Abstract
The Syd protein has been implicated in the Sec-dependent transport of polypeptides across the bacterial inner membrane. Using Nanodiscs, we here provide direct evidence that Syd binds the SecY complex, and we demonstrate that interaction involves the two electropositive and cytosolic loops of the SecY subunit. We solve the crystal structure of Syd and together with cysteine cross-link analysis, we show that a conserved concave and electronegative groove constitutes the SecY-binding site. At the membrane, Syd decreases the activity of the translocon containing loosely associated SecY-SecE subunits, whereas in detergent solution Syd disrupts the SecYEG heterotrimeric associations. These results support the role of Syd in proofreading the SecY complex biogenesis and point to the electrostatic nature of the Sec channel interaction with its cytosolic partners.
The membrane-embedded SecYEG heterotrimer, also called core translocon or SecY complex, is a conserved protein-conducting channel essential for the biogenesis of most of the secretory and integral membrane proteins (1). The SecY complex is a passive conduit and thus cooperates with cytosolic and membranous partners to drive efficient polypeptide transport into and across membrane. During co-translational transport, the SecY complex associates with the ribosome and elements of the signal recognition particle (2, 3). During post-translational translocation in bacteria, the SecY complex interacts with the SecA ATPase to push secretory proteins across the channel (4). In the membrane, the SecY complex interacts with itself to form oligomers and with other membrane-embedded components such as the SecFDyajC complex and the insertase YidC (5–7).
The core translocon has been reconstituted into a functional in vitro system, and the structure of the SecY channel, SecA, and the ribosome has been determined at the atomic level (8–11). The interactions between these components, along with the stoichiometry and dynamics of their associations, are now crucial for the complete understanding of the translocation reaction. Much remains to be learned about the other components also involved in the biogenesis, regulation, and modulation of the translocon activity. Syd is a nonessential and hydrophilic protein of 181 amino acid residues. Ito and co-workers (12, 13) originally identified its gene as a multicopy suppressor of the dominant-negative secY-d1 mutation (and thus termed Syd for suppressor of SecYdominance). Biochemical analysis then provided the first but indirect evidence for an interaction between Syd and the translocon (14). For example, Syd was shown to interfere with protein translocation, but only in cells in which SecY and SecE proteins interact weakly. The physiological role of Syd remains uncertain (the deletion of the gene causes no obvious phenotype, see Ref. 12), but it is anticipated that Syd should provide new clues about the nature of SecY interactions with its cytosolic partners.
We here report the crystal structure of Syd and provide direct evidence for its interaction with the SecY complex. We also chart the association and investigate the consequence of Syd binding on both activity and stability of the SecY channel. This comprehensive analysis provides a new framework for the understanding of the SecY channel interactome.
EXPERIMENTAL PROCEDURES
Plasmids and Biological Reagents—The Syd open reading frame was PCR-amplified from the Escherichia coli genome and cloned into the expression vector pET23a (Clontech) using the restriction sites NdeI and XhoI. The plasmid pBAD22-hisEYG and purification of the SecY complex were previously described (15). The deletion Δ251–258 and Δ354–357 into SecY and Δ7–67 into SecE was obtained by PCR amplification using primers introducing a BglII restriction site on each side of the deletion.
Purification of Syd—Plasmid pET23-Syd was transformed in E. coli strain BL21 (DE3). Overproduction of Syd was initiated at A600 nm ∼ 0.5 with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h. Cells were collected in TSG buffer (25 mm Tris-Cl, pH 7.5; 50 mm NaCl; 10% glycerol; no DTT4) and lysed with a French press (8,000 p.s.i., three passes). After centrifugation (100,000 × g, 1 h at 4 °C), the supernatant was applied onto a Ni2+-chelated Sepharose column (GE Healthcare) equilibrated in TSG buffer. Syd was eluted with 500 mm imidazole and applied onto a 5-ml Q-Sepharose Fast Flow column (GE Healthcare) equilibrated in TS buffer (50 mm Tris-Cl, pH 7.5; 50 mm NaCl; 1 mm DTT). Syd was eluted with 250 mm NaCl and concentrated to 40 mg/ml using an Amicon 5-kDa centrifugation device. For selenomethionine labeling, cells were grown in 9 liters of M9 media. At A600 ∼ 0.3, amino acids were added (l-lysine, l-phenylalanine, l-threonine, l-isoleucine, l-leucine, l-valine, and l-selenomethionine; each 50 mg/liter), and Syd expression was induced with 1.5 mm isopropyl 1-thio-β-d-galactopyranoside during 16 h at 30 °C. 125I labeling was performed using IODO-GEN-coated tubes (Pierce) containing 60 μg of Syd and 25 μCi of Na125I. The reaction was quenched with 5 mm DTT, and the protein was desalted through a G-25 spin column equilibrated in TSG buffer. 125I-Syd (∼2 × 105 cpm/μg) was stored at -80 °C and used within a month.
Crystallization and Structure Determination—Native E. coli Syd was crystallized using the hanging drop vapor diffusion method in 0.8–1.0 m sodium citrate, 0.2 m sodium chloride, and 0.1 m Tris, pH 7.0. The starting protein concentration was 20 mg/ml. Crystals reached dimensions of up to 0.45 × 0.40 × 0.25 mm. Selenomethionine E. coli Syd crystallized under the same conditions, although the resultant crystals were smaller (up to 0.2 × 0.18 × 0.1 mm). All crystals were cryo-cooled in the presence of 30% sodium malonate, pH 7.0, prior to x-ray data collection at the Stanford Synchrotron Radiation Laboratory (Stanford, CA). Details of data collection, processing, and structural refinement statistics are given in the supplemental material. Each dataset was integrated and scaled using the programs MOSFLM and SCALA, respectively (16). The structure of Syd was determined by the single wavelength anomalous diffraction method. Two selenomethionine sites, one each from the two Syd molecules in the asymmetric unit, were located and refined using the program SOLVE, and phases were subsequently improved by density modification using the program RESOLVE (17). The program ARP/WARP was used to build the initial model (18). The primary sequence used in structure refinement was that from GenBank™ (accession number ABE08615). The program REFMAC5 (16) was used to refine the initial model using the native Syd dataset. This involved iterative cycles of fitting and rebuilding using the COOT program (19). Further structural refinement was conducted using the native Syd dataset and CNS (20). Note that the 6-histidine tag attached to each Syd molecule in the asymmetric unit was not observed in electron density maps and therefore was not included in the structural model. Notably, the structure determination clearly indicates the presence of a disulfide bridge between cysteines 147 and 154 in Syd. These two cysteines are ideally positioned with respect to one another within the polypeptide chain fold to form this linkage. However, for one of the molecules of Syd in the asymmetric unit, there is some evidence from electron density maps that this disulfide bridge may be broken in ∼20% of the crystallized molecules. The other two cysteines present in Syd are too far removed from one another to form a disulfide bridge. Structures were visualized using PyMol (version 0.99), and electrostatic maps were obtained using the APBS plug-in (version 1.0.0). Atomic charges and radii were generated with the AMBER option at the Protein Data Bank code 2PQR on-line service.
Analytical Gel Filtration—Analytical gel filtration was performed using a Superdex 200 HR 10/30 column (Amersham Biosciences) connected in-line to miniDAWN multiangle light scattering equipment coupled to an interferometric refractometer (Wyatt Technologies). Data analysis was recorded in real time using the ASTRA software (Wyatt Technologies). Molecular masses were calculated using the Debye fit method.
Other Methods—The preparation of reagents such as Nd-SecYEG, SecA, 125I-proOmpA, and urea-stripped IMVs, as well as the conditions for in vitro translocation assays and native/blue-native gel electrophoresis were described previously (7, 21). Detection of Na125I-labeled proteins and densitometry scanning were performed using a phosphorimager scanner.
RESULTS AND DISCUSSION
A Complex of SecYEG and Syd—Nanodiscs permit investigation of the biochemistry of the SecY complex, without using liposome or detergent (21, 22). Each particle, also referred to as Nd-SecYEG or SecYEG-Nanodisc, is made of a single SecY complex embedded in a small patch of lipid bilayer supported by two membrane scaffold proteins. Although homogeneous and water-soluble, the Nd-SecYEG particles (∼125 kDa) appear smeared on native-PAGE (Fig. 1A). When incubated with the purified Syd protein, however, the mobility of the particles is greatly enhanced, possibly as a result of complex formation and modification of the electrophoretic characteristics. On its own, the 125I-Syd protein migrates at the bottom of the gel but dramatically shifts to the expected high molecular weight position when incubated with the Nd-SecYEG particles (Fig. 1B). When the SecY subunit carries small internal deletion in one or the other of its largest cytosolic loops, C4 and C5, respectively, the binding of Syd appears reduced (Fig. 1B). In the SecY crystal structure, these loops extend prominently from the surface of the cytoplasmic membrane and form binding sites for SecA or the ribosome (3, 10, 23, 24). The stability of the Nd-SecYEG-Syd complex was further assessed by ultracentrifugation. The 125I-Syd protein, which floats at the top of the sucrose gradient, sediments to a higher density in the presence of Nd-SecYEG (Fig. 1C). The sedimentation is only partial when the SecY subunit carries the internal deletion in either C4 or C5 loops (Fig. 1C). Interestingly, the absence of salt in the buffer increases the stability of the SecYEG-Syd complex (Fig. 1C, bottom panel), suggesting that electrostatic forces mediate the interaction.
FIGURE 1.
Binding of Syd onto the SecYEG-Nanodisc. A, indicated amount of Syd was incubated with 3 μg of Nd-SecYEG in TSG buffer (5 min at room temperature). The samples were analyzed by native-PAGE followed by Coomassie Blue staining of the gel. The molecular weight markers (MWM) are ferritin (440 kDa), catalase (232 kDa), and bovine serum albumin (66/132 kDa). The Nd-SecYEG preparation contains some free membrane scaffold protein (MSP) dimers that migrate near the 66-kDa marker. B, 125I-Syd (∼25,000 cpm; 150 ng) was incubated with Nd-SecYEG, either wild-type or carrying the indicated deletion in the SecY subunit. Samples were analyzed by native-PAGE and phosphorimaging. C, 125I-Syd (∼250,000 cpm; 1.25 μg) was analyzed by sucrose density centrifugation with the SecYEG-Nanodiscs (∼35 μg), either wild-type (WT) or carrying the indicated deletion in the SecY subunit. The gradient (6–13%) was prepared in 50 mm Tris, pH 7.9, 5% glycerol, 1 mm DTT containing 100 mm NaCl, or no salt as indicated. The centrifugation was at 197,000 × g for 16 h at 4 °C in a Beckman SW41 rotor. Equal fractions were collected and analyzed by SDS-PAGE and phosphorimaging.
The oligomeric state of Syd and the stoichiometry between Syd and Nd-SecYEG was assessed by analytical gel filtration. In the presence of an excess of Syd, most of the Nd-SecYEG is complexed as judged by the shift and the symmetry of the elution peak during gel filtration chromatography (Fig. 2A). Multiangle light scattering analyses indicate that the purified Nd-SecYEG-Syd complex is monodisperse with a measured molecular mass of 152 kDa ±3%. On their own, the Nd-SecYEG particles and the Syd protein are also homogeneous but with a measured molecular mass of ∼126 and ∼23 kDa, respectively (Fig. 2B). Thus, the protein Syd is monomeric in solution and forms a stable 1:1 stoichiometry with the SecYEG complex.
FIGURE 2.
Isolation of the Syd-SecYEG-Nanodisc complex and mass determination. A, SecYEG-Nanodisc (∼200 μg) incubated with a molar excess of Syd (∼100 μg) and then applied onto a Superdex 200 HR10/10 column equilibrated in TSG buffer. The fractions containing the complex Nd-SecYEG-Syd were pooled and concentrated to 1.2 mg/ml for subsequent multiangle light scattering analysis. B, multiangle light scattering of Syd, Nd-SecYEG, and Nd-SecYEG-Syd. In each case, about 100 μg of protein is loaded onto a Superdex 200 HR 10/30 column equilibrated in 10 mm HEPES, 50 mm NaCl, pH 7.4.
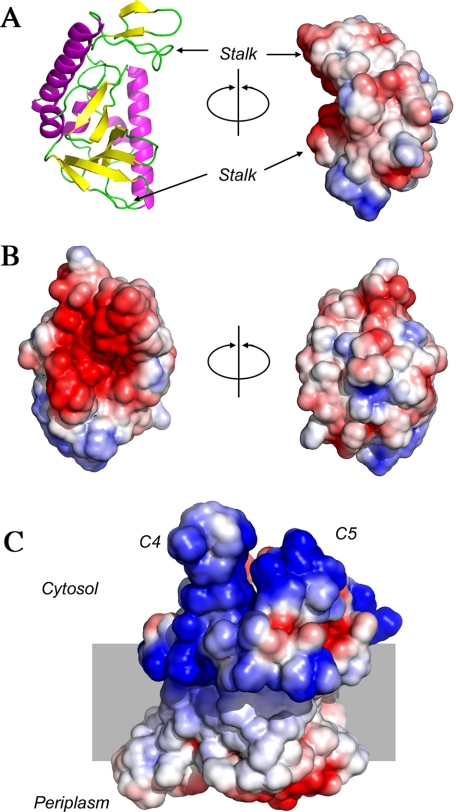
Atomic Structure of Syd—Syd was crystallized and its structure solved by single wavelength anomalous dispersion to 2.0 Å resolution (Fig. 3A). There are two molecules in the asymmetric unit, and their refined structures include all residues, except for the C-terminal 6-histidine tag used to purify the protein. The Syd structure is relatively compact and globular, being primarily comprised of a six-stranded antiparallel β-sheet and two long α-helices. A space-filling representation of Syd reveals that part of this β-sheet structure forms a concave cavity (or groove) on the protein surface. Sequence alignments show that the residues forming the concave surface of this groove are highly conserved (supplemental Fig. S1). The periphery of the observed groove is made of two protruding stalks located to the N terminus (residues 30–34; 45–48) and C terminus (residues 138–143) of the protein. Examination of surface electrostatics indicates that Syd has a distinct negatively charged patch localized on the concave groove surface and its surrounding stalks (Fig. 3B). A similar electrostatic analysis of the SecY complex shows that the two SecY loops involved in the binding of Syd are electropositive, because of several conserved basic residues in those regions (Fig. 3C). These observations strongly suggest that the negatively charged groove found in Syd forms the binding site for the SecY electropositive loops.
FIGURE 3.
Atomic structure and surface electrostatic of Syd and SecYEG. A, ribbon diagram representation of Syd (β-sheets, yellow; α-helices, magenta; loops, green) and space-filling representation showing the concave structure of the protein (180° rotation compared with right panel). Arrows denote the position of the “stalk” regions that consist of negatively charged protruding loops that delineate the concave region. B, electrostatic potential of the concave (left) and convex surface (right, 180° rotation). Blue and red represent electropositive and electronegative potential, respectively. The surface potential was set between -2.5 and +2.5 kT/e using the solvent-accessible area option of the software. C, electrostatic map of the archaeal SecY complex (Protein Data Bank code 1RH5) showing the position of the positively charged loops C4 and C5. The surface potential was generated using the parameters described in B. Positive charges are located near the predicted location of the phospholipids head group. Note that in E. coli, the loops C4 and C5 contain additional basic residues compared with Methanococcus jannaschii. In particular, three arginine residues are located at the tip of the loop C4 in the E. coli complex.
Interacting Surface between SecY and Syd and Exclusion of SecA— Single cysteine residues were introduced at different positions on the concave surface of Syd (Fig. 4A) and tested for reactivity with IMVs enriched for the SecY complex carrying the mutation M255A. The position SecY-255C is located at the tip of loop C4, and it is involved in the binding of SecA (23). None of the purified Syd variants formed cysteine-linked dimer in solution (data not shown) nor cross-reacted with the wild-type IMVs (Fig. 4B, top). In contrast, a strong SecY-Syd cysteine cross-link was detected when Syd-115C was incubated with the SecY-255C IMVs (Fig. 4B). This cross-link is specific and efficient because it appeared without the addition of oxidizing agent, and it engaged most of the SecY proteins in the corresponding titration experiment (Fig. 4B, bottom). Weaker cross-links were observed when the cysteine residue was located at other positions along the groove (90, 97, and 135, respectively; Fig. 4B, top), suggesting more distant or transient interactions with the position SecY-255.
FIGURE 4.
Contact surface between Syd and SecY and exclusion of SecA. A, location of the unique cysteine residues (90, 97, 115, and 135) introduced at the surface of Syd is indicated in yellow. B, purified Syd proteins (each 2 μg) were mixed with IMVs (5 μg) enriched for the SecY complex, either wild-type (WT) or carrying the cysteine mutation at position 255 (SecY255C) in 100 μl of TSG buffer (without DTT). After 5 min of incubation at room temperature, the disulfide bond formation was stopped with N-ethylmaleimide (NEM; 10 mm). Samples were analyzed by SDS-PAGE followed by immunostaining with a polyclonal antibody directed against SecY. The presence of N-ethylmaleimide during the incubation prevents the formation of the SecY-Syd cross-link (lower panel, left lane). C, IMVs SecY255C (5 μg) were incubated with a fixed amount of Syd-115C (2 μg) and a variable amount of SecA (0.0–5.0 μg) in 100 μl of TSG buffer without DTT. Samples were analyzed by SDS-PAGE followed by immunostaining with anti-SecY antibodies. D, about 3 μg of SecYEG-Nanodisc was incubated with a fixed amount of SecA (1 μg) and a variable amount of Syd (0.0–1.0 μg) in 50 μl of TSG buffer. After 5 min of incubation at room temperature, samples were analyzed by native-PAGE and Coomassie Blue staining of the gel.
Previous Scatchard analyses indicate that Syd does not interfere with the binding of SecA onto membrane-embedded SecYEG (14). We confirm here that SecA can displace Syd from the SecY complex. Indeed, the cysteine cross-link between Syd-115C and SecY-255C is almost completely abolished when the IMVs are incubated with an equimolar amount of SecA and Syd (Fig. 4C). Interestingly, when the SecYEG complex in Nanodiscs is incubated with an equimolar amount of Syd and SecA, most of the resulting complex consists of Syd only (Fig. 4D). Thus, Syd and SecA seem to possess different affinity for the SecY complex whether it is embedded in the membrane as an oligomer or integrated in Nanodiscs as a monomer. Further discussion will require knowledge of the Kd values, but the results already indicate that the binding of Syd and SecA is mutually exclusive. The conclusion is consistent with the overlap of the interaction site, along with the size consideration of SecA and Syd.
Effect of Syd on Translocation Efficiency and Stability of the SecY Complex—Search of genome data banks reveals that Syd exists only in Gram-negative bacteria and solely in those where SecE is comprised of three transmembrane segments (TMS). In general, SecE contains a single TMS except in about 30% of Gram- bacteria where two additional TMS are located at the N terminus of the protein. In E. coli, the two TMS are not essential for cell viability or translocation activity but seem important for the stability of the SecY complex (25, 26) (Fig. 5A). The SecE subunit is indeed critical for the biogenesis of SecY, and any unassembled SecY is rapidly eliminated by the Fts protease (27). The possible evolutive relationship between Syd and SecE prompted us to test the effect of Syd when the two TMS of SecE are deleted from the SecY complex (deletion SecEΔ7–67). As reported previously, in vitro protein translocation into wild-type IMVs is not appreciably affected by Syd (14) (Fig. 5B). In contrast, the concentration of Syd required to achieve half-inhibition of translocation appears ∼5 times lower for the mutant SecY complex carrying the deletion SecEΔ7–67 (Fig. 5B). A similar inhibition of translocation is also observed when the SecY complex carries the mutation G240D (or SecY24) (14). The mutation G240D is located at the interface between SecY and SecE, and it reduces the stability of the SecYE complex (10, 28). Together, the observations suggest a relationship between the activity of Syd and the strength of the SecYEG heterotrimeric associations. Syd seems to preferentially recognize mis-assembled SecYEG complexes. In the membrane, the two TMS of SecE stabilize the SecY complex and may also prevent the binding of Syd.
FIGURE 5.

Translocation activity of the SecY complex in the presence of Syd. A, SecY complex, wild-type (WT) or carrying the mutation secY24(G240D) or SecEΔ7-67, was overproduced in E. coli, and the IMVs were immunostained with anti-SecY antibodies. The SecY mutant complex cannot be overproduced to the same level as the wild type, probably due to a higher instability. B, IMVs were urea-stripped, and their concentration was adjusted so that a comparable amount of SecY complex is tested in the corresponding in vitro translocation assays. The IMVs were premixed with the indicated amount of purified Syd before addition of the translocation substrate 125I-proOmpA and SecA (0.25 μg). Translocation was initiated with ATP (1 mm) for 10 min at 37 °C. After proteinase K digestion (10 min on ice), samples were analyzed by SDS-PAGE and phosphorimaging. The results were quantified by densitometry and expressed on the graph curve (right panel).
We thus analyzed further the effect of Syd on the stability of the SecY complex in solution. The SecY complex in DDM migrates as a dimer during Blue-Native PAGE analysis; the ratio with monomers depending on the concentration of detergent (7). However, in the presence of Syd, the equilibrium is shifted, and most of the SecYEG dimers are dissociated in monomers (Fig. 6A). In other detergents such as Triton X-100, the SecY dimer is relatively labile and easily dissociates in single subunits (29). In the presence of Syd, the dissociation of the SecY complex is further enhanced (Fig. 6A). Thus, the SecYEG associations are weakened in detergent micelles, and Syd contributes to their further destabilization. These observations are consistent with a model in which Syd acts against a translocation channel in which the SecY-SecE interactions are mal-formed, unstable, or compromised (14).
FIGURE 6.
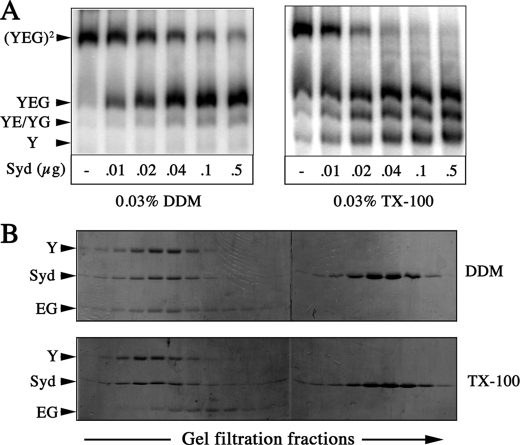
Stability of the SecY complex in the presence of Syd. A, 125I-SecY complex (∼75,000 cpm; 0.75 μg) was incubated with the indicated amount of purified Syd protein. The incubation was for 10 min at 22 °C in TSG buffer containing the indicated amount of detergent. Samples were analyzed by blue-native PAGE and phosphorimaging. B, detergent-soluble SecYEG complex (600 μg in 50 mm Tris, pH 7.9, 600 mm NaCl, 5% glycerol, 0.03% DDM) was mixed with a molar excess of Syd (450 μg in TSG buffer) and incubated for 5 min with 0.03% DDM at room temperature or with 0.1% Triton X-100 (TX-100) at 37 °C. The mixtures were applied onto a Superdex 200 HR10/30 gel filtration column equilibrated in 50 mm Tris, pH 7.9, 300 mm NaCl, 5% glycerol containing either 0.03% DDM or 0.1% Triton X-100 as indicated. The eluted fractions were analyzed by SDS-PAGE and Coomassie staining.
Although the effects observed are proportional to the concentration of Syd, we failed to detect the presence of this protein in the complexes revealed by Blue-Native PAGE (Fig. 6A), suggesting weaker interaction in detergent solution or the disruption of this interaction by the charged blue dye during the gel electrophoresis. To differentiate the two possibilities, the SecYEG complex was incubated with Syd in detergent solution and analyzed by size exclusion chromatography. In the DDM micelles, the SecYEG complex and Syd are co-eluted together in a seemingly stoichiometric ratio, whereas excess of Syd is eluted in the later fractions (Fig. 6B). In the Triton X-100 micelles, SecY and Syd are also co-eluted together, but the SecE/SecG proteins clearly appear in the later fractions (Fig. 6B). Thus, Syd and the SecY subunit alone have the capacity to form a stable complex, as seen in detergent solution.
Concluding Remarks—Syd is a monomeric and hydrophilic protein, with a conserved electronegative and concave surface. The simplicity of the protein makes its interaction with the SecY channel straightforward in comparison with SecA or the ribosome, both of which include multiple and complex functional domains. For all three partners however, the nature of all interactions with the SecY cytosolic loops may largely involve electrostatics. This is likely the case with Syd. In the case of the ribosome, cryo-electron microscopy studies indicate that the acidic phosphate backbone of the rRNA helices at the ribosome exit tunnel is establishing major contact points with the basic residues in the SecY loops (2, 3). In the case of SecA, the exact binding mode needs to be clarified, but one of the few electronegative patches at the surface of the protein would be prime candidate for the interaction.
The capacity of Syd to dissociate the dimeric and intramolecular SecYEG associations in detergent solution is remarkable. It is consistent with the Syd inhibitory activity against certain SecY mutant complexes presenting abnormal SecY-SecE associations (Refs. 12, 14 and this study). Together the results support a model in which Syd represents a quality control system for the correct assembly of the SecY channel. In the cell membrane, the mal-associated SecY complex would be preferentially recognized and further dissociated upon binding of Syd. The unassembled SecY subunit would then be targeted for degradation by the FtsH protease (27). The Syd protein possesses an unprecedented mode of action, contributing to an additional level of sophistication to the biogenesis and the function of the translocon.
Supplementary Material
The atomic coordinates and structure factors (code 3FFV) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by the Canadian Institutes of Health Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental figure, Table 1, and acknowledgments.
Footnotes
The abbreviations used are: DTT, dithiothreitol; DDM, dodecyl maltoside; IMVs, inner membrane vesicles; TMS, transmembrane segment.
References
- 1.Rapoport, T. A. (2007) Nature 450 663-669 [DOI] [PubMed] [Google Scholar]
- 2.Menetret, J. F., Hegde, R. S., Aguiar, M., Gygi, S. P., Park, E., Rapoport, T. A., and Akey, C. W. (2008) Structure (Lond.) 16 1126-1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menetret, J. F., Schaletzky, J., Clemons, W. M., Jr., Osborne, A. R., Skanland, S. S., Denison, C., Gygi, S. P., Kirkpatrick, D. S., Park, E., Ludtke, S. J., Rapoport, T. A., and Akey, C. W. (2007) Mol. Cell 28 1083-1092 [DOI] [PubMed] [Google Scholar]
- 4.Schiebel, E., Driessen, A. J., Hartl, F. U., and Wickner, W. (1991) Cell 64 927-939 [DOI] [PubMed] [Google Scholar]
- 5.Duong, F., and Wickner, W. (1997) EMBO J. 16 2756-2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuelson, J. C., Chen, M., Jiang, F., Moller, I., Wiedmann, M., Kuhn, A., Phillips, G. J., and Dalbey, R. E. (2000) Nature 406 637-641 [DOI] [PubMed] [Google Scholar]
- 7.Bessonneau, P., Besson, V., Collinson, I., and Duong, F. (2002) EMBO J. 21 995-1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cate, J. H., Yusupov, M. M., Yusupova, G. Z., Earnest, T. N., and Noller, H. F. (1999) Science 285 2095-2104 [DOI] [PubMed] [Google Scholar]
- 9.Clemons, W. M., Jr., May, J. L., Wimberly, B. T., McCutcheon, J. P., Capel, M. S., and Ramakrishnan, V. (1999) Nature 400 833-840 [DOI] [PubMed] [Google Scholar]
- 10.Van den Berg, B., Clemons, W. M., Jr., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C., and Rapoport, T. A. (2004) Nature 427 36-44 [DOI] [PubMed] [Google Scholar]
- 11.Papanikolau, Y., Papadovasilaki, M., Ravelli, R. B., McCarthy, A. A., Cusack, S., Economou, A., and Petratos, K. (2007) J. Mol. Biol. 366 1545-1557 [DOI] [PubMed] [Google Scholar]
- 12.Shimoike, T., Taura, T., Kihara, A., Yoshihisa, T., Akiyama, Y., Cannon, K., and Ito, K. (1995) J. Biol. Chem. 270 5519-5526 [DOI] [PubMed] [Google Scholar]
- 13.Matsuo, E., and Ito, K. (1998) Mol. Gen. Genet. 258 240-249 [DOI] [PubMed] [Google Scholar]
- 14.Matsuo, E., Mori, H., Shimoike, T., and Ito, K. (1998) J. Biol. Chem. 273 18835-18840 [DOI] [PubMed] [Google Scholar]
- 15.Collinson, I., Breyton, C., Duong, F., Tziatzios, C., Schubert, D., Or, E., Rapoport, T., and Kuhlbrandt, W. (2001) EMBO J. 20 2462-2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. Sect. D. Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 17.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. Sect. D. Biol. Crystallogr. 55 849-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 19.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D. Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 20.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D. Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 21.Alami, M., Dalal, K., Lelj-Garolla, B., Sligar, S. G., and Duong, F. (2007) EMBO J. 26 1995-2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denisov, I. G., Grinkova, Y. V., Lazarides, A. A., and Sligar, S. G. (2004) J. Am. Chem. Soc. 126 3477-3487 [DOI] [PubMed] [Google Scholar]
- 23.Osborne, A. R., and Rapoport, T. A. (2007) Cell 129 97-110 [DOI] [PubMed] [Google Scholar]
- 24.Karamanou, S., Bariami, V., Papanikou, E., Kalodimos, C. G., and Economou, A. (2008) Mol. Microbiol. 70 311-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schatz, P. J., Bieker, K. L., Ottemann, K. M., Silhavy, T. J., and Beckwith, J. (1991) EMBO J. 10 1749-1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishiyama, K., Suzuki, H., and Tokuda, H. (2000) Biosci. Biotechnol. Biochem. 64 2121-2127 [DOI] [PubMed] [Google Scholar]
- 27.Kihara, A., Akiyama, Y., and Ito, K. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 4532-4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taura, T., Akiyama, Y., and Ito, K. (1994) Mol. Gen. Genet. 243 261-269 [DOI] [PubMed] [Google Scholar]
- 29.Maillard, A. P., Lalani, S., Silva, F., Belin, D., and Duong, F. (2007) J. Biol. Chem. 282 1281-1287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.