Abstract
Histone methylation is associated with both transcription activation and repression. However, the functions of different states of methylation remain largely elusive. Here, using methyl-lysine analog technology, we demonstrate that the histone deacetylase complex, Rpd3S, can distinguish the nucleosomes methylated to different extents and that K36me2 is sufficient to target Rpd3S in vitro. Through a genome-wide survey, we identified a few mutants in which the level of K36me3 is significantly reduced, whereas the level of K36me2 is sustained. Transcription analysis and genome-wide histone modification studies on these mutants suggested that K36me2 is sufficient to target Rpd3S in vivo, thereby maintaining a functional Set2-Rpd3S pathway.
The histone N-terminal tails, as well as their globular domains, are subjected to various post-translational modifications, such as acetylation, methylation, phosphorylation, ubiquitination, etc. (1). It is commonly believed that histone modifications function in altering histone-DNA association and/or internucleosomal interactions or serve as a recognition platform for other chromatin-regulating proteins (2, 3). Histone methylation is a small modification that has no impact on the electrostatic charge of either lysine or arginine, and therefore, is unlikely to have a significant effect on nucleosome structure. This modification has, however, been shown to play an important role in signaling for a wide variety of physiological processes.
The functions of histone methylation were originally studied most extensively in transcriptional repression. Histone H3K9 methylation was first shown to specifically recruit heterochromatin protein 1 (HP1) via its chromodomain to silence gene expression (reviewed in Ref. 4). Later, it was discovered that H3K27me recruits the chromodomain-containing polycomb group complexes (5, 6), which are implicated in HOX gene silencing and X-inactivation (reviewed in Ref. 7). As for transcriptional activation, three major histone methylation marks, H3K4me, K36me, and K79me, are known to primarily associate with actively transcribed genes (1). Many protein domains within chromatin-associated complexes can selectively bind to H3K4 depending on its methylation status. The inhibitor of growth (ING) family of PHD domains can recognize H3K4me, which leads to the recruitment of histone deacetylase complexes, the NuA3 histone acetyltransferase complex, or chromatin-remodeling complexes (NuRD) (8, 9). By contrast, the PHD4 finger of BHC80 binds to unmethylated H3K4, and thereby, repels the demethylase LSD1 from its substrates, maintaining the level of H3K4 methylation (10). A more direct function of K4me in transcription was recently revealed by Sims et al. (11). The recognition of K4me3 by the chromodomain of CHD1 helps coordinate the RNA processing activity (11).
When compared with the diverse roles of K4me, the function of K36me appears relatively straightforward. Methylation at H3K36 is carried out by Set2, a conserved histone methyltransferase that specifically associates with elongating RNA polymerase II (reviewed in Ref. 12). K36me2 and K36me3 are highly enriched at the 3′ sides of the open reading frames (ORFs) and can be recognized by the chromodomain of Eaf3 with the help of the PHD domain of Rco1. Eaf3 and Rco1 are subunits of the Rpd3S histone deacetylase complex; they target this complex to the Lys-36 methylated coding regions of transcribed genes. The recruitment of Rpd3S is responsible for maintaining a hypoacetylated state at coding regions throughout the entire genome, which in turn suppresses cryptic transcription initiation from within the body of the genes (13-17). This pathway also represses meiotic recombination at certain hot spots in budding yeast (18).
Histone methylation not only occurs on different lysine residues but also takes place to different extents, namely at mono-, di-, and trimethylation. In some cases, all states of methylation are determined by a single modifying enzyme, such as yeast Set2 (H3K36), COMPASS (Set1) (H3K4), and Dot1 (H3K79); for others, multiple methyltransferases are required to reach different levels of methylation. For instance, in mouse and flies, HYPB, the yeast Set2 homologue, only controls H3K36 trimethylation, whereas Drosophila MES4 regulates the mono- and dimethylation of H3K36 (19, 20). More intriguingly, many histone demethylases also prefer substrates that are methylated to specific states. Yeast Rph1 is more active toward K36me2 and K36me3, whereas Jhd1 favors K36me2 and K36me1 (Klose et al. (33)). The fact that both modifying and demodifying enzymes are geared toward different methylation states underscores their importance. Moreover, the states of histone methylation are differentially regulated by other mechanisms as well. Histone H2B ubiquitination is only required for H3K4 di- and trimethylation but not for monomethylation (1). Recently, Youdell et al. (21) showed that Ctk1 and Spt6 are involved in regulating the level of Lys-36 trimethylation but not dimethylation when Set2 is overexpressed. Lastly, the genome-wide distribution pattern of different states of histone methylation is also very indicative of their potentially diverse functions. K36me2 and K36me3 are both enriched at coding regions. However, only the enrichment of K36me3 is proportional to the transcription frequency of the underlying genes, whereas the enrichment of K36me2 correlates with the overall on/off state of transcription (22, 23). As for K4me, the peak of K4me3 is located at the 5′ end of ORFs, whereas the enrichment of K4me2 and K4me1 is within the coding regions but more toward the 3′ end. Although all K4me states are found at the coding regions, only the enrichment of K4me3 displays a strong correlation with transcription potency (reviewed in Ref. 24). On the other hand, it was postulated that all three states of K79me contribute equally to its function, based on the distributive catalytic nature of Dot1, which methylates Lys-79, and evidence that the sum of all methylation states of Lys-79 correlate with silencing activity (25).
Despite much reasonable speculation regarding whether different methylation states of histones play distinct physiological roles, this question has not been carefully addressed experimentally. With our long term interests on the function of H3K36me, we set out to test the hypothesis above by asking whether the Rpd3S complex can distinguish nucleosomes that are methylated at lysine 36 to different extents. Using a recently developed methyl-lysine analogs (MLA) technology, we discovered that Rpd3S binds to H3K36 di- and trimethylated at a much higher affinity than monomethylated or unmodified nucleosomes in vitro. We then carried out a global proteomic analysis (GPS) screen to look for the mutants that only display defects on K36me3 but not K36me2. Gene expression analysis and genome-wide chromatin landscape surveys on those mutants demonstrate that K36me2 is indeed sufficient for recruiting Rpd3S in vivo and for maintaining a functional Set2-Rpd3S pathway.
EXPERIMENTAL PROCEDURES
MLA Histone Preparation, Nucleosome Manipulation, and Gel Shift Assays—Recombinant Xenopus histones (H3K36C, H4, H2A, and H2B) were individually expressed in BL21 codon plus-RIL (Stratagene) cells and purified as described (26). Methyl-lysine analog histones were prepared using the previous protocol (27), and histone octamers were isolated through a Superdex 200 gel filtration column. Mononucleosomes were then reconstituted using a 216-bp DNA probe containing 601 sequence via the serial dilution method (16), and the resulting nucleosomes were gel-purified prior to the electrophoretic mobility shift assay experiments as described (16). Short oligonucleosomes (SON) used in Fig. 1B were reconstituted through the octamer transfer method using extracted native HeLa nucleosomes and the 32P-labeled 216-bp 601-DNA probe (28).
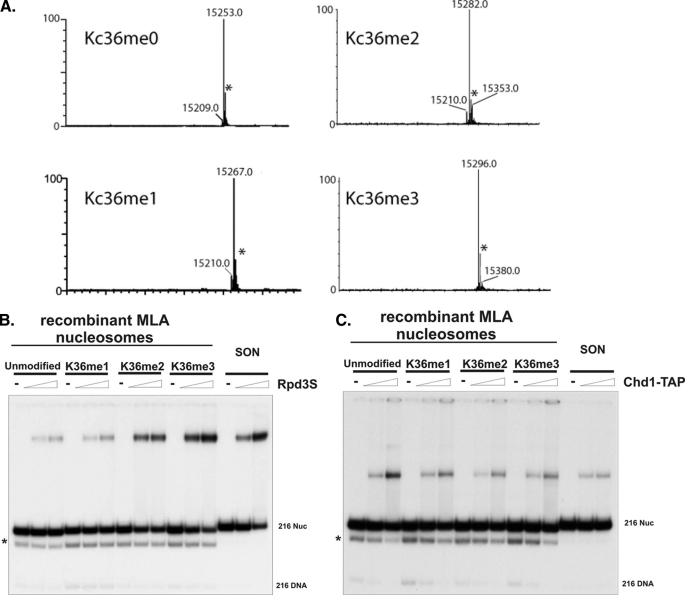
FIGURE 1.
Rpd3S preferentially binds to H3K36 di- and trimethylated nucleosomes. A, mass spectra (ESI-oaTOF) analysis demonstrates that the majority of end products are correct forms of Lys-36 methyl-lysine mimic histones. The y axis denotes relative intensity. The * denotes a mass spectrometry artifact peak at +41 that does not appear on other spectrometers and is present in both the starting material and the methylated histone products. B, Rpd3S preferentially binds to the di- and trimethylated nucleosomes. Recombinant MLA nucleosomes are reconstituted through the serial dilution method followed by gel purification. SON, short oligonucleosomes, represents the nucleosomes made through the octamer transfer method using extracted native HeLa nucleosomes (Nuc). C, Chd1-TAP served as a negative control that cannot distinguish nucleosomes methylated at Lys-36 or that are unmethylated. The * marks represent the partially dissociated nucleosome population.
Functional Genomics—The GPS screen was performed essentially as described in Ref. 29 with antibodies raised against dimethyl and trimethyl H3K36 peptides.
ChIP-on-Chip Assay and Data Analysis—Chromatin immunoprecipitation was carried out as described previously using anti-acetylated H4 antibody (Upstate Biotechnology) (26, 30). Approximately 10-30 ng of chromatin was immunoprecipitated, and whole cell extract DNA was amplified according to the round A/B/C random amplification of DNA protocol (26). Amplified DNA from round B was purified by a QIAquick mini-elute PCR purification kit (catalog number 28004) and quantified with the ND-1000 spectrophotometer. From this point on, all procedures were carried out in an ozone-free atmosphere to preserve the integrity of the dye coupling. 2.5 μg of the round B product was then labeled with the ULS array CGH labeling kit (BioMicro EA-005). Dye coupling efficiency was quantified with the ND-1000 and the DoL (degree of labeling or amount of dyes per 100 is normally between 0.5 and 2.5.). 2 μg of Cy3 labeled DNA and 2 μg of Cy5 labeled DNA were then hybridized to one array of an Agilent 4X44K yeast whole genome ChIP-on-chip array (catalog number G4493A). Arrays were hybridized at 20 RPM for 24 h at 65 °C and were washed according to the manufacturer's protocol (Version 9.2). Finally, arrays were scanned with the Agilent G2505B scanner with the extended dynamic range option (80 photomultiplier tube high and 20 photomultiplier tube low). Data were extracted with Agilent feature extraction version 9.5.3.1. All experiments were performed in duplicates with Pearson correlation exceeding 0.85. The average gene analysis was described in Ref. 16.
RESULTS
Rpd3S Recognizes Nucleosomes Methylated at Different States—We have demonstrated that the histone deacetylase complex Rpd3S preferentially binds to nucleosomes that are methylated at lysine 36 in a defined biochemical assay (16). However, because modification reactions were carried out enzymatically using recombinant Set2, the end products were primarily the mixture of di- and trimethylated nucleosomes. Therefore, it was difficult to utilize this method to obtain a homogenous population of nucleosomes that are only methylated to certain states. To overcome this problem, we took advantage of a recently developed technology in which recombinant histones are chemically modified to synthesize uniform and functional methyl-lysine analogs (27). Mass spectra analysis showed that the majority of the end products are the correct forms of Lys-36 methyl-lysine mimic histones (Fig. 1A). We then reconstituted these MLA histone octamers into mononucleosomes with a 216-bp DNA segment that contains a 601-nucleosome positioning sequence. The gel shift experiments were performed using a native Rpd3S complex that was purified through the tandem affinity purification (TAP) approach from yeast. Indeed, we found that Rpd3S robustly binds to both K36me2 and K36me3 nucleosomes, with K36me3 having a slightly higher affinity (Fig. 1B). In contrast, monomethylated and unmodified nucleosomes only display background levels of binding. To demonstrate the specificity of interactions between MLA nucleosomes and Rpd3S, we also showed that, unlike Rpd3S, Chd1-TAP could not discriminate differently modified K36me nucleosomes (Fig. 1C). This result demonstrates that K36me2 is sufficient to recruit Rpd3S in vitro, whereas K36me1 is not adequate.
Identification of Genes That Specifically Regulate H3K36 Trimethylation—Based on our biochemical evidence above, we speculated that K36me2 might be sufficient to target Rpd3S. However, to test this hypothesis in vivo, we needed mutants that show defects in H3K36me3 but not in K36me2. To this end, we systematically surveyed the entire non-essential gene knock-out collection using antibodies against H3K36me2 and H3K36me3. Our initial screen scored a few potential candidates, such as Paf1, a component of a large complex that participates in transcription elongation (Fig. 2), and Lge1, a protein associated with Bre1 ubiquitin-protein isopeptide ligase (E3) ligase for H2B ubiquitination (GPS data not shown). To further confirm these candidates, we performed a comprehensive titration analysis. As shown in Fig. 3A, we found that in the Δpaf1 mutant, K36me3 was completely eliminated, but a significant amount of K36me2 remained. We also observed a clear reduction in the levels of K36me3 in the Δlge1 mutant, whereas K36me2 remains within the normal levels (Fig. 3A). Surprisingly, we did not observe a similar reduction of K36me3 in the Δrad6 and Δbre1 mutant, in which H2B ubiquitination defects are supposed to be more pronounced (1). This suggests that H2B ubiquitination may not directly relate to the regulation of K36me3 and that Lge1 may have an independent role in this process. (Fig. 3B).
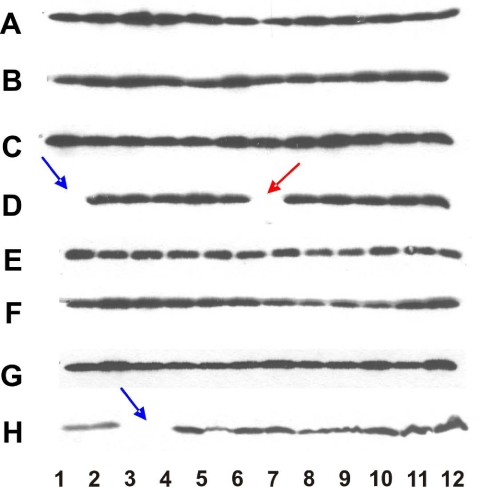
FIGURE 2.
Identification of Paf1 gene product to be required for histone H3 lysine 36 trimethylation by GPS. Yeast whole cell extracts were made from each of the non-essential gene deletion mutants of the Saccharomyces cerevisiae genome and were resolved on SDS-PAGE. The histone H3K36me3 antibody was used to carry out the GPS screen. Blue arrows indicate empty wells as place markers. The red arrow indicates the position of the PAF1 deletion mutant at plate location D7.
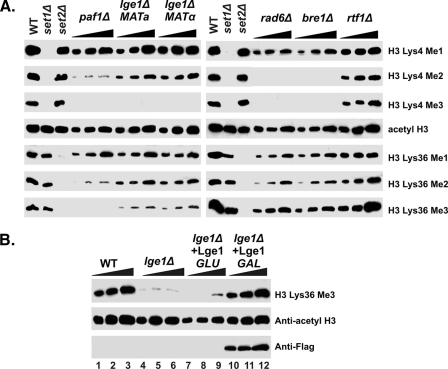
FIGURE 3.
Multiple genes affect the states of methylation at lysine 36 of histone H3. A, roles of the Rad6-Bre1 and Paf1 complexes (A) in H3 Lys-36 trimethylation. Cell extracts were obtained from several components of each complex and subjected to SDS-PAGE followed by Western blotting using antibodies specifically against different methylation states of histone H3. Anti-acetyl H3 was also applied as a load control. WT, wild type. B, Lge1 is required for proper H3 Lys-36 trimethylation. Lys-36 trimethylation was restored after transforming of a plasmid carrying LGE1, which contains a FLAG tag at their C terminus and is under the control of the GAL1 promoter, to strains that are lacking Lge1. Cell extracts were subjected to SDS-PAGE and Western blotting using the indicated antibodies. Anti-acetyl H3 was used as a loading control, and anti-FLAG was used to detect expression of targeted proteins.
To rule out the possibility of secondary mutations spontaneously arising in our initial mutants, we examined the mutants in a complementation assay. We transformed a plasmid carrying the LGE1 gene under the control of the GAL1 promoter into Δlge1. Indeed, when the level of Lge1 is restored in the mutant under the inducible condition (Fig. 3B), K36me3 is recovered accordingly.
At this point, we do not know whether these proteins play any direct roles in regulating the level of K36me3. However, given that these mutations result in specific defects on K36me3 but not on K36me2, we decided to use these reagents to test whether K36me2 can function alone in the absence of K36me3 in the Set2-Rpd3S pathway.
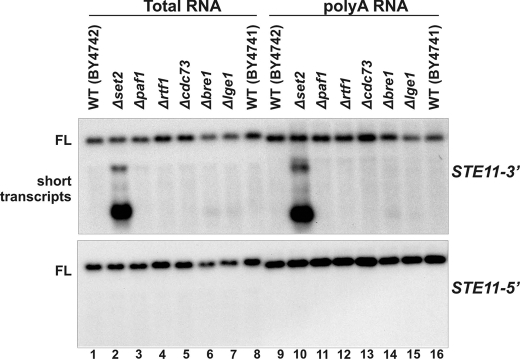
H3K36me2 Is Sufficient for Maintaining a Functional Set2-Rpd3S Pathway—Our previous studies have demonstrated that defects in the Set2-Rpd3S pathway inevitably lead to the generation of spurious transcripts at the STE11 gene (13). Thus, we performed Northern blot analysis using 3′ and 5′ probes of STE11 on a collection of mutants discovered above. As shown in Fig. 4A, unlike Δset2, all of the PAF complex mutants, Δbre1 and Δlge1, do not display any cryptic transcription phenotype. This result indicates that K36me2 within those mutants is sufficient to repress unwanted transcription at these genes.
FIGURE 4.
Dimethylation of H3K36 in various mutants is sufficient for normal suppression of cryptic promoters at the STE11 gene. Yeast strains grown exponentially were subjected to Northern blot analysis using probes against STE11 (5′ ORF or 3′ ORF). FL denotes full-length transcript; short transcripts refer to transcripts initiated from cryptic promoters. WT, wild type.
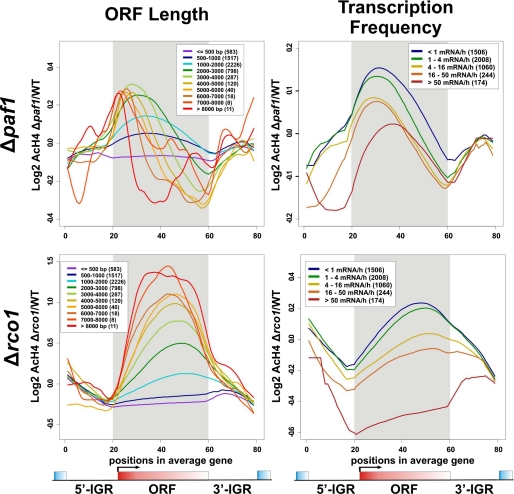
To further evaluate the effect of K36me2 on a genome-wide scale, we sought to understand whether K36me2 alone, as in Δpaf1, could maintain a hypoacetylated state at the coding regions. We have previously shown that deletion of SET2 or RCO1 results in a genome-wide increase of acetylated H4 (AcH4) at coding regions, which peaks toward the 3′ end of the ORFs (17). This change is likely due to the loss of Lys-36 methylation because the profile of distribution changes resembles that of K36me3 in the wild type. With this in mind, we decided to examine AcH4 distribution in the Δpaf1 mutant using ChIP-chip technology with Agilent high resolution yeast tiling arrays. When the genome-wide average of distribution changes was plotted, we noticed that the deletion of PAF1 causes an apparent AcH4 surge (data not shown). However, this increase seems to locate more toward the 5′ end of the ORF relative to that seen in Δrco1 (data not shown). To further dissect this trend, we divided the entire genome into multiple subgroups based on their ORF length (Fig. 5, left side) or transcription frequency (Fig. 5, right side) and then plotted the average values of each group against their relative position to the ORFs (Fig. 5). Although the overall changes as illustrated in Fig. 5 are similar to what we have seen in the genome-wide average pattern, we noticed that in the Δpaf1 mutant, particularly for long genes, AcH4 increases primarily at the promoter proximal regions (Fig. 5, upper left). This change is dramatically different from that in the Set2-Rpd3S mutant (Δrco1) where acetylation increases largely at the main body of the ORFs (Fig. 5, lower left). We reasoned that perhaps the remaining K36me2 in Δpaf1 is sufficient for recruiting Rpd3S to the coding regions and maintaining a functional Set2-Rpd3S pathway, which governs the level of acetylation at regions that are away from the promoters. This conclusion is also consistent with our earlier results (Fig. 4) showing that no cryptic transcript phenotype is observed in Δpaf1. Although at the moment we do not understand the molecular mechanism by which the deletion of PAF1 leads to acetylation increases at promoter proximal areas, we speculate that drastically altered K4 methylation and ubiquitination at the same location in the Δpaf1 mutant contribute.
FIGURE 5.
Deletion of PAF1 does not lead to genome-wide increase of histone H4 acetylation level at the 3′ end of coding region, a typical phenotype seen in the Set2-Rpd3S defective mutants. ChIP-chip was performed using Agilent high resolution tiling arrays. The log2 ratio of acetylation of H4 (AcH4) in mutants over AcH4 in wild type was subjected to a modified average gene analysis (17). All genes were divided into multiple subclasses based on either their ORF length or their transcription frequency in complete mediums. The averages of each subclass were plotted, with the number of genes in each class being indicated within the parentheses in the white box.
DISCUSSION
Many lysine residues of histones are methylated to different states. An obvious assumption in the field is that these different states of methylation may somehow play distinct roles in regulating chromatin dynamics. However, so far, very few studies have been reported to directly test this hypothesis, particularly in the context of chromatin. In this study, we set out to measure the role of H3K36 mono-, di-, and trimethylation in regulating the association of the native Rpd3S histone deacetylase complex with nucleosomes. We found that both di-Lys-36 methylated and tri-Lys-36 methylated nucleosomes strongly bind to Rpd3S, with K36me3 having the highest affinity. By contrast, we found that the affinity of monomethylated nucleosomes to Rpd3S was as low as that of the unmodified ones (Fig. 1B), implying that the gap between mono- and dimethylation may be critical for Rpd3S binding and, consequently, the Set2-Rpd3S pathway. Thus, it appears that K36me2 is sufficient to recruit Rpd3S in vitro. To test whether this is also true in vivo, we needed mutants that contained K36me2 but lacked K36me3. A K36me2 and K36me3 GPS screen was subsequently carried out, aiming to search for such specific mutants. This screen led us to two potential candidates: PAF1 and LGE1. In an independent study, Chu et al. (31) also discovered that the deletion of multiple subunits of the PAF1 complex individually resulted in specific defects on K36me3 but not K36me2. We then tested the cryptic transcription phenotype and global acetylation status in the Δpaf1 mutant. It turned out that the remaining level of K36me2 in Δpaf1 is sufficient for a functional Set2-Rpd3S pathway. This conclusion is in good agreement with another study in which they demonstrated that K36me2 is capable of directing histone deacetylation and repressing spurious transcript using a different genetic system (21).
Our study suggests that K36me2 and K36me3 may play redundant roles in recruiting Rpd3S, whereas K36me1 fails to do so. This notion echoed a recent finding that di- and tri-, but not monomethylation, on H3K36 marks actively transcribed genes in Arabidopsis thaliana (32). It is possible that K36me1 may not make enough contact with the chromodomain of Eaf3 to sustain a strong interaction. Detailed structural studies in the future might shed light on this aspect.
Although we suggest here that K36me2 and K36me3 have some redundant functions, this does not necessarily mean that their true biological roles in cells are identical. In fact, only K36me3 correlates with transcription frequency, whereas K36me2 is only linked to the on/off states of transcription. So perhaps the stronger interaction between K36me3 and Rpd3S (Fig. 1B) may be more advantageous to recruit Rpd3S at highly transcribed genes where nucleosome density is low. In conclusion, we believe that, as far as K36me is concerned, different states of methylation potentially play very different biological roles. This observation is quite different from the proposed function of H3K79 methylation in silencing. Frederiks et al. (25) recently proposed that all three states of H3K79 methylation might function in silencing. This conclusion was primarily based on genetic evidence showing that the total amount of Lys-79 methylation is proportional to its silencing activity. The rationale is that K79me may serve as a signal to repel some chromatin binding factors (the SIR complex for instance) and that Dot1 is a non-processive enzyme; thus, any methyl state may function in silencing. Thus, the methylation of states of particular lysines may differ in their functional readout in gene regulation pathways.
Acknowledgments
We thank members of the Workman laboratory and Shilatifard laboratory for useful discussions and technical suggestions and Laura Shilatifard for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM047867 through the NIGMS (to J. L. W.) and National Institutes of Health Grant 2R01GM069905 (to A. S.). This work was also supported by funding from the Stowers Institute for Medical Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PHD, plant homeodomain; ORF, open reading frame; MLA, methyl-lysine analogs; ChIP, chromatin immunoprecipitation; TAP, tandem affinity purification; GPS, global proteomic analysis; AcH4, acetylated H4.
References
- 1.Shilatifard, A. (2006) Annu. Rev. Biochem. 75 243-269 [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein, T., and Allis, C. D. (2001) Science 293 1074-1080 [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides, T. (2007) Cell 128 693-705 [DOI] [PubMed] [Google Scholar]
- 4.Hediger, F., and Gasser, S. M. (2006) Curr. Opin. Genet. Dev. 16 143-150 [DOI] [PubMed] [Google Scholar]
- 5.Fischle, W., Wang, Y., Jacobs, S. A., Kim, Y., Allis, C. D., and Khorasanizadeh, S. (2003) Genes Dev. 17 1870-1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Min, J., Zhang, Y., and Xu, R. M. (2003) Genes Dev. 17 1823-1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Royce-Tolland, M., and Panning, B. (2008) Curr. Biol. 18 R255-256 [DOI] [PubMed] [Google Scholar]
- 8.Zhang, Y. (2006) Nat. Struct. Mol. Biol. 13 572-574 [DOI] [PubMed] [Google Scholar]
- 9.Martin, D. G., Baetz, K., Shi, X., Walter, K. L., Macdonald, V. E., Wlodarski, M. J., Gozani, O., Hieter, P., and Howe, L. (2006) Mol. Cell Biol. 26 7871-7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lan, F., Collins, R. E., De Cegli, R., Alpatov, R., Horton, J. R., Shi, X., Gozani, O., Cheng, X., and Shi, Y. (2007) Nature 448 718-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sims, R. J., III, Millhouse, S., Chen, C. F., Lewis, B. A., Erdjument-Bromage, H., Tempst, P., Manley, J. L., and Reinberg, D. (2007) Mol. Cell 28 665-676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, B., Carey, M., and Workman, J. L. (2007) Cell 128 707-719 [DOI] [PubMed] [Google Scholar]
- 13.Carrozza, M. J., Li, B., Florens, L., Suganuma, T., Swanson, S. K., Lee, K. K., Shia, W. J., Anderson, S., Yates, J., Washburn, M. P., and Workman, J. L. (2005) Cell 123 581-592 [DOI] [PubMed] [Google Scholar]
- 14.Joshi, A. A., and Struhl, K. (2005) Mol. Cell 20 971-978 [DOI] [PubMed] [Google Scholar]
- 15.Keogh, M. C., Kurdistani, S. K., Morris, S. A., Ahn, S. H., Podolny, V., Collins, S. R., Schuldiner, M., Chin, K., Punna, T., Thompson, N. J., Boone, C., Emili, A., Weissman, J. S., Hughes, T. R., Strahl, B. D., Grunstein, M., Greenblatt, J. F., Buratowski, S., and Krogan, N. J. (2005) Cell 123 593-605 [DOI] [PubMed] [Google Scholar]
- 16.Li, B., Gogol, M., Carey, M., Lee, D., Seidel, C., and Workman, J. L. (2007) Science 316 1050-1054 [DOI] [PubMed] [Google Scholar]
- 17.Li, B., Gogol, M., Carey, M., Pattenden, S. G., Seidel, C., and Workman, J. L. (2007) Genes Dev. 21 1422-1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merker, J. D., Dominska, M., Greenwell, P. W., Rinella, E., Bouck, D. C., Shibata, Y., Strahl, B. D., Mieczkowski, P., and Petes, T. D. (2008) DNA Repair (Amst.) 7 1298-1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell, O., Wirbelauer, C., Hild, M., Scharf, A. N., Schwaiger, M., MacAlpine, D. M., Zilbermann, F., van Leeuwen, F., Bell, S. P., Imhof, A., Garza, D., Peters, A. H., and Schubeler, D. (2007) EMBO J. 26 4974-4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmunds, J. W., Mahadevan, L. C., and Clayton, A. L. (2008) EMBO J. 27 406-420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Youdell, M. L., Kizer, K. O., Kisseleva-Romanova, E., Fuchs, S. M., Duro, E., Strahl, B. D., and Mellor, J. (2008) Mol. Cell Biol. 28 4915-4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao, B., Shibata, Y., Strahl, B. D., and Lieb, J. D. (2005) Mol. Cell Biol. 25 9447-9459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pokholok, D. K., Harbison, C. T., Levine, S., Cole, M., Hannett, N. M., Lee, T. I., Bell, G. W., Walker, K., Rolfe, P. A., Herbolsheimer, E., Zeitlinger, J., Lewitter, F., Gifford, D. K., and Young, R. A. (2005) Cell 122 517-527 [DOI] [PubMed] [Google Scholar]
- 24.Shilatifard, A. (2008) Curr. Opin. Cell Biol. 20 341-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frederiks, F., Tzouros, M., Oudgenoeg, G., van Welsem, T., Fornerod, M., Krijgsveld, J., and van Leeuwen, F. (2008) Nat. Struct. Mol. Biol. 15 550-557 [DOI] [PubMed] [Google Scholar]
- 26.Li, B., Pattenden, S. G., Lee, D., Gutierrez, J., Chen, J., Seidel, C., Gerton, J., and Workman, J. L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18385-18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon, M. D., Chu, F., Racki, L. R., de la Cruz, C. C., Burlingame, A. L., Panning, B., Narlikar, G. J., and Shokat, K. M. (2007) Cell 128 1003-1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steger, D. J., Owen-Hughes, T., John, S., and Workman, J. L. (1997) Methods (Amst.) 12 276-285 [DOI] [PubMed] [Google Scholar]
- 29.Schneider, J., Dover, J., Johnston, M., and Shilatifard, A. (2004) Methods Enzymol. 377 227-234 [DOI] [PubMed] [Google Scholar]
- 30.Li, B., and Reese, J. C. (2001) J. Biol. Chem. 276 33788-33797 [DOI] [PubMed] [Google Scholar]
- 31.Chu, Y., Simic, R., Warner, M. H., Arndt, K. M., and Prelich, G. (2007) EMBO J. 26 4646-4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu, L., Zhao, Z., Dong, A., Soubigou-Taconnat, L., Renou, J. P., Steinmetz, A., and Shen, W. H. (2008) Mol. Cell Biol. 28 1348-1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klose, R. J., Yamane, K., Bae, Y., Zhang, D., Erdjument-Bromage, H., Tempst, P., Wong, J., and Zhang, Y. (2006) Nature 442 312-316 [DOI] [PubMed] [Google Scholar]