bellin, M. F., Khayat, D., Opolon, P., and Poynard, T. (1998) Aliment Pharmacol. Ther. 12 111-126 [DOI] [PubMed] [Google Scholar]
- 34.Buiakova, O. I., Xu, J., Lutsenko, S., Zeitlin, S., Das, K., Das, S., Ross, B. M., Mekios, C., Scheinberg, I. H., and Gilliam, T. C. (1999) Hum. Mol. Genet. 8 1665-1671 [DOI] [PubMed] [Google Scholar]
- 35.Seglen, P. O. (1973) Exp. Cell Res. 82 391-398 [DOI] [PubMed] [Google Scholar]
- 36.Gebhardt, R., Jung, W., and Robenek, H. (1982) Eur. J. Cell Biol. 29 68-76 [PubMed] [Google Scholar]
- 37.Kruger, N. J. (1994) Methods Mol. Biol. 32 9-15 [DOI] [PubMed] [Google Scholar]
- 38.Bartee, M. Y., and Lutsenko, S. (2007) Biometals 20 627-637 [DOI] [PubMed] [Google Scholar]
- 39.Huster, D., and Lutsenko, S. (2003) J. Biol. Chem. 278 32212-32218 [DOI] [PubMed] [Google Scholar]
- 40.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265-275 [PubMed] [Google Scholar]
- 41.Gmaj, P., Zurini, M., Murer, H., and Carafoli, E. (1983) Eur. J. Biochem. 136 71-76 [DOI] [PubMed] [Google Scholar]
- 42.Cater, M. A., La Fontaine, S., Shield, K., Deal, Y., and Mercer, J. F. (2006) Gastroenterology 130 493-506 [DOI] [PubMed] [Google Scholar]
- 43.Vanderwerf, S. M., Cooper, M. J., Stetsenko, I. V., and Lutsenko, S. (2001) J. Biol. Chem. 276 36289-36294 [DOI] [PubMed] [Google Scholar]
- 44.Forbes, J. R., and Cox, D. W. (1998) Am. J. Hum. Genet. 63 1663-1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forbes, J. R., and Cox, D. W. (2000) Hum. Mol. Genet. 9 1927-1935 [DOI] [PubMed] [Google Scholar]
- 46.Hatori, Y., Hirata, A., Toyoshima, C., Lewis, D., Pilankatta, R., and Inesi, G. (2008) J. Biol. Chem. 283 22541-22549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hatori, Y., Majima, E., Tsuda, T., and Toyoshima, C. (2007) J. Biol. Chem. 282 25213-25221 [DOI] [PubMed] [Google Scholar]
- 48.Uthus, E. O., Reeves, P. G., and Saari, J. T. (2007) J. Nutr. 137 1370-1374 [DOI] [PubMed] [Google Scholar]
- 49.Zisowsky, J., Koegel, S., Leyers, S., Devarakonda, K., Kassack, M. U., Osmak, M., and Jaehde, U. (2007) Biochem. Pharmacol. 73 298-307 [DOI] [PubMed] [Google Scholar]
- 50.Huster, D., Purnat, T. D., Burkhead, J. L., Ralle, M., Fiehn, O., Stuckert, F., Olson, N. E., Teupser, D., and Lutsenko, S. (2007) J. Biol. Chem. 282 8343-8355 [DOI] [PubMed] [Google Scholar]
- 51.Zelazowski, A. J., Garvey, J. S., and Hoeschele, J. D. (1984) Arch. Biochem. Biophys. 229 246-252 [DOI] [PubMed] [Google Scholar]
- 52.Hamilton, T. C., Winker, M. A., Louie, K. G., Batist, G., Behrens, B. C., Tsuruo, T., Grotzinger, K. R., McKoy, W. M., Young, R. C., and Ozols, R. F. (1985) Biochem. Pharmacol. 34 2583-2586 [DOI] [PubMed] [Google Scholar]
- 53.Bier, H., Bergler, W., Mende, S., and Ganzer, U. (1988) Arch. Otorhinolaryngol. 245 166-169 [DOI] [PubMed] [Google Scholar]
- 54.DiDonato, M., Narindrasorasak, S., Forbes, J. R., Cox, D. W., and Sarkar, B. (1997) J. Biol. Chem. 272 33279-33282 [DOI] [PubMed] [Google Scholar]
- 55.Dolgova, N. V., Olson, D., Lutsenko, S., and Dmitriev, O. Y. (2009) Biochem. J., in press DOI: 10.1042/BJ20081359 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.