Abstract
The mammalian target of rapamycin (mTOR) kinase is the catalytic subunit of two functionally distinct complexes, mTORC1 and mTORC2, that coordinately promote cell growth, proliferation, and survival. Rapamycin is a potent allosteric mTORC1 inhibitor with clinical applications as an immunosuppressant and anti-cancer agent. Here we find that Torin1, a highly potent and selective ATP-competitive mTOR inhibitor that directly inhibits both complexes, impairs cell growth and proliferation to a far greater degree than rapamycin. Surprisingly, these effects are independent of mTORC2 inhibition and are instead because of suppression of rapamycin-resistant functions of mTORC1 that are necessary for cap-dependent translation and suppression of autophagy. These effects are at least partly mediated by mTORC1-dependent and rapamycin-resistant phosphorylation of 4E-BP1. Our findings challenge the assumption that rapamycin completely inhibits mTORC1 and indicate that direct inhibitors of mTORC1 kinase activity may be more successful than rapamycin at inhibiting tumors that depend on mTORC1.
The mammalian target of rapamycin (mTOR)3 pathway is considered a major regulator of cell growth. The mTOR serine/threonine kinase is the founding component of the pathway and the catalytic subunit of two functionally distinct protein complexes, mTORC1 and mTORC2. mTORC1 contains the large protein Raptor, as well as mLST8/GβL and PRAS40, whereas mTORC2 is defined by the protein Rictor and also includes Sin1, Protor, and mLST8/GβL (1). Growth factors, such as insulin and IGF, activate both complexes, and they are important downstream effectors of the PI3K/PTEN signaling network (2). Additionally, the availability of nutrients, like amino acids and glucose, regulates mTORC1.
Many insights into mTOR signaling have come from investigations into the mechanism of action of rapamycin, a bacterially produced macrolide inhibitor of mTOR that has diverse clinical applications as an anti-fungal, immunosuppressant, and anti-cancer drug (3). Rapamycin acts through an unusual allosteric mechanism that requires binding to its intracellular receptor, FKBP12, for inhibition of its target. Under acute treatment, rapamycin is thought to selectively inhibit mTORC1, which is often referred to as the rapamycin-sensitive complex. Conversely, mTORC2 is considered rapamycin-insensitive, although its assembly can be inhibited by prolonged rapamycin treatment in some cell types (4). Because of its perceived potency and selectivity, rapamycin is commonly used in research experiments as a test of the involvement of mTORC1 in a particular process.
Two downstream mTORC1 substrates that were identified, in part, by their sensitivity to rapamycin are the S6 kinases (S6K1 and S6K2) and the translational inhibitor 4E-BP1. Both proteins mediate important links between mTORC1 and the cell growth machinery, largely through their influence on cap-dependent translation (reviewed in Ref. 5). All nuclear-encoded mRNAs possess a 5′,7-methyl guanosine cap, which is recognized and bound by the small protein eIF-4E. Under growth-promoting conditions, eIF-4E also associates with the large scaffolding protein eIF-4G, the eIF-4A helicase, and the eIF-4B regulatory protein, together forming the eIF-4F complex. This complex, in conjunction with the eIF3 preinitiation complex, delivers the mRNA to the 40 S ribosomal subunit and primes the translational apparatus. 4E-BP1 interferes with this process by binding to eIF-4E and preventing the formation of a functional eIF-4F complex. However, its ability to do this is blocked by phosphorylation at four sites, two of which are considered rapamycin-sensitive. S6K1 also plays a role in regulating translational initiation by phosphorylating the S6 protein of the 40 S ribosomal subunit and by stimulating eIF-4A helicase activity (6-8).
Despite the connections of mTORC1 to the translational machinery, the effects of rapamycin on mammalian cell growth and proliferation are, oddly, less severe than its effects in yeast. In Saccharomyces cerevisiae, rapamycin treatment induces a starvation-like state that includes a severe G1/S cell cycle arrest and suppression of translation initiation to levels below 20% of nontreated cells (9). Moreover, in yeast rapamycin strongly promotes induction of autophagy (self-eating), a process by which cells consume cytoplasmic proteins, ribosomes, and organelles, such as mitochondria, to maintain a sufficient supply of amino acids and other nutrients (10).
The effects of rapamycin in mammalian cells are similar to those in yeast, but typically much less dramatic and highly dependent on cell type. For instance, rapamycin only causes cell cycle arrest in a limited number of cell types and has modest effects on protein synthesis (11-13). Moreover, rapamycin is a relatively poor inducer of autophagy, and it is often used in combination with LY294002, an inhibitor of PI3K and mTOR (14). These inconsistent effects may explain why, despite high expectations, rapamycin has had only limited success as a clinical anti-cancer therapeutic. We have hypothesized that the effectiveness of rapamycin against a particular cancer might be determined by its ability to inhibit mTORC2 in addition to mTORC1 (15). To test this hypothesis, we developed the ATP-competitive inhibitor Torin1 that suppresses both complexes. In contrast to rapamycin, Torin1 treatment recapitulates in mammalian cells many of the phenotypes caused by TOR inhibition in yeast. Surprisingly, however, we find that these effects are independent of mTORC2 and are instead caused by inhibition of rapamycin-resistant functions of mTORC1.
EXPERIMENTAL PROCEDURES
Materials—Reagents were obtained from the following sources: antibodies to phospho-Thr-389 S6K, phospho-Ser-473 Akt, phospho-Thr-308 Akt, pan-Akt, phospho-Thr-36/47 4E-BP1, phospho-Ser-65 4E-BP1, phospho-Thr-70 4E-BP1, 4E-BP1, α-tubulin, Raptor, eIF-4E, phospho-S51 eIF2α, cyclin D1, cyclin D3 and p27/Kip1 from Cell Signaling Technology (note: we have not confirmed that the phospho-Thr-70 4E-BP1 antibody does not detect unphosphorylated 4E-BP1); antibodies to mTOR, S6K, and horseradish peroxidase-labeled anti-mouse, anti-goat, and anti-rabbit secondary antibodies from Santa Cruz Biotechnology; anti-Rictor antibodies from Bethyl Laboratories; FuGENE 6 and Complete Protease Mixture from Roche Applied Science; FLAG M2 antibody, FLAG M2-agarose, and ATP from Sigma; 7-methyl-GTP-Sepharose from GE Healthcare; PI-103 from Calbiochem; NVP-BEZ235 from Axon Medchem; rapamycin from LC Laboratories; PI3K-α from Millipore/Upstate; CellTiter-Glo, DNA-PK, and DNA-PK peptide substrate from Promega; phosphatidylinositol and phosphatidylserine from Avanti Polar Lipids; EasyTag™ EXPRESS 35S protein labeling mix and ATP [γ-32P] EasyTide from PerkinElmer Life Sciences; Dulbecco's modified Eagle's medium from SAFC Biosciences; inactivated fetal calf serum from Invitrogen. p53-/-/TSC2-/- MEFs as well as p53-/-/TSC2+/+ MEFs were kindly provided by David Kwiatkowski (Harvard Medical School) and cultured in Dulbecco's modified Eagle's medium with 10% inactivated fetal calf serum. p53-/-/mLST8-/- and p53-/-/Rictor-/- MEFs have been described (16). Torin1 was synthesized and purified in the Gray Laboratory and is available upon request.
Cell Lysis—Cells rinsed once with ice-cold PBS were lysed in ice-cold lysis buffer (40 mm HEPES, pH 7.4, 2 mm EDTA, 10 mm pyrophosphate, 10 mm glycerophosphate, and 0.3% CHAPS or 1% Triton X-100, and 1 tablet of EDTA-free protease inhibitors per 25 ml). The soluble fractions of cell lysates were isolated by centrifugation at 13,000 rpm for 10 min in a microcentrifuge.
Mammalian Lentiviral shRNAs—All shRNA vectors were obtained from the collection of The RNAi Consortium at the Broad Institute (17). These shRNAs are named with the numbers found at the RNAi Consortium public website: mouse Raptor shRNA, TRCN0000077472, NM_028898.1-3729s1c1; and mouse Rictor shRNA, TRCT0000037708, NM_030168.2-867s1c1. shRNA-encoding plasmids were co-transfected with the Δ VPR envelope and vesicular stomatitis virus G packaging plasmids into actively growing HEK-293T using FuGENE 6 transfection reagent as described previously (18, 19). Viruscontaining supernatants were collected at 48 h after transfection and filtered to eliminate cells, and target cells were infected in the presence of 8 μg/ml Polybrene. 24 h later, cells were selected with puromycin and analyzed on the 4th day after infection.
Metabolic Labeling—Cells were seeded in 6-well plates and grown overnight. Cells were then treated with appropriate compounds for 2.5 h, washed one time with cysteine/methionine-free Dulbecco's modified Eagle's medium, and then incubated in 2 ml of cysteine/methionine-free Dulbecco's modified Eagle's medium, 10% dialyzed inactivated fetal calf serum, compound, and 165 μCi (15 μl, 11 mCi/μl) of EasyTag™ EXPRESS 35S protein labeling mix. After 30 min, cells were lysed, and soluble fractions were isolated by centrifugation at 13,000 rpm for 10 min. To precipitate protein, lysates were spotted on Whatman filter paper, precipitated with 5% trichloroacetic acid, washed two times for 5 min in cold 10% trichloroacetic acid, washed two times for 2 min in cold ethanol, washed one time for 2 min in acetone, and air-dried at room temperature. The amount of 35S incorporated into protein was measured using a Beckman LS6500 Scintillation Counter.
mTORC1 and mTORC2 in Vitro Kinase Assays—To produce soluble mTORC1, we generated HEK-293T cell lines that stably express N-terminally FLAG-tagged Raptor using vesicular stomatitis virus G-pseudotyped MSCV retrovirus. For mTORC2, we similarly generated HeLa cells that stably express N-terminally FLAG-tagged Protor-1. Both complexes were purified by lysing cells in 50 mm HEPES, pH 7.4, 10 mm sodium pyrophosphate, 10 mm sodium β-glycerophosphate, 100 mm NaCl, 2 mm EDTA, 0.3% CHAPS. Cells were lysed at 4 °C for 30 min, and the insoluble fraction was removed by microcentrifugation at 13,000 rpm for 10 min. Supernatants were incubated with FLAG-M2 monoclonal antibody-agarose for 1 h and then washed three times with lysis buffer and once with lysis buffer containing a final concentration of 0.5 m NaCl. Purified mTORC1 was eluted with 100 μg/ml 3× FLAG peptide in 50 mm HEPES, pH 7.4, 100 mm NaCl. Eluate can be aliquoted and stored at -80 °C. Substrates S6K1 and Akt1 were purified as described previously (16, 20). Kinase assays were performed for 20 min at 30 °C in a final volume of 20 μl consisting of the kinase buffer (25 mm HEPES, pH 7.4, 50 mm KCl, 10 mm MgCl2, 500 μm ATP) and 150 ng of inactive S6K1 or Akt1 as substrates. Reactions were stopped by the addition of 80 μl of sample buffer and boiled for 5 min. Samples were subsequently analyzed by SDS-PAGE and immunoblotting.
PI3K and hVps34 Assays—Cellular IC50 values for PI3Kα were determined using p53-/-/mLST8-/- MEFs. Cells were treated with vehicle or increasing concentrations of compound for 1 h and then lysed. Phosphorylation of Akt Thr-308 was monitored by immunoblotting using a phospho-specific antibody. In vitro IC50 values for PI3Kα were determined as described previously (21). Briefly, chloroform stocks of phosphatidylinositol (PI) and phosphatidylserine were combined in equimolar ratios, dried under nitrogen gas, resuspended in 50 mm HEPES, pH 7.4, 100 mm KCl, sonicated to clarity using a bath sonicator, and aliquoted and stored at -80 °C. For kinase assays, purified PI3Kα was combined with 100 μm phosphatidylserine/phosphatidylinositol, compound, and 10 μCi of [γ-32P]ATP (100 μm final concentration) in kinase buffer and incubated at 37 °C for 20 min. Reactions were stopped with 1 n HCl. Lipid was extracted with a 1:1 mixture of chloroform: methanol and separated on silica TLC plates. 32P-Labeled phosphatidylinositol 3-phosphate was quantitated by PhosphorImager. hVps34 was purified as a glutathione S-transferase fusion protein from HEK-293T cells(22) and assayed using the same procedure.
ATM and DNA-PK—For DNA-PK kinase assays, purified DNA-PK was combined with DNA-PK peptide substrate (derived from the N-terminal sequence of p53), compound, and 10 μCi/reaction [γ-32P]ATP (100 μm final concentration) in kinase buffer and incubated for 10 min at 37 °C. Reactions were stopped with 1 n HCl and spotted onto P81 phosphocellulose squares. P81 squares were washed three times for 5 min in 0.75% phosphoric acid, and one time for 5 min in acetone, dried, and measured by scintillation counter. ATM in vitro kinase assays were performed according to previously published protocols (21).
Cell Size Determinations—Cells were seeded in 10-cm culture dishes, grown overnight, and subjected to appropriate treatment. 24 h later, cells were harvested by trypsinization in a 5-ml volume, diluted 1:20 with counting solution (Isoton II Diluent, Beckman Coulter), and cell diameters determined using a particle size counter (Coulter Z2, Beckman Coulter) with Coulter Z2 AccuComp software.
Cell Proliferation/Viability Assay—Cell viability was assessed with the CellTiter-Glo Luminescent Cell Viability Assay. On Day 0, 96-well plates were seeded with 500 cells per well and grown overnight. On Day 1, cells were treated with the appropriate compounds and subsequently analyzed on Days 3-5. For analysis, plates were incubated for 60 min at room temperature; 50 μl of CellTiter-Glo reagent was added to each well, and plates were mixed on an orbital shaker for 12 min. Luminescence was quantified on a standard plate luminometer.
Cell Cycle Analysis—Cells were seeded in 15-cm plates and grown overnight. Cells were then subjected to the appropriate treatment for 48 h and then trypsinized, washed twice in PBS + 2% FBS, and then fixed overnight at 4 °C in ethanol. Cells were then washed three times in PBS + 1% BSA and incubated in PBS, 1% BSA, 50 μg/ml propidium iodide, and 100 μg/ml RNase at 37 °C for 30 min. Cells were then washed 1× in PBS + 1% BSA, resuspended in 1 ml PBS, and analyzed using a FACS-Calibur flow cytometer (BD Biosciences). Cell cycle distribution was determined using the ModFit LT software package.
RESULTS
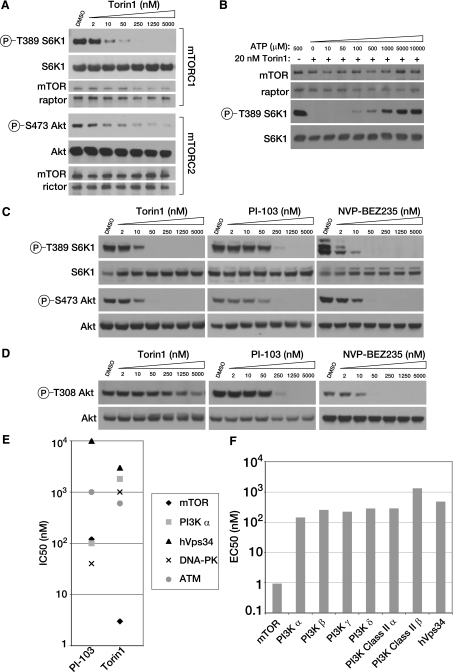
Torin1 Is a Potent and Selective mTOR Inhibitor—To identify small molecule ATP-competitive inhibitors of mTOR, we conducted a biochemical screen for inhibitors of mTOR kinase activity in a library of heterocyclic chemical compounds. From this screen, we identified a lead compound that was further elaborated through a medicinal chemistry effort to produce Torin1, a member of the pyridinonequinoline class of kinase inhibitors.4 In in vitro kinase assays using immuno-purified mTORC1 or mTORC2, Torin1 inhibits both mTOR-containing complexes with IC50 values between 2 and 10 nm (Fig. 1A) and acts through an ATP-competitive mechanism (Fig. 1B). We also measured the potency of Torin1 in cells. MEFs were treated with increasing amounts of Torin1 or the dual mTOR/PI3K inhibitors PI-103 and NVP-BEZ235, and the activity of each complex was determined by monitoring the phosphorylation status of S6K at Thr-389 and Akt at Ser-473, mTORC1 and mTORC2 substrates, respectively (Fig. 1C). As in vitro, the IC50 for Torin1 in cells is also between 2 and 10 nm. Unlike rapamycin, Torin1 had no effect on the stability of either mTORC1 or mTORC2.
FIGURE 1.
Torin1 is a potent and selective mTOR inhibitor. A, Torin1 inhibits mTORC1 and mTORC2 in vitro. mTORC1 and mTORC2 were purified from HEK-293T stably expressing FLAG-Raptor and HeLa cells expressing FLAG-Protor-1, respectively. Following FLAG purification, each complex was subjected to in vitro kinase assays using S6K1 as a substrate for mTORC1 and Akt1 as a substrate for mTORC2. Assays were then analyzed by immunoblotting for the indicated proteins and phosphorylation states. B, Torin1 is an ATP-competitive inhibitor. The in vitro kinase activity of purified mTORC1 toward S6K1 was assayed in the presence of 20 nm Torin1 and increasing concentrations of ATP, as indicated. Assays were then analyzed by immunoblotting for the indicated proteins and phosphorylation states. C, Torin1 is a potent mTORC1 and mTORC2 inhibitor in cells. MEFs (p53-/-) were treated with increasing concentrations of Torin1 or dual mTOR/PI3K inhibitors PI-103 and BEZ-235 for 1 h and then analyzed by immunoblotting for the indicated proteins and phosphorylation states. D, Torin1 has little effect on PI3K at concentrations where mTOR is completely inhibited. The experiment was performed as in C using mLST8-null MEFs and phosphorylation of Akt at Thr-308 was determined by immunoblotting. In mLST8-null MEFs, mTORC2 is inactive and Akt Ser-473 is constitutively dephosphorylated and so PDK1-mediated phosphorylation of Thr-308 only reflects PI3K activity. E, Torin1 is selective for mTOR over related kinases. IC50 values for Torin1 were determined using in vitro kinase assays for mTOR (3 nm), hVps34 (3 μm), PI3K-α (1.8 μm), DNA-PK (1.0 μm), and ATM (0.6 μm). IC50 values for PI-103 for mTOR (120 nm), PI3K-α (100 nm), DNA-PK (40 nm) were determined by the same assays. IC50 values for PI-103 for hVps34 (10 μM) and ATM (1.0 μm) were determined previously (21). F, Torin1 is selective for mTOR over other PI3K isoforms. EC50 values were determined for the indicated PI3K isoforms using the Invitrogen Adapta platform. The EC50 for mTOR was determined using the cell-based LanthaScreen platform.
We next determined the selectivity of Torin1 for mTOR over other kinases. Because mTOR belongs to the PI3K-like kinase family, a family of protein kinases that is defined by a high degree of homology to PI3K within the catalytic domain, many inhibitors of PI3K, such as wortmannin, LY294002, PI-103, and BEZ-235, are also reasonable mTOR inhibitors (21, 23, 24). To measure PI3K inhibition in cells, we made use of the observation that the phosphorylation of Akt at Thr-308 depends on two processes that directly reflect PI3K activity: phosphatidylinositol 3,4,5-triphosphate-dependent targeting of Akt to the plasma membrane and activation of PDK1, the kinase that directly phosphorylates this site. In wild-type cells, phosphorylation of Thr-308 is also influenced by phosphorylation at Ser-473 (19, 25, 26). To remove this latter variable, we tested compounds in MEFs where mLST8, an essential mTORC2 component, is deleted and Akt Ser-473 is constitutively dephosphorylated. Because Ser-473 is fixed in a single state in these cell lines, phosphorylation at Thr-308 only reflects PI3K activity. Using this system, we determined the cellular IC50 of Torin1 for PI3K to be ∼1.8 μm (Fig. 1D), nearly identical to our in vitro measurement of the IC50 for PI3Kα (Fig. 1E). We also profiled our compound against other PI3K isoforms using the Adapta in vitro assay method, which confirmed a high degree of selectivity for mTOR (Fig. 1F).
Compounds that inhibit PI3K and mTOR also have the potential to inhibit other PI3K-like kinases, including the DNA-damage response kinases ATM and DNA-PK. For DNA-PK and ATM, we measured the IC50 of Torin1 using in vitro assays (Fig. 1E). We also measured inhibition of the Class III PI3K hVps34. Some reports have proposed that hVps34 acts upstream of mTORC1, and we wanted to be sure that cross-reactivity with this kinase was not indirectly influencing mTORC1 activity in cells (22). Torin1 was at least 200-fold selective for mTOR over each of these kinases. Finally, we screened Torin1 at a concentration of 10 μm against a panel of 353 diverse kinases using the Ambit Biosciences KinomeScan screening platform, which measures the relative binding of the target molecule to each kinase, and we found no indication of significant off-target effects (data shown in supplemental material). These results suggest that Torin1 is a highly selective inhibitor of mTOR when profiled against an extensive panel of serine/threonine, tyrosine, and lipid kinases.
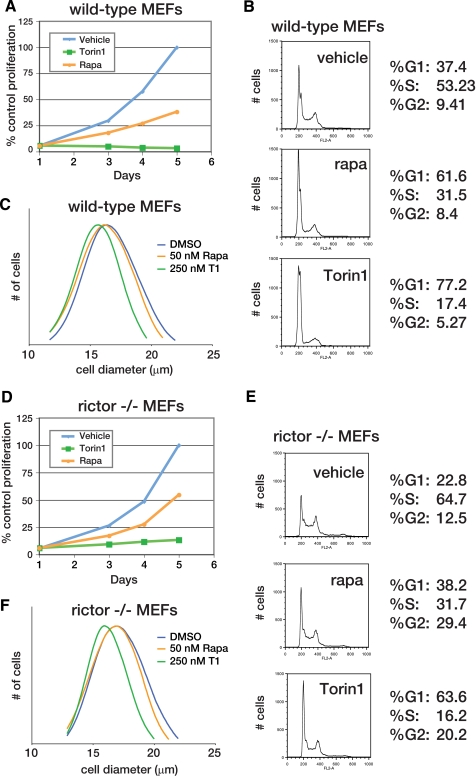
Torin1 Causes Cell Cycle Arrest through a Rapamycin-resistant Mechanism That Is Also Independent of mTORC2—Our next goal was to test the role of mTOR signaling in normally growing cells. Rapamycin-mediated mTORC1 inhibition slows cell proliferation and reduces cell size, and so we suspected that dual mTORC1/2 inhibition would have similar but more severe effects (22). Indeed, wild-type MEFs treated with up to 500 nm rapamycin continued to proliferate, albeit at a reduced rate (Fig. 2A and supplemental Fig. S2). In contrast, 250 nm Torin1 completely inhibited proliferation (Fig. 2A and supplemental Fig. S2) and caused a G1/S cell cycle arrest (Fig. 2B). Moreover, 250 nm Torin1 decreased cell size to a greater degree than 50 nm rapamycin (Fig. 2C). Based on the assumption that rapamycin completely disables mTORC1 kinase activity, we hypothesized that the enhanced effect of Torin1 was because of mTORC2 inhibition.
FIGURE 2.
mTOR inhibition prevents cell growth and proliferation through an mTORC2-independent mechanism. A, mTOR inhibition by Torin1 but not rapamycin prevents the proliferation of wild-type MEFs. MEF (p53-/-) cells were grown in the presence of vehicle (blue), 50 nm rapamycin (orange), or 250 nm Torin1 (green) for 4 days. Cell proliferation was measured in triplicate at indicated time points using the CellTiterGlo viability assay. B, Torin1 causes a G1/S cell cycle arrest in wild-type MEFs. MEF (p53-/-) cells were treated with vehicle (DMSO), 50 nm rapamycin (rapa), or 250 nm Torin1 for 48 h. Cells were then harvested, stained with propidium iodide, and analyzed by flow cytometry. C, normalized cell size distributions for Torin1 and rapamycin-treated wild-type MEFs. MEF (p53-/-) cells were treated with vehicle (blue, mean 17.81 μm), 50 nm rapamycin (orange, mean 17.58), or 250 nm Torin1 (green, mean 16.46 μm) for 24 h. Cell sizes were measured using a particle counter and are displayed as a histogram. D, experiment was performed as in A using Rictor-/-, p53-/- MEFs. E, experiment was performed as in B using Rictor-/-, p53-/- MEFs. F, experiment was performed as in C using Rictor-/-, p53-/- MEFs. Cells were treated with vehicle (blue, mean diameter 17.85 μm), 50 nm rapamycin (orange, mean diameter 17.33 μm), or 250 nm Torin1 (green, mean diameter 16.24 μm).
To test this hypothesis, we conducted identical experiments using MEFs that lack mTORC2 activity because Rictor has been deleted (16). We reasoned that Torin1 should have the same effect as rapamycin on the proliferation and growth of these cells because mTORC2 is already inhibited. As in wild-type MEFs, rapamycin reduced but did not prevent proliferation (Fig. 2D). However, we were surprised to find that Torin1 continued to dramatically suppress proliferation and diminish cell size (Fig. 2, D-F), indicating that the differential effects of this compound with respect to rapamycin were not due to mTORC2 inhibition. Thus, mTOR has functions that are absolutely required for cell growth and proliferation and that are kinase-dependent, rapamycin-resistant, and independent of mTORC2.
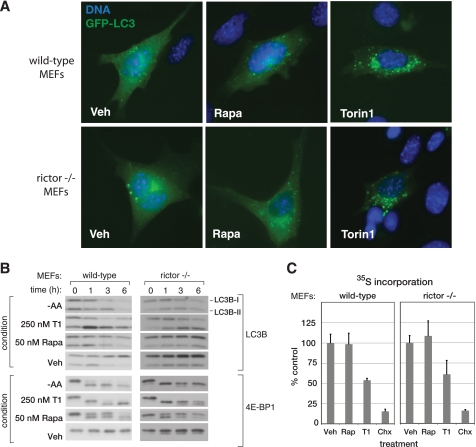
Torin1 Disrupts mTORC1-dependent Phenotypes More Completely than Rapamycin—Despite the widely held assumption to the contrary, one explanation for our results is that rapamycin inhibits some but not all of the functions of mTORC1. To explore this possibility, we examined the effects of Torin1 on other processes besides growth and proliferation that are commonly associated with mTORC1 signaling. One such process is macroautophagy, often referred to simply as autophagy. Normally considered a response to starvation conditions, autophagy involves the formation of large double-membrane enclosed vesicles that engulf cytoplasmic contents, including both proteins and organelles (reviewed in Ref. 27). These vesicles then fuse with lysosomes to form autophagosomes that digest their contents, providing the cell with a source of amino acids and other nutrients when these are not available from the environment.
In yeast, rapamycin is a potent activator of autophagy (10). The situation is less clear in mammalian systems, where rapamycin alone is, at best, an inconsistent activator of autophagy and frequently requires combination with other PI3K/mTOR inhibitors, such as LY294002, or concomitant starvation for nutrients. We suspected that autophagy might also be regulated in part by rapamycin-resistant functions of mTORC1. A commonly used marker of autophagy is the protein light chain 3 (LC3), which translocates from the cytoplasm to autophagosomes where it is degraded when autophagy is induced (28). Using a green fluorescent protein-tagged LC3 construct, we found that Torin1 causes a strong re-localization of LC3 from the cytoplasm to autophagosomes in both wild-type and Rictor-/- MEFs, whereas rapamycin caused only a minor change (Fig. 3A). Furthermore, we found that Torin1 treatment, like amino acid starvation, causes degradation of LC3B (LC3B-I) and transient accumulation of the faster running lipidated form (LC3B-II) in both MEFs and HeLa cells (Fig. 3B and supplemental Fig. S4A). An RNA interference-induced decrease in Raptor expression affected LC3 in a similar fashion as Torin1 treatment (supplemental Fig. S4B). Collectively, these results suggest that mTORC1 inhibition is sufficient to induce autophagy. Although the signaling mechanisms that connect mTORC1 to autophagy are currently unclear, ATP-competitive inhibitors, like Torin1, will likely reveal specific roles for mTORC1 that have been missed because of their insensitivity to rapamycin.
FIGURE 3.
Torin1 inhibits mTORC1-dependent processes that are resistant to rapamycin. A, Torin1 but not rapamycin (Rapa) causes LC3 to relocalize from the cytoplasm to autophagosomes. Wild-type (p53-/-) or Rictor-null (p53-/-) MEFs were transiently transfected with GFP-LC3 and treated with vehicle (Veh) (DMSO), 50 nm rapamycin, or 250 nm Torin1 for 3 h before being fixed and processed. Cells were also stained with Hoechst to visualize nuclei and imaged at ×63. B, amino acid starvation and Torin1, but not rapamycin, cause LC3 degradation. Wild-type (p53-/-) and Rictor-null (p53-/-) MEFs were treated with vehicle (DMSO), 50 nm rapamycin, 250 nm Torin1 or grown in amino acid (AA)-free conditions for 0, 1, 3, or 6 h. Cells were lysed at the indicated time points and analyzed by immunoblotting. Induction of autophagy causes the degradation of the native LC3B (LC3B-I) protein and the transient accumulation of the faster running lipidated version (LC3B-II). C, Torin1 suppresses global protein synthesis through a rapamycin-resistant and mTORC2-independent process. Wild-type (p53-/-) and Rictor-null (p53-/-) MEFs were treated with vehicle (DMSO), 50 nm rapamycin (Rap), 250 nm Torin1, or 10 μg/ml cycloheximide (Chx) for 2.5 h and then pulsed with 35S-labeled methionine and cysteine for 30 min. The amount of 35S incorporation was determined by scintillation counting. Measurements were made in triplicate, and error bars indicate standard deviation.
The mTORC1 pathway also has many connections to the regulation of cap-dependent translation. However, rapamycin often has only modest effects on rates of protein synthesis. To test whether Torin1 might inhibit protein synthesis more completely, we metabolically labeled cells using 35S methionine/cysteine in the presence of either Torin1 or rapamycin. Surprisingly, whereas rapamycin had very little effect, Torin1 caused a nearly 50% decline in total protein synthesis in both wild-type and Rictor-/- MEFs (Fig. 3C). As with autophagy, these results indicate that mTORC1 is a far more important regulator of protein synthesis than experiments with rapamycin have indicated.
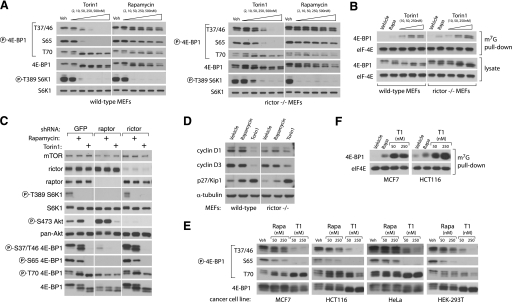
Rapamycin-resistant Functions of mTORC1 Are Required for Cap-dependent Translation—Because known mTORC1 substrates, S6K and 4E-BP1, are important regulators of mRNA translation, we next considered whether either is involved in the transduction of mTORC1-dependent but rapamycin-resistant functions. S6K activity has been shown to be completely inhibited by rapamycin treatment, and therefore we considered it unlikely to be the target of any rapamycin-resistant activity of mTORC1. 4E-BP1, however, is subject to a more complex regulatory process. The ability of 4E-BP1 to bind and inhibit eIF-4E is primarily regulated by the phosphorylation of four residues: Thr-37, Thr-46, Ser-65, and Thr-70. Phosphorylation of Thr-37 and Thr-46 is thought to be a priming event that permits the phosphorylation of the other two, thereby promoting dissociation from eIF-4E and permitting the formation of a functional eIF-4F complex (29). mTORC1 has been implicated in the regulation of 4E-BP1, but there are conflicting accounts of the importance of this connection as well as the underlying mechanism. For instance, mTORC1 phosphorylates the Thr-37 and Thr-46 sites in vitro, but these sites are considered rapamycin-insensitive in cells (30-32). Conversely, mTORC1 has little effect in vitro on the phosphorylation of sites that are considered rapamycin-sensitive, Ser-65 and Thr-70. Moreover, a C-terminal motif in 4E-BP1, known as the TOR signaling motif and believed to mediate binding to mTORC1, and the N-terminal RAIP motif are required for phosphorylation of all sites (33-35). Finally, although rapamycin causes a substantial decrease in overall protein translation in some cell types (36), it has very little effect in others (13). A possible explanation is simply that rapamycin cannot completely inhibit mTORC1-dependent phosphorylation of 4E-BP1.
To test this hypothesis, we treated MEFs with increasing concentrations of either Torin1 or rapamycin and assessed the phosphorylation status of Thr-36, Thr-47, Ser-65, and Thr-70 by immunoblotting (Fig. 4A). Rapamycin completely prevented phosphorylation of S6K1 and caused a slight decrease in the phosphorylation of Ser-65 of 4E-BP1, but it had little effect on the phosphorylation of either Thr-37/46 or Thr-70 even at concentrations as high as 500 nm, over 500 times greater than its IC50 value for inhibition of mTORC1 (Fig. 4A). In striking contrast, Torin1 substantially suppressed phosphorylation of Thr-37/46 and Ser-65 at concentrations as low as 10 nm and abolished it completely at 250 nm (Fig. 4A). Torin1 had nearly identical effects in Rictor-null MEFs, consistent with the hypothesis that these effects are because of inhibition of mTORC1 (Fig. 4A). Surprisingly, Thr-70 was unaffected by either Torin1 or rapamycin, supporting earlier predictions that it may be the target of a different kinase, such as Erk2 (37). Alternatively, it is possible that the Thr-70 4E-BP1 antibody is not phospho-specific. The dual-PI3K/mTOR inhibitors PI-103 and NVP-BEZ235 caused similar effects as Torin1 on 4E-BP1 phosphorylation (supplemental Fig. S3). Additionally, Torin1 had much greater effects than rapamycin on 4E-BP1 phosphorylation in a variety of human tumor cell lines, indicating that rapamycin resistance of mTORC1 is likely a general feature of most if not all mammalian systems (Fig. 4E). We next asked whether the increased dephosphorylation of 4E-BP1 by Torin1 led to increased association with eIF-4E. Using 7-methyl-GTP-Sepharose to purify eIF-4E from cell lysates, we found that Torin1 causes substantially more binding of 4E-BP1 to eIF-4E than does rapamycin (Fig. 4, B and F). Torin1 did not affect the phosphorylation of eIF2 (supplemental Fig. S5).
FIGURE 4.
mTORC1 regulation of 4E-BP1 phosphorylation and binding to eIF-4E reveals rapamycin-resistant functions. A, phosphorylation of 4E-BP1 at Thr-37/46 and Ser-65 is dependent on mTORC1 but resistant to rapamycin. Wild-type (p53-/-) and Rictor-null (p53-/-) MEFs were treated with the indicated concentrations of Torin1 or rapamycin for 1 h and then lysed. Cell lysates were analyzed by immunoblot using antibodies specific for the indicated proteins or phosphorylation states. B, Torin1 increases the amount of 4E-BP1 bound to eIF-4E to a degree that far exceeds the effects of rapamycin. Wild-type (p53-/-) and Rictor-null (p53-/-) MEFs were treated with vehicle (DMSO), 50 nm rapamycin, or 250 nm Torin1 for 1 h before lysis. eIF-4E was purified from lysates using 7-methyl-GTP-Sepharose and analyzed by immunoblotting for the indicated proteins. C, phosphorylation of Thr-36/47 on 4E-BP1 requires Raptor but not Rictor. MEFs (p53-/-) were infected with lentivirus expressing either control, Raptor-specific, or Rictor-specific shRNAs. Cells were grown for 4 days and then treated with vehicle (DMSO), 50 nm rapamycin, or 250 nm Torin1 for 1 h. Cell lysates were then analyzed by immunoblot using antibodies specific for the indicated proteins or phosphorylation states. D, prolonged mTOR inhibition alters the expression of key cell cycle regulators. Wild-type (p53-/-) and Rictor-null (p53-/-) MEFs were treated with vehicle (DMSO), 50 nm rapamycin, or 250 nm Torin1 for 48 h. Cell lysates were then analyzed by immunoblotting using antibodies specific for the indicated proteins. E, Torin1 prevents phosphorylation of rapamycin-resistant sites in human cancer cell lines. MCF7, HCT116, HeLa, and HEK-293T cell lines were treated with vehicle (Veh), rapamycin (Rap) (50 or 250 nm), or Torin1 (50 or 250 nm) for 1 h and then analyzed by immunoblotting for the indicated proteins and phosphorylation states. F, Torin1 increases the amount of 4E-BP1 bound to eIF-4E in human cancer cell lines. MCF7 and HCT116 cells were treated with vehicle (DMSO), 50 nm rapamycin, 50 nm Torin1, or 250 nm Torin1 for 48 h before lysis. eIF-4E was purified from lysates using 7-methyl-GTP-Sepharose and analyzed by immunoblotting for the indicated proteins.
Because the effects of Torin1 were nearly equivalent in wild-type and Rictor-null MEFs, we concluded that they could not be dependent on mTORC2. However, it remained possible that mTOR alone or an unidentified mTORC3 were responsible. To show that mTORC1 inhibition is sufficient to explain the effects of Torin1 on 4E-BP1 phosphorylation, we used RNA interference to knock down Raptor, an obligatory mTORC1 component, in wild-type MEFs (Fig. 4C). Depletion of Raptor in these cells suppressed Thr-37/46 and Ser-65 phosphorylation and 4E-BP1 mobility to a degree that equaled the effects of Torin1 and exceeded those of rapamycin, thereby supporting the conclusion that mTORC1, or at least a Raptor-containing mTOR complex, regulates 4E-BP1 phosphorylation through a rapamycin-insensitive kinase-dependent mechanism.
Defects in cap-dependent translation are also known to cause cell cycle arrest. This is thought to occur primarily through decreased translation of cap-dependent mRNAs that encode factors that promote cell cycle progression, such as cyclin D1 and cyclin D3, and increased translation of cap-independent mRNAs that encode factors that suppress it, such as p27Kip1 (38-40). Moreover, recent work has shown that the depletion of cyclin D1 that is caused by amino acid starvation and rapamycin treated is mediated by 4E-BP1 (41). We suspected that the cell cycle arrest caused by Torin1 might be explained by changes in the abundance of these factors. Consistent with this, both wild-type and Rictor-null MEFs treated for 48 h with Torin1, but not rapamycin, had greatly depleted levels of cyclin D1 and D3, and a strong induction of p27/Kip1 (Fig. 4D). The ability of cells to recover from this arrest upon the removal of Torin1 was highly dependent on cell type (data not shown).
DISCUSSION
Rapamycin has been an indispensable tool throughout the history of TOR research and remains widely employed as a “complete” mTORC1 inhibitor in both research and clinical settings. Indeed, in yeast, it is a convincing mimic of the genetic inactivation of TORC1. In mammalian systems, most known mTOR substrates were discovered and validated using rapamycin as a pharmacological probe. Rapamycin forms a complex with the intracellular protein FKBP12, which then binds to the FRB domain of mTOR and inhibits phosphorylation of substrates through a poorly characterized mechanism. Although structural information is available for rapamycin in a complex with FKBP12 and the FRB domain of mTOR, it remains unclear how this prevents phosphorylation of direct mTOR kinase substrates (42). A model to explain our findings is that rapamycin blocks access to only a specific subset of mTORC1 substrates, whereas Torin1, because of its ATP-competitive mode of action, blocks phosphorylation of all. Additionally, as Torin1 is much smaller than FKBP12-rapamycin, it likely accesses its target site in mTOR-containing complexes more easily than FKBP12-rapamycin.
Re-interpretations of several recent studies support the notion that considerable mTORC1 functionality is resistant to rapamycin. Shor et al. (13) found that high concentrations (10 μm) of rapamycin inhibit mTOR directly through an FKBP12-independent mechanism, suppressing both mTORC1 and mTORC2. Unlike the commonly used “low dose” (10-50 nm) and similarly to Torin1, “high-dose” rapamycin potently suppresses cap-dependent translation and inhibits proliferation in a wide variety of tumor cell lines. Although these authors concluded that these effects are because of mTORC2 inhibition, our findings indicate that they are more likely because of inhibition of rapamycin-resistant mTORC1-dependent functions. A study from Averous et al. (41) found that amino acid starvation caused a more complete depletion of cyclin D1 than rapamycin treatment and that this effect was mediated through 4E-BP1. Based on the assumption that rapamycin completely disables mTORC1, these authors concluded that amino acid starvation signals to 4E-BP1 through additional pathways besides mTORC1. We would suggest that it is more likely that amino acid starvation leads to a more complete inhibition of mTORC1 functions than does rapamycin. Finally, Choo et al. (43) found that phosphorylation sites on 4E-BP1 that are acutely sensitive to rapamycin become re-phosphorylated in some cell lines after long periods of rapamycin treatment. Moreover, the recovery of 4E-BP1 phosphorylation depends on the mTORC1 component Raptor, leading the authors to conclude that prolonged rapamycin treatment confers on mTORC1 the capacity to phosphorylate 4E-BP1 in a rapamycin-resistant fashion. We find that mTORC1 likely has rapamycin-resistant functions in all cell lines (Fig. 4E). Because prolonged rapamycin treatment is known to hyperactivate the PI3K pathway, which is upstream of mTORC1, one possible explanation for the results of Choo et al. (43) is that rapamycin leads to the hyperactivation of the rapamycin-resistant functionality of mTORC1, effectively overcoming the partial inhibition caused by rapamycin.
Because many important features of TOR signaling are conserved between yeast and mammals, our finding that mTORC1 possesses cell-essential but rapamycin-resistant functions is unexpected. At the same time, our results indicate that the requirements for TORC1 signaling in maintaining protein synthesis and promoting cell division are more similar between yeast and mammalian systems than had been appreciated. Although we have focused on the rapamycin-insensitive regulation of 4E-BP1, we consider it likely that other similar mTORC1 substrates exist, particularly among the regulators of autophagy. The future combined use of Torin1 and phosphoproteomics will likely permit a more comprehensive assessment of all mTOR substrates. Given the current enthusiasm for rapamycin as a potential therapeutic, it is likely that ATP-competitive inhibitors of mTOR will have clinical utility as well.
Supplementary Material
Acknowledgments
We thank D. Kwiatowski (Harvard Medical School) for p53-/-/TSC2-/- and p53-/-/TSC2+/+ MEFs; Christian Reinhardt (Massachusetts Institute of Technology) for p53 reagents; and Chris Armstong (Invitrogen) for kinase profiling assistance. We also thank members of the Sabatini and Gray laboratories for helpful discussions and Ambit Biosciences for performing KinomeScan profiling.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI47389 and R01 CA103866. This work was also supported by start-up funding from the Dana Farber Cancer Institute, the Barr Foundation, and the Damon Runyon Cancer Research Foundation (to N. S. G.), Keck Foundation, LAM Foundation, and Department of Defense Grant W81XWH-07-1-0448 (to D. M. S.), and a fellowship from the American Cancer Society (to S. A. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. S1-S5, and an additional reference.
Footnotes
The abbreviations used are: mTOR, mammalian target of rapamycin; Raptor, regulatory associated protein of mTOR; Rictor, rapamycin-insensitive companion of mTOR; PI3K, phosphatidylinositol 3-kinase; LC3, light chain 3; 4E-BP1, eIF4E-binding protein 1; eIF4E, eukaryotic initiation factor 4E; GβL, G-β subunit-like; MEF, mouse embryonic fibroblast; mTORC1, mTOR complex 1; mTORC2, mTOR complex 2; HEK, human embryonic kidney; DMSO, dimethyl sulfoxide; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; ATM, ataxia telangiectasia, mutated; PI, phosphatidylinositol; BSA, bovine serum albumin; PBS, phosphate-buffered saline; shRNA, short hairpin RNA.
N. S. Gray, manuscript in preparation.
References
- 1.Guertin, D. A., and Sabatini, D. M. (2007) Cancer Cell 12 9-22 [DOI] [PubMed] [Google Scholar]
- 2.Manning, B. D., and Cantley, L. C. (2007) Cell 129 1261-1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sehgal, S. N. (2003) Transplant. Proc. 35 S7-S14 [Google Scholar]
- 4.Sarbassov, D. D., Ali, S. M., Sengupta, S., Sheen, J. H., Hsu, P. P., Bagley, A. F., Markhard, A. L., and Sabatini, D. M. (2006) Mol. Cell 22 159-168 [DOI] [PubMed] [Google Scholar]
- 5.Richter, J. D., and Sonenberg, N. (2005) Nature 433 477-480 [DOI] [PubMed] [Google Scholar]
- 6.Holz, M. K., Ballif, B. A., Gygi, S. P., and Blenis, J. (2005) Cell 123 569-580 [DOI] [PubMed] [Google Scholar]
- 7.Raught, B., Peiretti, F., Gingras, A. C., Livingstone, M., Shahbazian, D., Mayeur, G. L., Polakiewicz, R. D., Sonenberg, N., and Hershey, J. W. (2004) EMBO J. 23 1761-1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shahbazian, D., Roux, P. P., Mieulet, V., Cohen, M. S., Raught, B., Taunton, J., Hershey, J. W., Blenis, J., Pende, M., and Sonenberg, N. (2006) EMBO J. 25 2781-2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbet, N. C., Schneider, U., Helliwell, S. B., Stansfield, I., Tuite, M. F., and Hall, M. N. (1996) Mol. Biol. Cell 7 25-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noda, T., and Ohsumi, Y. (1998) J. Biol. Chem. 273 3963-3966 [DOI] [PubMed] [Google Scholar]
- 11.Neshat, M. S., Mellinghoff, I. K., Tran, C., Stiles, B., Thomas, G., Petersen, R., Frost, P., Gibbons, J. J., Wu, H., and Sawyers, C. L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10314-10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedersen, S., Celis, J. E., Nielsen, J., Christiansen, J., and Nielsen, F. C. (1997) Eur. J. Biochem. 247 449-456 [DOI] [PubMed] [Google Scholar]
- 13.Shor, B., Zhang, W. G., Toral-Barza, L., Lucas, J., Abraham, R. T., Gibbons, J. J., and Yu, K. (2008) Cancer Res. 68 2934-2943 [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi, H., Kondo, Y., Fujiwara, K., Kanzawa, T., Aoki, H., Mills, G. B., and Kondo, S. (2005) Cancer Res. 65 3336-3346 [DOI] [PubMed] [Google Scholar]
- 15.Sabatini, D. M. (2006) Nat. Rev. Cancer 6 729-734 [DOI] [PubMed] [Google Scholar]
- 16.Guertin, D. A., Stevens, D. M., Thoreen, C. C., Burds, A. A., Kalaany, N. Y., Moffat, J., Brown, M., Fitzgerald, K. J., and Sabatini, D. M. (2006) Dev. Cell 11 859-871 [DOI] [PubMed] [Google Scholar]
- 17.Moffat, J., Grueneberg, D. A., Yang, X., Kim, S. Y., Kloepfer, A. M., Hinkle, G., Piqani, B., Eisenhaure, T. M., Luo, B., Grenier, J. K., Carpenter, A. E., Foo, S. Y., Stewart, S. A., Stockwell, B. R., Hacohen, N., Hahn, W. C., Lander, E. S., Sabatini, D. M., and Root, D. E. (2006) Cell 124 1283-1298 [DOI] [PubMed] [Google Scholar]
- 18.Ali, S. M., and Sabatini, D. M. (2005) J. Biol. Chem. 280 19445-19448 [DOI] [PubMed] [Google Scholar]
- 19.Sarbassov, D. D., Guertin, D. A., Ali, S. M., and Sabatini, D. M. (2005) Science 307 1098-1101 [DOI] [PubMed] [Google Scholar]
- 20.Sancak, Y., Thoreen, C. C., Peterson, T. R., Lindquist, R. A., Kang, S. A., Spooner, E., Carr, S. A., and Sabatini, D. M. (2007) Mol. Cell 25 903-915 [DOI] [PubMed] [Google Scholar]
- 21.Knight, Z. A., Gonzalez, B., Feldman, M. E., Zunder, E. R., Goldenberg, D. D., Williams, O., Loewith, R., Stokoe, D., Balla, A., Toth, B., Balla, T., Weiss, W. A., Williams, R. L., and Shokat, K. M. (2006) Cell 125 733-747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nobukuni, T., Joaquin, M., Roccio, M., Dann, S. G., Kim, S. Y., Gulati, P., Byfield, M. P., Backer, J. M., Natt, F., Bos, J. L., Zwartkruis, F. J., and Thomas, G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14238-14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunn, G. J., Williams, J., Sabers, C., Wiederrecht, G., Lawrence, J. C., Jr., and Abraham, R. T. (1996) EMBO J. 15 5256-5267 [PMC free article] [PubMed] [Google Scholar]
- 24.Maira, S. M., Stauffer, F., Brueggen, J., Furet, P., Schnell, C., Fritsch, C., Brachmann, S., Chene, P., De Pover, A., Schoemaker, K., Fabbro, D., Gabriel, D., Simonen, M., Murphy, L., Finan, P., Sellers, W., and Garcia-Echeverria, C. (2008) Mol. Cancer Ther. 7 1851-1863 [DOI] [PubMed] [Google Scholar]
- 25.Biondi, R. M., Kieloch, A., Currie, R. A., Deak, M., and Alessi, D. R. (2001) EMBO J. 20 4380-4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheid, M. P., Marignani, P. A., and Woodgett, J. R. (2002) Mol. Cell. Biol. 22 6247-6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima, N., Levine, B., Cuervo, A. M., and Klionsky, D. J. (2008) Nature 451 1069-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., Kominami, E., Ohsumi, Y., and Yoshimori, T. (2000) EMBO J. 19 5720-5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gingras, A. C., Gygi, S. P., Raught, B., Polakiewicz, R. D., Abraham, R. T., Hoekstra, M. F., Aebersold, R., and Sonenberg, N. (1999) Genes Dev. 13 1422-1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnett, P. E., Barrow, R. K., Cohen, N. A., Snyder, S. H., and Sabatini, D. M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 1432-1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fingar, D. C., Richardson, C. J., Tee, A. R., Cheatham, L., Tsou, C., and Blenis, J. (2004) Mol. Cell. Biol. 24 200-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, X., Beugnet, A., Murakami, M., Yamanaka, S., and Proud, C. G. (2005) Mol. Cell. Biol. 25 2558-2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi, K. M., McMahon, L. P., and Lawrence, J. C., Jr. (2003) J. Biol. Chem. 278 19667-19673 [DOI] [PubMed] [Google Scholar]
- 34.Schalm, S. S., Fingar, D. C., Sabatini, D. M., and Blenis, J. (2003) Curr. Biol. 13 797-806 [DOI] [PubMed] [Google Scholar]
- 35.Tee, A. R., and Proud, C. G. (2002) Mol. Cell. Biol. 22 1674-1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beretta, L., Gingras, A. C., Svitkin, Y. V., Hall, M. N., and Sonenberg, N. (1996) EMBO J. 15 658-664 [PMC free article] [PubMed] [Google Scholar]
- 37.Herbert, T. P., Tee, A. R., and Proud, C. G. (2002) J. Biol. Chem. 277 11591-11596 [DOI] [PubMed] [Google Scholar]
- 38.Albers, M. W., Williams, R. T., Brown, E. J., Tanaka, A., Hall, F. L., and Schreiber, S. L. (1993) J. Biol. Chem. 268 22825-22829 [PubMed] [Google Scholar]
- 39.Jiang, H., Coleman, J., Miskimins, R., and Miskimins, W. K. (2003) Cancer Cell Int. 3 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenwald, I. B., Lazaris-Karatzas, A., Sonenberg, N., and Schmidt, E. V. (1993) Mol. Cell. Biol. 13 7358-7363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Averous, J., Fonseca, B. D., and Proud, C. G. (2008) Oncogene 27 1106-1113 [DOI] [PubMed] [Google Scholar]
- 42.Choi, J., Chen, J., Schreiber, S. L., and Clardy, J. (1996) Science 273 239-242 [DOI] [PubMed] [Google Scholar]
- 43.Choo, A. Y., Yoon, S. O., Kim, S. G., Roux, P. P., and Blenis, J. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 17414-17419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.