Abstract
The catalytic mechanism of Propionibacterium acnes polyunsaturated fatty acid isomerase (PAI) is explored by kinetic, spectroscopic, and thermodynamic studies. The PAI-catalyzed double bond isomerization takes place by selective removal of the pro-R hydrogen from C-11 followed by suprafacial transfer of this hydrogen to C-9 as shown by conversion of C-9-deuterated substrate isotopologs. Data on the midpoint potential, photoreduction, and cofactor replacement suggest PAI to operate via an ionic mechanism with the formation of FADH2 and linoleic acid carbocation as intermediates. In line with this proposal, no radical intermediates were detected neither by stopped flow absorption nor by EPR spectroscopy. The substrate preference toward free fatty acids is determined by the interaction between Arg-88 and Phe-193, and the reaction rate is strongly affected by replacement of these amino acids, indicating that the efficiency of the hydrogen transfer relies on a fixed distance between the free carboxyl group and the N-5 atom of FAD. Combining data obtained for PAI from the structural studies and experiments described here suggests that at least two different prototypical active site geometries exist among polyunsaturated fatty acid double bond isomerases.
Conjugated linoleic acid (CLA)3 refers to the isomers of linoleic acid (LA, 18:2Δ9Z,12Z; x:yΔz denotes a fatty acid with x carbons and y double bonds in position z counting from the carboxyl end) with two conjugated double bonds in various geometrical configurations. It originates from metabolism of anaerobic ruminal bacteria (1-3). Chemically, isomerization of LA to CLA is overall a non-redox process and does not involve oxygen, implying an unique mechanism for all polyunsaturated fatty acid (PUFA) double bond isomerases (4). The first of such isomerases was characterized from Butiryvibrio fibrisolvens by Tove and co-workers (1, 2), who demonstrated that this enzyme prefers C-18 fatty acids and produces (9Z,11E)-CLA. Here the abstraction of a hydrogen atom occurs at position C-11 of LA during catalysis, and one solvent-derived proton is found at position C-13 in the product (2). A mechanistically related eukaryotic PUFA isomerase from marine algae Ptilota filicina has been also described (5, 6), but the identity of the product derived from LA has not yet been reported, because this enzyme prefers C-20 PUFA.
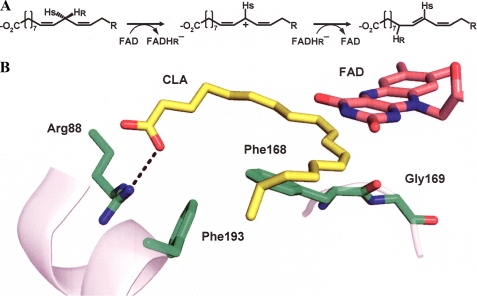
Recently, we reported the crystal structure of a FAD-dependent PUFA isomerase from Propionibacterium acnes (PAI) in complex with (10E,12Z)-CLA (7) and proposed a viable reaction mechanism involving the transient oxidation of LA by FAD (Fig. 1A). Thus, PAI belongs to a functionally diverse group of flavoenzymes catalyzing reactions with no net redox change (8). Similar to other PUFA isomerases described so far, PAI prefers free fatty acids as substrates (9), which can be rationalized by the crystal structure (7, 10). The carboxylate group of the fatty acid is hydrogen bonded to Arg-88, which itself interacts via π-stacking with Phe-193 (Fig. 1B). This Arg/Phe lock ensures selectivity for fatty acids, and the interaction with unpolar and bulky head groups such as fatty acid methyl esters, triacylglycerols, or phospholipids is sterically and electrostatically unfavorable. Moreover, the structural model of PAI reaction suggested that the direction of the allylic shift during LA isomerization is governed by the position of Phe-168 relative to FAD (Fig. 1B). However, for biotechnological purposes, a modified PAI accepting fatty acid esters as substrates would be preferable because these are much more readily accessible than the free fatty acids.
FIGURE 1.
Scheme of PAI reaction. A, presumable stereochemistry of hydrogen transfer and redox transitions during the reaction. B, drawing of PAI active site based on Ref. 9. Key residues involved in substrate binding and catalysis are shown. FAD is represented as gray aromatic rings behind the plane of LA.
In the present work we describe the stereochemical analysis of hydrogen transfer during LA isomerization and the redox properties of free PAI and the enzyme-product complex. The roles of Arg-88, Phe-193, and Phe-168 for substrate recognition and catalysis are demonstrated by site-directed mutagenesis and analysis of kinetic properties of the mutant enzymes. Exchange of Arg-88 and Phe-193 to less polar/smaller residues did not increase activity of PAI toward complex lipids, indicating that the geometry of the fatty acid binding channel is crucial to optimal substrate positioning and selectivity. Furthermore, the potential occurrence of transient radical intermediates has been tested by rapid scan electronic absorption spectroscopy and EPR spectroscopy.
EXPERIMENTAL PROCEDURES
Chemicals—Restriction enzymes and DNA-modifying enzymes were obtained from MBI Fermentas (St. Leon-Rot, Germany). Standards of fatty acids as well as all other chemicals were purchased from Sigma; methanol, hexane, and 2-propanol (all HPLC grade) were from Baker.
PCR-based Mutagenesis—The cloning of PAI open reading frame into bacterial expression vector has been described elsewhere (9). The plasmid pGEX-6P-1-PAI was used as a template for PCR-based mutagenesis with QuikChange® site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The following oligonucleotides were used to introduce point mutations: R88A_NruI_a, 5′-cgacgggccgaaactgcgtagcgagttcctgcacgagg-3′; R88A_NruI_b, 5′-cctcgtgcaggaactcgctacgcagtttcggcccgtcg-3′; R88S_NruI_a, 5′-cgacgggccgaaactgcgtagcgagttcctgcacgagg-3′; R88S_NruI_b, 5′-cctcgtgcaggaactcgctacgcagtttcggcccgtcg-3′; F193A_BgIII_a, 5′-cttcgtcaccatgatgtccgcggccaagggagacctg-3′; F193A_BgIII_b, 5′-caggtctcccttggccgcggacatcatggtgacgaag-3′; F168L_Eco47III_a, 5′-ggatcaaccccttcacagcgctcggctacgggcacttcgac-3′; F168L_Eco47III_b, 5′-gtcgaagtgcccgtagccgagcgctgtgaaggggttgatcc-3′; G168F169_PstI_a, 5′-tggatcaaccccttcactgcaggcttctacgggcacttcgacaacg-3′; and G168F169_PstI_b, 5′-cgttgtcgaagtgcccgtagaagcctgcagtgaaggggttgatcca-3′. Amplification program consisted of 2 min of denaturation at 94 °C, followed by 18 cycles of 30 s at 94 °C, 30 s at 60 °C, and 6.5 min at 72 °C.
Protein Purification—PAI wild type protein and variants were overproduced in Escherichia coli and purified as described in Ref. 7.
Labeled Substrate Synthesis—[11,11-2H2]LA was prepared by organic synthesis using [1,1-2H2]1-bromo-2-octyne as the labeled synthon. The chemical purity according to GC-MS was more than 98%, and the isotope composition was 94.3% dideuterated, 5.1% monodeuterated, and 0.6% undeuterated molecules. [(11S)-2H]LA was prepared by enzymatic desaturation of [(11S)-2H]stearic acid using methodology described in detail (11): chemical purity, >98%; isotope enrichment, >98.0% monodeuterated molecules; stereochemical purity of deuterium label, >90%.
[(9S)-2H]Stearic acid was prepared from a sample of (9S)-hydroxy LA (40 mg) by a sequence involving methyl esterification (diazomethane), catalytic hydrogenation (platinum catalyst; solvent, methanol), conversion to the p-toluenesulfonate ester (p-toluenesulfonyl chloride in pyridine), lithium aluminum deuteride reduction, and oxidation (Jones reagent; solvent, acetone). The product was purified by reverse phase HPLC to afford 11 mg of the labeled stearic acid. Its isotope composition was 98.9% monodeuterated and 1.1% undeuterated molecules as determined by GC-MS analysis of the methyl ester derivative.
Another sample of deuterated stearic acid was prepared from CLA (4 mg) produced by action of PAI on [11,11-2H2]LA. A solution of labeled CLA in toluene (1.5 ml) containing Wilkinson's catalyst (4 mg) was stirred at 57 °C for 1 h under an atmosphere of hydrogen gas. Following purification by reverse phase HPLC, 1.2 mg of deuterated stearic acid was obtained having an isotopic composition of 61.1% dideuterated and 35.6% monodeuterated, and 3.3% undeuterated molecules.
Activity Assays and Analysis of Reaction Products—Typically, 10 μg of substrate was incubated with 1 μg of purified PAI protein in 100 mm Tris, pH 7.5 (final volume, 1 ml). The products were extracted according to Ref. 12, and the fatty acids were dissolved in methanol and converted to methyl esters with (trimethylsilyl)-diazomethane. Esterified fatty acids were transmethylated with sodium methoxide in methanol as described (13). To trace the fate of isotopic label, fatty acids were converted to 4,4-dimethyloxazoline derivatives as described (14) and analyzed mass spectrometrically according to (14), using the 6890 gas chromatograph/5973 mass selective detector system (Agilent). Otherwise, GC and GC-MS analysis was performed as in Ref. 9. The relative percentage of LA methyl ester isomerization by PAI mutants compared with wt enzyme was estimated by GC based on the ratio of CLA to LA peak areas for each protein.
Kinetic Assays—All of the assays were performed in 100 mm Tris/HCl, pH 7.5, at 25 °C. FA stock solutions were prepared in ethanol in the concentration range of 0.1-50 mm. For every measurement 5 μl of FA stock solution was mixed with buffer, 0.5-4 μg of purified wt or mutant PAI protein added, and absorbance of the solution at 234 nm was recorded with an Amersham Biosciences Ultrospec 1100 pro spectrophotometer for 1 min. The final reaction volume was 500 μl, with the ethanol concentration thus not exceeding 1%, and the molar excess of the substrate over the enzyme ranged from 50- to 500-fold. The concentration of CLA was calculated using ε234 = 24000 m-1 cm-1. Three independent measurements per data point were averaged, and a Michaelis-Menten equation was fit using Origin 6.1 software.
Cofactor Exchange—5-Deaza-5-carba-FAD was a generous gift of Prof. S. Ghisla (University of Konstanz, Konstanz, Germany). 200 μg of purified PAI was diluted to 50 μg/ml in 6 m guanidium chloride, 100 mm Tris/HCl, pH 7.5, 10 mm dithiothreitol. This solution was passed through a 25-ml G25 desalting column (GE Healthcare) equilibrated with 100 mm Tris/HCl, pH 7.5, 1 mm dithiothreitol to remove FAD and refold PAI. The protein solution was concentrated in Vivaspin concentrators (molecular mass cut-off = 30 kDa), and 5× molar excess of 5-deaza-FAD was added; protein diluted 10-fold with 100 mm Tris/HCl, pH 7.5, and concentrated again. Washing step was repeated twice; final protein concentration was about 1 mg/ml or 20 μm. With this preparation UV-visible spectra were recorded and activity tests performed.
Incubations with Tetrahymena pyriformis—The sodium salt of the labeled stearates (0.5 mg) was added to 20 ml of growth medium followed by 0.5 ml of a suspension of an actively growing culture of T. pyriformis phenoset 30039 (purchased from American Type Culture Collection) (11). Cells were collected by centrifugation after a growth period of 48 h at 25 °C. The fatty acid fraction obtained following alkaline hydrolysis was methyl-esterified and analyzed by GC-MS.
Anaerobic Photoreduction and Redox Potential Measurement—For photoreduction and redox potential determination, the method described by Massey and Hemmerich (16) was used. The buffer in all of the experiments was 100 mm Tris, pH 7.0, 10 mm EDTA, and protein concentration was 30-40 μm. For the photoreduction of free PAI and PAI in complex with (10E,12Z)-CLA, 2 μm 5-deaza-FAD and 1 μm of the following redox dyes were added: methyl viologen (E = -430 mV), 2-hydroxy-1,4-napthaquinone (E = -145 mV), and phenazine methosulfate (E = 80 mV). The sample was transferred to the cuvette and rendered anaerobic by 20-30 cycles of vacuum application and flushing with oxygen-free nitrogen. The cuvette was illuminated in the ice bath for 2-min intervals using a 200 W slide projector lamp and allowed to equilibrate at 20 °C until no changes in the UV-visible spectrum were observed, after which the final spectrum was recorded. For redox potential determination the reaction was prepared as above except that phenazine methosulfate and 2-hydroxy-1,4-napthoquinone were omitted and 30 μm anthraquinone-2-sulfonate (E = -225 mV), 2-hydroxy-1,4-napthaquinone or safranine O (Em = -290 mV) was used as the reference dye. For the PAI- (10E,12Z)-CLA complex, an equimolar amount of the substrate LA was included in the reaction. LA was quantitatively converted to (10E,12Z)-CLA during the course of the measurements as confirmed by GC-MS (data not shown).
Rapid Scan Stopped Flow UV-visible Absorption Measurements—The time-resolved absorbance spectra of PAI during turnover of its substrate LA were recorded with a SX20 stopped flow spectrophotometer equipped with diode array acquisition (Applied Photophysics Ltd.). PAI (1.6 mg/ml in 0.1 m Tris/HCl, pH 7.5) was mixed with varied concentrations of LA (same buffer) ranging from 50 to 250 μm in 1 + 1 mixing ratio at 25 °C. All of the measurements were carried out using an optical cell with a path length of 10 mm. A total of 100 spectra were recorded successively in two different time regimes (50 spectra from 2 to 100 ms, 50 spectra from 100 ms to 100 s).
RESULTS
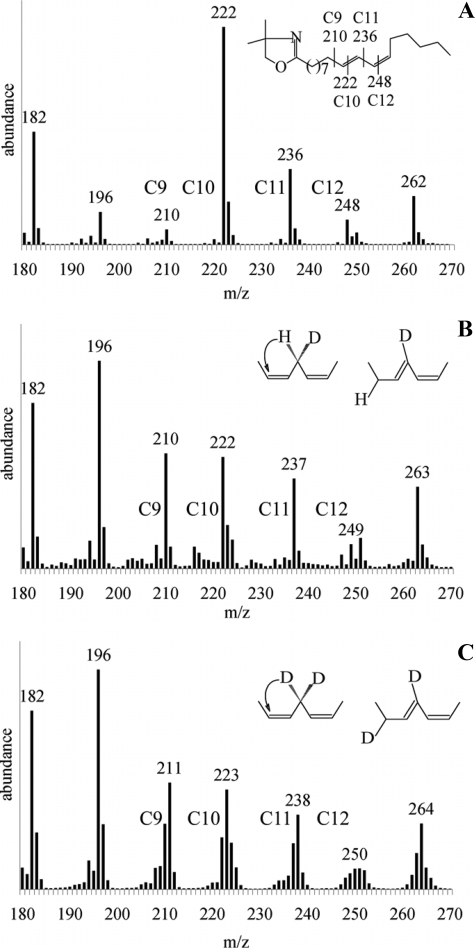
Hydrogen Transfer during the Isomerization—To establish the stereochemistry of hydrogen transfer during PAI-catalyzed isomerization, the reaction products deriving from LA (isotopically labeled with deuterium at C-11) were analyzed by GC-MS after conversion to 4,4-dimethyloxazoline derivatives. The pattern of 4,4-dimethyloxazoline-(10E,12Z)-CLA fragmentation is shown in Fig. 2A (here only the region of interest containing peaks of high m/z is included). In nonlabeled (10E,12Z)-CLA, gaps of 12 atomic mass units between the fragments C-9-C-10 (m/z = 210 and 222) and C-12-C-13 (m/z = 236 and 248) enable localization of double bonds at positions 10 and 12; fragment C-11 and M+ have masses of m/z = 236 and 333, respectively. When [(11S)-2H]linoleic acid was used as a substrate, deuterium remained at position C-11, as indicated by +1-atomic mass unit increase in the C-11 fragment (observed masses for C-9-C-12 fragments are 210, 222, 237, 249, and M+ = 334; Fig. 2B), which is in line with the mechanism based on the PAI crystal structure (7). In case of [11,11-2H2]linoleic acid, the transfer of one deuterium from C-11 to C-9 is evident (observed masses for C-9-C-12 fragments are 211, 223, 238, 250, and M+ = 335; Fig. 2C), i.e. fragments C-9 and C-10 have +1-atomic mass unit mass increase compared with nonlabeled (10E,12Z)-CLA, and all fragments higher than C-10 have a +2-atomic mass unit increase. These results prove that replacement of 1H for 2H at the C-11 position of LA does not render the enzyme inactive but allows for kinetic isotope effect studies (see below). The isotopic ratio between hydrogen and deuterium was similar for the substrate and product, indicating no loss of deuterium during the isomerization reaction. The reaction in 2H2O did not lead to deuterium incorporation in the product, ruling out both direct involvement of solvent in the reaction and hydrogen exchange between N-5 of reduced FAD and water. This result is buttressed by crystal structure analysis where there was no water detectable in the PAI active site (7).
FIGURE 2.
Analysis of isotopic label migration in (10E,12Z)-CLA. A, mass spectrum of (10E,12Z)-CLA derived from nonlabeled LA. Peaks corresponding to fragments C-9-C12 are marked, and the fragmentation pattern of 4,4-dimethyloxazoline-(10E,12Z)-CLA is shown (inset). B, mass spectrum of (10E,12Z)-CLA derived from (11S)-deuterio-LA. Inset, 11-proS-hydrogen is not abstracted by PAI. C, same as B, but 11-dideuterio-LA was used as the substrate. Inset, scheme of deuterium transfer by PAI.
The fact that even after prolonged incubation of PAI with (10E,12Z)-CLA, no traces of LA were detected by GC (data not shown) confirms that isomerization of LA by PAI is a quasi-irreversible process. Assuming that the initial PAI·LA complex is irreversibly converted to the PAI· (10E,12Z)-CLA complex at the rate determined by k3, the classical Michaelis-Menten kinetic scheme consisting of two steps can be applied to PAI reaction. Obviously, in our scheme k3 combines several kinetically distinct steps, including hydrogen transfer from LA to FAD and from reduced FAD to the fatty acid intermediate as well as dissociation of (10E,12Z)-CLA from the active site upon completed isomerization.
Enzymatic isomerization of [11,11-2H2]LA led to the formation of CLA, which retained one of the deuterium atoms at C-11, whereas the other was transferred to C-9 (supplemental Fig. S1). To determine the absolute configuration of the C-9 deuterium atom of CLA, a sample of [9,11-2H2](10E,12Z)-CLA was hydrogenated, and the resulting dideuterated stearate was incubated with a growing culture of T. pyriformis.
This microorganism actively desaturates stearic acid to yield a mixture of oleic acid, LA, and γ-linolenic acid by sequential introduction of double bonds at Δ9, Δ12, and Δ6 (11). As first shown by Schroepfer and Bloch (17) using the bacterium Corynebacterium diphteriae, such double bond-forming processes proceed stereospecifically, e.g. the Δ9 double bond of oleate was introduced by selective removal of the pro-R hydrogens at C-9 and C-10 of the parent stearate. We prepared a sample of [(9S)-2H]stearic acid from the easily accessible (9S)-HOD and found that γ-linolenic acid generated from this stearate upon incubation with Tetrahymena largely retained the deuterium label (data not shown). Therefore, the Tetrahymena Δ9-desaturase, like the corresponding enzyme in Corynebacterium, selectively removed the pro-R hydrogen from C-9 when creating the Δ9 double bond. Incubation of the dideuterated stearic acid with Tetrahymena prepared from [9,11-2H2](10E,12Z)-CLA likewise generated γ-linolenic acid, which retained the deuterium label (data not shown), thus demonstrating that the configuration of the C-9 deuterium of the 9,11-dideuterated CLA was “R” (supplemental Fig. S1). As seen, PAI-catalyzed double bond isomerization takes place by stereoselective abstraction of the pro-R hydrogen from C-11 followed by a suprafacial transfer of this hydrogen to C-9 (Fig. 1A).
To analyze the role of hydrogen abstraction at C-11 for overall catalysis, we carried out kinetic isotope effect (KIE) studies with the same substrates as above (data are summarized in Table 1). Briefly, KIE values of 5.26 and 1.25 were observed for [11,11-2H2]LA and [(11S)-2H]LA, respectively, based on Vmax values. Unfortunately, (11R)-deuterated LA was not available as a substrate for KIE studies; however, a theoretical KIE of [(11R)-2H]LA conversion can be calculated according to the “rule of the geometric mean” (no isotope effects on isotope effects) and would thus amount to 4.21. This value is typical for a primary isotope effect and clearly identifies the (11R) deuteron to undergo transfer to C-9 in the course of isomerization. The small KIE of 1.25 for turnover of [(11S)-2H]LA corresponds well with an α-secondary isotope effect.
TABLE 1.
Kinetic parameters of PAI wt and mutant forms
Km = (k2 + k3)/k1 (see Fig. 3), k3 = Vmax/[Et]. Et is total enzyme concentration, which was 10 nm for PAI wt, and other data were normalized to this value.
| Protein | Substrate | Km | Vmax | k3 |
|---|---|---|---|---|
| μm | nm/s | s−1 | ||
| PAI wt | LA | 4.82 ± 0.84 | 43.67 ± 3.40 | 4.37 ± 0.34 |
| PAI wt | [(11S)-2H]LA | 4.08 ± 0.52 | 35.09 ± 2.14 | 3.51 ± 0.21 |
| PAI wt | [11, 11-2H2]LA | 2.28 ± 0.55 | 8.27 ± 0.79 | 0.83 ± 0.08 |
| PAI | LA | 65.70 ± 8.04 | 29.28 ± 1.23 | 2.93 ± 0.12 |
| R88S | ||||
| PAI | LA | 37.94 ± 7.88 | 32.70 ± 2.58 | 0.33 ± 0.26 |
| F193A | ||||
| PAI | LA | 25.77 ± 4.13 | 15.52 ± 0.60 | 0.16 ± 0.06 |
| R88S/F193A | ||||
| PAI | LA | All mutants inactive | ||
| R88A | ||||
| F168L | ||||
| F168G/G169F |
The estimated Km values for LA and [(11S)-2H]LA are identical within experimental error. However, the Km value for [11,11-2H2]LA is 2-fold decreased compared with that of LA (2.28 ± 0.55 and 4.82 ± 0.84 μm, respectively), indicating that substrate binding occurs significantly faster than the subsequent hydrogen transfer steps. In addition, hydrogen abstraction at C-11 (11R) appears to be rate-limiting for PAI reaction, consistent with pronounced KIE calculated for conversion of [(11R)-2H]LA.
Mutation of Key Residues Involved in Substrate Alignment—To study the substrate specificity of PAI with respect to fatty acids and FA esters, the roles of the gating residues Arg-88, Phe-193, and Phe-168 in substrate recognition and catalysis were analyzed. The two gating residues, Arg-88 and Phe-193, were mutated to small noncharged amino acids, and the steadystate kinetic parameters of the mutant enzymes with LA as substrate were estimated (Table 1). Mutation of Arg-88 to Ala inactivated PAI to an extent that activity could not be measured spectrophotometrically, and only residual amounts of (10E,12Z)-CLA were detectable by GC. Single exchanges of Arg-88 to Ser and of Phe-193 to Ala, as well as the combination of both, caused pronounced increases in Km ranging from 5- to 13-fold. Thus, substrate binding is compromised in the absence of the gating residues. The Vmax was decreased 25% for Ser-88 and 30% for Ala-193 mutation, respectively, and it was ∼60% lower for the double variant compared with wt PAI. Moreover, mutations of Arg-88 and Phe-193 slightly improved PAI activity toward esterified fatty acids (Table 2). A clear difference was observed for LA methyl ester, which was converted to (10E,12Z)-CLA methyl ester by the Ser-88 mutant 4-fold more efficiently than by wt PAI, based on product analysis by GC.
TABLE 2.
Reaction products of PAI wt and selected mutants with LA analogs and lipids
| Protein | Substrate | Product |
|---|---|---|
| PAI wt | Methyl ester LA | 10,12-CLA (traces) |
| R88S, F193A, R88S/F193A | 10,12-CLA (3-5%) | |
| PAI wt | Linoleyl-CoA | 10,12-CLA-CoA |
| R88S, F193A, R88S/F193A | 10,12-CLA-CoA | |
| PAI wt | Tri-linoleyl-glycerol | None |
| R88S, F193A, R88S/F193A | None | |
| PAI wt | Di-linoleyl-phosphatidylcholine | None |
| R88S, F193A, R88S/F193A | None |
The structural model of PAI reaction suggested that the direction of the allylic shift during LA isomerization is governed by the position of Phe-168 relative to FAD (7). When Phe-168 was switched in position with neighboring Gly-169 or replaced by Ile in PAI, the activity was abolished (Table 1), although the enzyme seemed to be properly folded with FAD incorporated as has been verified by spectroscopic properties of the variants as shown for the Phe-168 mutant (supplemental Fig. S2). These experiments highlight the crucial role of the aromatic ring in Phe-168 and its proper position relative to FAD for PAI activity.
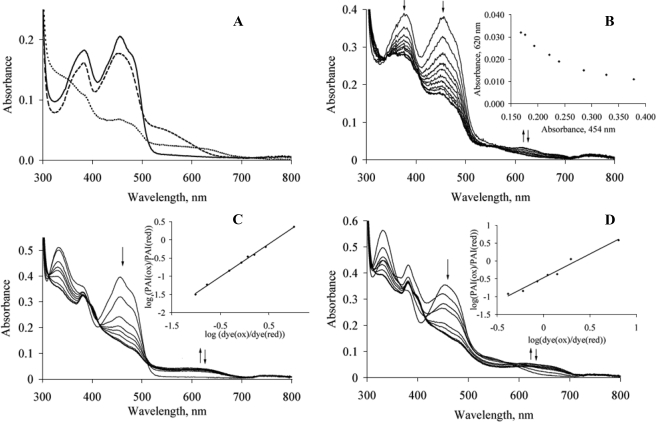
Substrate Binding and Anaerobic Photoreduction—For an unambiguous identification of FAD semiquinone and calculation of equilibrium constants for semiquinone formation, the redox properties of free PAI and the enzyme-product complex were analyzed by photoreduction. The changes in PAI spectrum upon substrate binding and anaerobic photoreduction are shown in Fig. 3A. The addition of LA (both aerobic and anaerobic) turned the color of PAI solution from yellow to orangered, and an additional shoulder appeared in the spectrum around 545 nm. The color returned to yellow by repeated cycles of dilution-ultrafiltration because of the washout of LA. Neither oleic acid (18:1Δ9Z) nor stearic acid (18:0) produced similar spectral changes upon mixing with PAI, showing that two double bonds in substrate or product are essential for this effect. Thus, these spectral changes could be attributed to the charge-transfer interaction between LA or (10E,12Z)-CLA and oxidized FAD. Flavoenzymes often form charge-transfer complexes with electron-rich substrates, e.g. this type of interaction was extensively studied for acyl-CoA oxidases and modified acyl-CoA species (18). Because the complete transfer of an π-electron from LA and (10E,12Z)-CLA to FAD is unlikely, the spectral changes probably reflect a transient state resulting from a π-stacking interaction between Phe-168, the double bonds of the fatty acids, and the isoalloxazine ring of FAD. Anaerobic photoreduction of PAI resulted in decreased absorbance at 380 and 454 nm and in the appearance of a broad spectral peak in the range 500-700 nm with the isosbestic points at 325 and 525 nm (Fig. 3C). Apparently, the same species was formed upon photoreduction of the PAI-(10E,12Z)-CLA complex (Fig. 3B) with the second isosbestic point shifted from 525 to 560 nm. These broad peaks could be attributed to neutral flavin semiquinone, which absorbs in the range 580-600 nm with an extinction coefficient of ∼4000 m-1 cm-1 (19). However, under our experimental conditions, even after extensive irradiation of both the PAI-(10E,12Z)-CLA complex and the unreacted protein, the long wavelength peak was not disappearing to the base level as would be expected for the reduction of flavin semiquinone to hydroquinone (Fig. 3B, inset, and data not shown). After exposure to air, the initial spectrum was restored (Fig. 3B, bold line), indicating that the absorbance increase at 620 nm was due to the reduced FAD but the spectral data could not be used for the unambiguous identification of FAD semiquinone and calculation of equilibrium constants for semiquinone formation and decay.
FIGURE 3.
A, substrate complex formation. Bold line, PAI; dashed line, PAI upon 10× excess of LA; dotted line, photoreduced PAI, semiquinone. B, photoreduction of PAI. The arrows indicate absorbance changes at 380, 454, and 620 nm. On the inset, plot of absorbance changes at 620 versus 450 nm shows that no decay of reduced FAD species occurs upon extensive photoreduction. C, determination of PAI midpoint potential. Protein was photoreduced in the presence of reference dye, anthraquinone-2-sulfonate. The arrows indicate absorbance changes during the reduction. Insets, changes of redox states for the dye and protein plotted according to Minnaert (21). D, same as C, but for the PAI-(10E,12Z)-CLA complex.
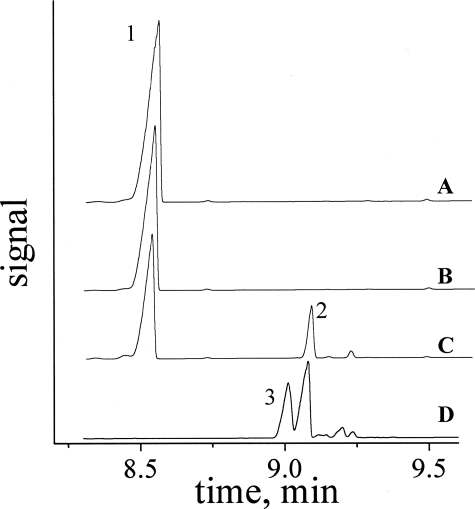
Cofactor Replacement—To further examine the nature of the reduced FAD species involved in PAI turnover, the activity of PAI reconstituted with the flavin analog 5-deaza-5-carba-FAD was probed. In contrast to FAD and FMN, the latter compound acts exclusively as a two-electron acceptor and is therefore commonly employed as a probe for the discrimination between radical and ionic mechanisms in flavoenzymes (20). Refolding of PAI with 6 m guanidium chloride by a gel filtration method was used to produce apo-PAI with about 30% yield (as determined by A280 for protein samples before and after refolding). The incorporation of 5-deaza-5-carba-FAD in apo-PAI was confirmed by UV-visible spectroscopy of the reconstituted enzyme. As the activity of PAI reconstituted with 5-deaza-5-carba-FAD was not detectable spectrophotometrically, GC analysis of potential reaction products was performed (Fig. 4). The amount of injected sample was intentionally set very high, so that even trace amounts of CLA could have been detected. Although, as expected, the apo-enzyme was inactive toward LA isomerization, reconstitution with FAD restored the activity (Fig. 4, trace C). Most importantly, the addition of LA to PAI reconstituted with 5-deaza-5-carba-FAD gave no detectable amount of (10E,12Z)-CLA (Fig. 4, traces A and B, respectively). This result suggests that the reaction may not proceed via ionic intermediates as required by simultaneous two-electron transfer from LA to 5-deaza-5-carba-FAD. Therefore, as an alternative to the ionic mechanism, homolytic C-H bond cleavage resulting in flavin semiquinone and LA radical might be considered as the first catalytic step in PAI reaction. However, the more negative redox potential of 5-deaza-5-carba-FAD versus FAD (-310 and -208 mV, respectively (19)) may be insufficient for the abstraction of the hydrogen at C-11 of LA. For acyl-CoA dehydrogenases this option was tested experimentally, because the reduction of the double bond in 2-enoyl-CoA can still be catalyzed by the reduced 5-deaza-5-carba-FAD (18). In the case of PAI, the isomerization of the Δ9 double bond rather than oxidation occurs, and therefore the reverse reaction also requires an oxidized cofactor, excluding the above mentioned analysis.
FIGURE 4.
GC analysis of reconstitution of PAI with 5-deaza-5-carba-FAD. A, Apo PAI. B, Apo-PAI + 5-deaza-5-carba-FAD. C, Apo-PAI + FAD. D, CLA standard. Peak 1, LA. Peak 2, 10,12-CLA. Peak 3, 9,11-CLA.
Redox Potential—To further characterize redox properties of PAI-bound FAD, midpoint potentials of free PAI and of the complex with (10E,12Z)-CLA were determined by pho-toreduction in the presence of anthraquinone-2-sulfonate as a reference dye (Fig. 3, C and D, respectively). The reduction of anthraquinone-2-sulfonate was monitored at 325 nm, which is the isosbestic point of PAI. Anthraquinone-2-sulfonate has the isosbestic point at 365 nm (Fig. 3D). Plotting the data at 365 and 454 nm according to Minnaert (21) gave similar values of Eox/red for free PAI (-210 and -208 mV, respectively). Thus, the Eox/red = -211 mV was calculated for the PAI-(10E,12Z)-CLA complex based on absorbance changes at 454 nm. The slopes of the Minnaert plots were 0.92 and 1.33 for the free PAI and the complex, respectively, close to the theoretical value of 1 for the equilibrium with the two-electron dye. It was observed that the protein is prone to aggregation in the presence of LA, which may be the reason for less accurate data in the latter case. To confirm the obtained Eox/red values, the experiments were carried out with 2-hydroxy-1,4-napthaquinone and safranine O as reference dyes, which were reduced before and after PAI, respectively (data not shown). Similar to the photoreduction without a dye, the absorbance increase at 620 nm was observed, and it declined only partially at the late stages of reduction (Fig. 3, C and D, bold lines); therefore no quantification of putative semiquinone intermediate could be performed.
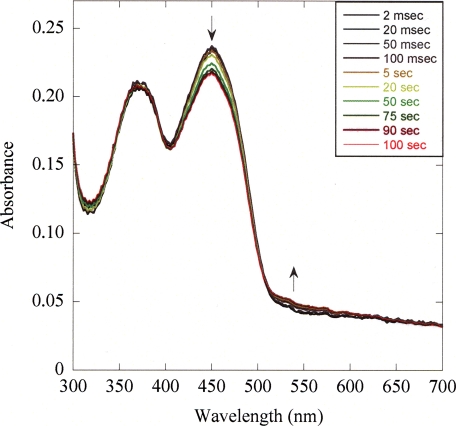
Rapid Scan Stopped Flow Experiments—To test the potential occurrence of transient FAD radicals (semiquinone), the reaction of PAI with LA was analyzed by stopped flow kinetics coupled to diode array acquisition (Fig. 5). The time-resolved spectra of the enzyme-bound flavin cofactor collected in the course of substrate processing do not evidence formation of a FAD semiquinone as a transitory state. This observation holds true for all deployed LA concentrations. The only spectral change is apparent in the 550-nm region, which we ascribe to formation of a charge-transfer complex between substrate or product and FAD. These results are consistent with an ionic mechanism; they can, however, not ultimately rule out a radical mechanism involving a FAD-LA radical pair, because the putative radical pair could be extremely short-lived and thus escape spectroscopic detection.
FIGURE 5.
Time-resolved absorbance spectra of PAI-bound FAD in the course of LA turnover. A solution of 1.6 mg/ml PAI was reacted with 100 μm LA in 1 + 1 mixing ration in 0.1 m Tris/HCl buffer, pH 7.5, at 25 °C and using an optical path length of 10 mm. The spectra shown are a selection and are corrected for buffer contributions.
DISCUSSION
Despite the intense studies related to the nutritional benefits and physiological effects of CLA consumption, the biosynthesis of these PUFA isomers is poorly understood. Our knowledge is mainly based on the results obtained for three PUFA double bond isomerases, namely the proteins from B. fibrisolvens, P. filicina, and P. acnes (4). The common feature of all these enzymes is the strict requirement for a free carboxylate in the substrate and the cleavage of a C-H bond at the position C-11 of FA during the catalysis (1, 2, 5-7, 9). In contrast to PAI, the reaction of the proteins from B. fibrisolvens and P. filicina involves a proton exchange with the solvent and the direction of the allylic shift is opposite. Recently, two studies reported that (9Z,11E)-CLA and (10E,12Z)-CLA are produced by mixed communities of ruminal bacteria via distinct routes, based on the incorporation of isotopic labels (22, 23). Thus, PUFA double bond isomerases can be divided in two classes according to their biochemical mechanism (4). Our previous work defined the key determinants of free carboxylate recognition and suggested two possible scenarios for the isomerization mechanism of (10E,12Z)-CLA-forming enzymes based on the PAI crystal structure (Fig. 1).
The decreased Vmax values for all mutants likely reflect that the proper alignment of LA in the active site critically depends on the fixed position of the free carboxyl group. This precise substrate positioning is in turn crucial for the efficient hydrogen transfer, as well as stabilizing the interaction between the pentadienyl moiety of LA and Phe-168.
The transfer of 11-(pro-R)-hydrogen by PAI to the position C-9 again in R configuration fully agrees with both structure-based reaction mechanisms. Moreover, selective mono- and dideuteration at C-11 strongly influences the reaction rate as evident from the experimentally determined 5-fold KIE for [11,11-2H2]LA. A somewhat lower KIE of 1.25 was estimated for conversion of [(11S)-2H]LA indicating an α-secondary isotope effect. When considering the rule of the geometric mean, the calculated primary KIE of 4.21 for turnover of [(11R)-2H]LA identifies the 11-(pro-R)-hydrogen as the primary side of reaction and further suggests that cleavage of the 9C-(pro-R)-H bond is the rate-limiting step of catalysis. The lower Km value for 11-dideutero-LA can be attributed to a decreased k3, and it suggests that the isomerization step is significantly slower than substrate binding. In similar experiments with the isomerase from B. fibrisolvens, it was found that [11,11-2H2]LA acid exhibits a 2.5-fold KIE but does not affect Km (2), in this case the higher Km value (12 μm) compared with PAI possibly indicates that the substrate binding rather than the isomerization itself is the rate-limiting step. In contrast to PAI, the latter enzyme transfers the proton from the solvent to the product (9Z,11E)-CLA, which probably reflects different hydrogen transfer mechanisms used by the two enzymes (2).
The dependence of the reaction rate on the correct substrate positioning by Arg-88 and Phe-193 as well as on the presence of Phe-168 invites speculation that hydrogen tunneling might ensure efficient cleavage of nonactivated C-H bond by PAI. The contribution of quantum chemical tunneling to the flavin reduction upon heterolytic cleavage of C-H bond has been demonstrated for morphinone reductase (24) and trimethylamine dehydrogenase (25). Unfortunately, a detailed evaluation of tunneling effects during the hydrogen transfer steps by PAI is complicated because of the complex synthetic scheme for preparation of stereospecifically labeled fatty acid derivatives, and thus tritiated LA derivatives are not available.
Our attempts to identify the redox state of FAD produced by interaction with LA were in part inconclusive. Apparently, a small amount of neutral FAD semiquinone is formed during the photoreduction of free PAI and PAI-(10E,12Z)-CLA complex, but its absorbance seems to be masked by a broad spectral band because of reduced FADH2 or a charge-transfer complex between FADH2 and either Phe-168 or the substrate. The inactivation of PAI by 5-deaza-5-carba-FAD is consistent with a radical mechanism, but the low midpoint potential of 5-deaza-5-carba-FAD might thermodynamically prevent hydrogen transfer from LA to the flavin. Eox/red of free PAI and its substrate complex are similar to that of free FAD, and as such slightly lower than for other flavin-dependent double bond isomerases such as 4-hydroxy butyryl-CoA dehydratase and isopentenyl diphosphate:dimethylallyl diphosphate isomerase (-180 and -197 mV, respectively) (26, 27). The substrate-induced modulation of flavin redox properties are exploited by many flavoenzymes, e.g. it was shown that substrate binding significantly increases the amount of neutral flavin semiquinone during the reduction of 4-hydroxy butyryl-CoA dehydratase and isopentenyl diphosphate:dimethylallyl diphosphate isomerase (26-28), and the Eox/red of acyl-CoA dehydrogenase is increased by 110 mV in the presence of substrate-product complex (29). Binding of the reaction product (10E,12Z)-CLA to PAI does not induce significant changes in the midpoint potential; neither does it increase the amount of FAD semiquinone upon photoreduction. It is still possible that these effects occur upon LA binding, but because of the low solubility of free fatty acids and relatively high turnover rate (4 s-1; Table 1), they cannot be easily monitored under equilibrium conditions. However, unlike the strictly anaerobic enzymes 4-hydroxy butyryl-CoA dehydratase and isopentenyl diphosphate:dimethylallyl diphosphate isomerase, PAI is not sensitive to molecular oxygen as is the family of dehydrogenases family of flavoenzymes (15), which operate via two-electron transfer. This observation, along with the absence of distinct semiquinone signal in the photoreduction experiments, favor an ionic mechanism of LA isomerization by PAI. Two independent lines of evidence further support this hypothesis. First, no transient FAD radicals are detectable by stopped flow UV-visible absorption measurements. Second, we have found that no paramagnetic intermediates were detectable by continuous wave or pulsed EPR spectroscopy when freeze quenching the reaction of PAI with LA or deuterated LA in the second time scale (supplemental Fig. S3).
To summarize, our results delineate the regio- and stereospecific course of hydrogen transfer during LA isomerization to (10E,12Z)-CLA as predicted by the crystal structure. Taken together, our data on the midpoint potential, photoreduction, cofactor replacement, and potential radical intermediates by EPR and UV-visible spectroscopy suggest that PAI presumably acts via an ionic mechanism with the formation of FADH2 and LA carbocation as intermediates. The substrate preference toward free fatty acids is determined by interaction between Arg-88 and Phe-193, and the reaction rate is strongly affected by replacement of these amino acids, indicating that the efficiency of the hydrogen transfer relies on a fixed distance between the free carboxylate group and the N-5 atom of FAD. Further studies are necessary to rule out the radical mechanism and evaluate the rates of individual hydrogen transfer steps during the LA isomerization.
Supplementary Material
Acknowledgments
We thank Prof. O. Einsle (Goettingen, Germany) for the facilities for anaerobic experiments and the critical discussion of results, M.-T. Türke and Dr. M. Bennati (Goettingen, Germany) for EPR measurements, and Prof. S. Ghisla (Konstanz, Germany) for providing us with 5-deaza-5-carba-FAD.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains Figs. S1-S3.
Footnotes
The abbreviations used are: CLA, conjugated linoleic acid; GC-MS, gas chromatography mass spectrometry; KIE, kinetic isotope effect; LA, linoleic acid; PAI, PUFA isomerase from P. acnes; PUFA, polyunsaturated fatty acid; HPLC, high pressure liquid chromatography; wt, wild type; FA, fatty acid.
References
- 1.Kepler, C. R., Tucker, W. P., and Tove, S. B. (1970) J. Biol. Chem. 245 3612-3620 [PubMed] [Google Scholar]
- 2.Kepler, C. R., Tucker, W. P., and Tove, S. B. (1971) J. Biol. Chem. 246 2765-2771 [PubMed] [Google Scholar]
- 3.Ogawa, J., Kishino, S., Ando, A., Sugimoto, S., Mihara, K., and Shimizu, S. (2005) J. Biosci. Bioeng. 100 355-364 [DOI] [PubMed] [Google Scholar]
- 4.Liavonchanka, A., and Feussner, I. (2008) ChemBioChem 9 1867-1872 [DOI] [PubMed] [Google Scholar]
- 5.Wise, M. L., Hamberg, M., and Gerwick, W. H. (1994) Biochemistry 33 15223-15232 [DOI] [PubMed] [Google Scholar]
- 6.Wise, M. L., Rossi, J., and Gerwick, W. H. (1997) Biochemistry 36 2985-2992 [DOI] [PubMed] [Google Scholar]
- 7.Liavonchanka, A., Hornung, E., Feussner, I., and Rudolph, M. G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2576-2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bornemann, S. (2002) Nat. Prod. Rep. 19 761-772 [DOI] [PubMed] [Google Scholar]
- 9.Hornung, E., Krueger, C., Pernstich, C., Gipmans, M., Porzel, A., and Feussner, I. (2005) Biochim. Biophys. Acta 1738 105-114 [DOI] [PubMed] [Google Scholar]
- 10.Liavonchanka, A., Hornung, E., Feussner, I., and Rudolph, M. (2006) Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 62 153-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamberg, M. (2005) FEBS J. 272 736-743 [DOI] [PubMed] [Google Scholar]
- 12.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37 911-917 [DOI] [PubMed] [Google Scholar]
- 13.Hornung, E., Pernstich, C., and Feussner, I. (2002) Eur. J. Biochem. 269 4852-4859 [DOI] [PubMed] [Google Scholar]
- 14.Christie, W. W. (1998) Lipids 33 343-353 [DOI] [PubMed] [Google Scholar]
- 15.Massey, V. (1994) J. Biol. Chem. 269 22459-22462 [PubMed] [Google Scholar]
- 16.Massey, V., and Hemmerich, P. (1978) Biochemistry 17 9-16 [DOI] [PubMed] [Google Scholar]
- 17.Schroepfer, G. J., and Bloch, K. (1965) J. Biol. Chem. 240 54-63 [PubMed] [Google Scholar]
- 18.Ghisla, S., and Thorpe, C. (2004) Eur. J. Biochem. 271 494-508 [DOI] [PubMed] [Google Scholar]
- 19.Chapman, S. K., and Reid G. A. (eds) (1999) Flavoprotein Protocols, Humana Press, New Jersey
- 20.Murthy, Y. V., and Massey, V. (1997) Methods Enzymol. 280 436-460 [DOI] [PubMed] [Google Scholar]
- 21.Minnaert, K. (1965) Biochim. Biophys. Acta 110 42-56 [DOI] [PubMed] [Google Scholar]
- 22.Devillard, E., McIntosh, F. M., Duncan, S. H., and Wallace, R. J. (2007) J. Bacteriol. 189 2566-2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace, R. J., McKain, N., Shingfield, K. J., and Devillard, E. (2007) J. Lipid Res. 48 2247-2254 [DOI] [PubMed] [Google Scholar]
- 24.Basran, J., Harris, R. J., Sutcliffe, M. J., and Scrutton, N. S. (2003) J. Biol. Chem. 278 43973-43982 [DOI] [PubMed] [Google Scholar]
- 25.Basran, J., Sutcliffe, M. J., and Scrutton, N. S. (2001) J. Biol. Chem. 276 24581-24587 [DOI] [PubMed] [Google Scholar]
- 26.Muh, U., Cinkaya, I., Albracht, S. P., and Buckel, W. (1996) Biochemistry 35 11710-11718 [DOI] [PubMed] [Google Scholar]
- 27.Kittleman, W., Thibodeaux, C. J., Liu, Y. N., Zhang, H., and Liu, H. W. (2007) Biochemistry 46 8401-8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothman, S. C., Helm, T. R., and Poulter, C. D. (2007) Biochemistry 46 5437-5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saenger, A. K., Nguyen, T. V., Vockley, J., and Stankovich, M. T. (2005) Biochemistry 44 16043-16053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.