Abstract
Platelet-derived growth factor (PDGF) β receptor activation inhibits N-methyl-d-aspartate (NMDA)-evoked currents in hippocampal and cortical neurons via the activation of phospholipase Cγ, PKC, the release of intracellular calcium, and a rearrangement of the actin cytoskeleton. In the hippocampus, the majority of NMDA receptors are heteromeric; most are composed of 2 NR1 subunits and 2 NR2A or 2 NR2B subunits. Using NR2B- and NR2A-specific antagonists, we demonstrate that PDGF-BB treatment preferentially inhibits NR2B-containing NMDA receptor currents in CA1 hippocampal neurons and enhances long-term depression in an NR2B subunit-dependent manner. Furthermore, treatment of hippocampal slices or cultures with PDGF-BB decreases the surface localization of NR2B but not of NR2A subunits. PDGFβ receptors colocalize to a higher degree with NR2B subunits than with NR2A subunits. After neuronal injury, PDGFβ receptors and PDGF-BB are up-regulated and PDGFβ receptor activation is neuroprotective against glutamate-induced neuronal damage in cultured neurons. We demonstrate that the neuroprotective effects of PDGF-BB are occluded by the NR2B antagonist, Ro25-6981, and that PDGF-BB promotes NMDA signaling to CREB and ERK1/2. We conclude that PDGFβR signaling, by preferentially targeting NR2B receptors, provides an important mechanism for neuroprotection by growth factors in the central nervous system.
N-Methyl-d-aspartate (NMDA)2 receptors are tetrameric, non-selective cation channels composed of two obligate NR1 and two variably expressed NR2 subunits (NR2A-D) although trimeric receptors (2NR1/NR2A/NR2B) are also likely to be present (1). In the hippocampus, NR2A and NR2B are the dominant NR2 forms (2, 3). The majority of synaptic receptors appear to contain the NR2A subunit, whereas NR2B-containing receptors are in greater density extrasynaptically (2, 4, 5), however, this paradigm has recently been challenged (6). Extrasynaptic NMDA receptors or, alternatively, the NR2B-containing NMDA receptors have been directly implicated in the Ca2+ influx that causes excitotoxic cell death (7-10), whereas the synaptic, NR2A-containing receptors have paradoxically been linked to neuroprotection in ischemia (11, 12).
Platelet-derived growth factor (PDGF) receptors are highly expressed in CA1 hippocampal neurons and are developmentally important but their functions in the mature CNS are largely unknown (13). There are two major PDGF receptor isoforms (PDGFα and PDGFβ receptors) found in the hippocampus (14). The most intense staining for PDGFβ receptors is found in the pyramidal cells of the hippocampus (15), whereas PDGFα receptors are located primarily on glial cells in this region (16). We have previously demonstrated that the PDGFβ receptor ligand PDGF-BB inhibits synaptic NMDA receptors at CA1 synapses (17) as well as NMDA-evoked currents of neurons acutely isolated from the CA1 region of the hippocampus (17, 18) or from the prefrontal cortex (19).
PDGFβ receptors have been implicated in neuroprotection following ischemic events (20). For example, focal ischemia in rat brain causes a rapid increase in PDGF-B chain isoform mRNA transcripts that peaks at 24 h (21, 22). Furthermore, PDGFβ receptor expression rises rapidly after ischemia in rat brain (22) and 24-h applications of PDGF-BB is neuroprotective against glutamate- or NMDA-induced death of cultured hippocampal neurons (23). These results lead us to hypothesize that PDGFβ receptor-mediated neuroprotection involves the inhibition of NMDA receptor signaling, specifically the inhibition of NR2B-containing NMDA receptors that are associated with glutamate-induced toxicity.
We demonstrate that PDGF-BB preferentially inhibits NR2B- and not NR2A-containing subtypes of NMDA receptors. In addition, treatment of hippocampal neurons with PDGF-BB selectively decreases the surface expression of NR2B subunits and their phosphorylation level. Furthermore, we found that PDGFβ receptors are predominantly localized extrasynaptically and they colocalize with NR2B subunits. PDGF-BB application also altered the ability of NMDA to signal to transcription factors in hippocampal neurons. This work illustrates that the neuroprotective effects of PDGF-BB treatment of hippocampal neurons are due at least in part to a preferential decrease in the activity of NR2B-containing NMDA receptors.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—PDGF-BB was purchased from R&D Systems (Minneapolis, MN). Ro25-6981 was purchased from Tocris Bioscience (Bristol, UK). NVPAAM077 was a generous gift from Novartis (Basel, Switzerland). NMDA, glycine, kainate, and all other chemical reagents were purchased from Sigma. Antibodies used include those raised against PSD-95 (Abcam, Cambridge, MA), PDGFβ receptor (Epitomics, CA), phosphotyrosine 1021 PDGFβ receptor (Santa Cruz, Santa Cruz, CA), phospho-ERK1/2 and ERK1/2 (Cell Signaling, Danvers, MA), NR1, NR2B, and NR2A antibodies from Chemicon (Temecula, CA) were used for biotinylation and Western blotting, and NR2A (Covance, Berkeley, CA) and NR2B antibodies (Zymed Laboratories Inc., San Francisco, CA) were used for immunocytochemistry. Anti-α-actinin was purchased from Sigma. For immunoprecipitation and phosphorylation experiments, anti-phosphotyrosine 1472 (Biomol), anti-NR2B (Santa Cruz), anti-phosphotyrosine (Santa Cruz), anti-NR1 phosphoserine 896 and 897, and anti-NR1 (Upstate) antibodies were employed as well as anti-PDGFβ receptor antibodies precoupled to agarose beads (Santa Cruz). All secondary antibodies including Alexa 488, Alexa 568, and streptavidin-conjugated Alexa 568 were obtained from Molecular Probes (Eugene, OR).
Cell Isolation and Whole Cell Recording—CA1 neurons were isolated from hippocampal slices of postnatal day 14-21 Wistar rats as previously described (24). The extracellular solution was composed of 140 mm NaCl, 1.3 mm CaCl2, 25 mm HEPES, 33 mm glucose, 5.4 mm KCl, 0.5 μm tetrodotoxin, and 0.5 μm glycine, with pH 7.3-7.4 and osmolarity ranging from 320 to 330 mosmol. Recordings were done at room temperature. The membrane potential was held at -60 mV throughout the recordings and a voltage step of 10 mV was applied prior to NMDA application to monitor series resistance. The intracellular solution consisted of 11 mm EGTA as intracellular calcium chelating buffer, 10 mm HEPES, 2 mm MgCl2, 2 mm tetraethylammonium chloride to block K+ channel, 1 mm CaCl2, 140 mm cesium fluoride, and 4 mm K2ATP. NMDA currents were evoked by rapid application of NMDA solution delivered from a multibarreled fast perfusion system for 3 s every minute. The solution was delivered at a rate of ∼1 ml/min.
Hippocampal Slice Recording—Hippocampal slices were prepared from 3- to 4-week-old male Wistar rats and placed in a holding chamber for at least 1 h prior to recording. A single slice was transferred to a recording chamber and superfused with artificial cerebral spinal fluid (5 ml/min) composed of (in mm) 120 NaCl, 4 KCl, 1.25 NaH2PO4, 1.3 MgCl2, 2.6 CaCl2, 26 NaHCO3, 10 d-glucose, saturated with 95% O2, 5% CO2 at room temperature. Synaptic responses were evoked with bipolar tungsten electrodes located about 50 μm from the cell body layer in CA1. Field potential recordings were made with glass micropipettes filled with artificial cerebral spinal fluid placed in the stratum radiatum 60-80 μm from the cell body layer. Test stimuli were evoked at 0.033 Hz with the stimulus intensity set to 70% of that which produced maximum synaptic responses. Long-term depression was induced by a 1-Hz, 15-min low-frequency stimulation delivered through the stimulation electrode. The value of fEPSP amplitude from the 10-min period immediately before tetanus was defined as baseline (100%). The magnitude of LTD was determined by averaging the amplitude of fEPSPs collected 50-60 min after the low-frequency stimulation. Data were recorded with an Axoclamp 2B amplifier and sampled at 10 kHz by computer. Statistical significance of data were determined using Student's t test or analysis of variance with a level of significance, p < 0.05.
Surface Biotinylation Assay—The CA1 region of the hippocampus was microdissected from day 14-22 Wistar rat pups. CA1 slices (5-10 per condition) were incubated for 10 min with control or PDGF-BB. Slices were washed in ice-cold ECF and incubated with 0.5 mg/ml Sulfo-NHS-LC-biotin (Pierce) for 30 min. The biotin reaction was quenched by washing with 10 mm glycine. Slices were washed twice more and homogenized in solubilization buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 30 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1% Triton, 1 mm Na3VO4, and MINI mixture (Roche) supplemented with 1% SDS. Total protein concentration was determined by BCA Protein Assay (Pierce). Lysate protein concentrations were normalized and lysates were incubated with streptavidin beads overnight at 4 °C (Sigma). Beads were washed three times in lysis buffer and boiled in loading buffer for 5 min before separation by SDS-PAGE. A similar protocol was followed to examine the cell surface expression of NR2B and PDGFβ receptors in cultured hippocampal neurons (15-23 days in vitro) except the 10-min drug treatment was at 37 °C.
Immunoprecipitation—CA1 slices were homogenized in immunoprecipitation buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 30 mm Na pyrophosphate, 1 mm β-glycerophosphate, 1% Nonidet P-40, 1 mm Na3VO4, and MINI mixture). Lysate protein concentrations were normalized after protein determination and 1 mg of total protein was incubated overnight with anti-PDGFβ receptor conjugated to agarose beads. Beads were washed and boiled in loading buffer prior to SDS-PAGE. For NR2B or NR2A immunoprecipitation, crude membrane fractions were obtained by centrifugation at 14,000 × g for 20 min, followed by solubilization of the pellet in 1% SDS buffer, and 5-fold dilution with 1% Triton buffer.
Western Blot and Analysis—For transcription factor blots, hippocampal cultures were lysed in loading buffer and samples were directly loaded onto gels. Homogenates were subjected to SDS-PAGE using 10 or 7.5% gels. Proteins were transferred to nitrocellulose membranes, blocked with 5% nonfat dry milk in Tris-buffered saline for 1 h at room temperature or overnight at 4 °C, and incubated in primary antibodies for 1 h or overnight at 4 °C. Membranes were washed three times in Tris-buffered saline with 0.1% Tween 20, incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h, washed again, and bound antibodies were visualized by the enhanced chemiluminescence method. Densitometric analysis of Western blots was performed using the Kodak Image Station 2000R software.
Immunocytochemistry—Day 14-21 hippocampal-cultured neurons were washed with PBS twice, then fixed in 4% paraformaldehyde and 4% sucrose in phosphate-buffered saline (PBS) for 20 min. The cultures were rinsed in PBS and permeabilized in 0.25% Triton X-100 in PBS (PBST) for 30 min. The cultures were incubated with PBS blocking buffer containing 10% normal goat serum and then incubated in the first primary antibody for 48 h at 4 °C. The coverslips were washed in three times PBST incubated with secondary antibody: Alexa 488 or Alexa 568 (Molecular Probes, 1:1000) for 1 h in PBS with 1% normal goat serum. Coverslips were washed again and subsequently incubated with the second primary antibody. After washing and incubation with a second secondary antibody, the coverslips were mounted with Prolong Gold antifade solution (Invitrogen). Fluorescent images were visualized on Leica TCS SL confocal microscope with a ×63 oil-based objective.
Image Analysis—A quantitative analysis was performed to compare the level of colocalization between NR2A containing NMDA receptors and NR2B containing NMDA receptors with respect to PDGFβ receptor or between PDGFβ receptor and PSD-95. A total of three sets of coverslips were used per condition, five neurons were imaged from each coverslip, and nine images from processes were analyzed. Thus at least 180 images were evaluated per condition. Background signaling was subtracted from each image and percentage of colocalization was calculated using a modified intensity correlation analysis with ImageJ (24).
Total Versus Extrasynaptic NMDA Receptor Activation—To differentiate between activation of the entire pool of NMDA receptors and the extrasynaptic pool, we used a protocol initially developed by Hardingham's group (7, 25). Briefly, for total NMDA receptor activation, cells were incubated for 3 min with 100 μm NMDA, 10 μm glycine, 1 μm tetrodotoxin, 40 μm 6-cyano-7-nitroquinoxaline-2,3-dione, and 5 μm nifedipine after pretreatment with vehicle or 10 ng/ml PDGF-BB for 10 min. To activate extrasynaptic NMDA receptors exclusively, we added 50 μm MK-801, 10 μm bicuculline, 10 μm glycine, and 5 μm nifedipine during the last 2 min of the vehicle/PDGF-BB pretreatment. After stimulation, cultures were lysed directly in loading buffer, samples were resolved by SDS-PAGE, and incubated with antibodies as indicated above.
Cell Death Assay—We adapted the cell death assay protocol from Carrier et al. (26). Briefly, 18-21-day-old hippocampal neurons on 12-well plates were pretreated for 10 min with drugs, washed, and incubated with vehicle or NMDA/glycine for 3 min. The cells were washed and returned to culture medium overnight. After 24 h, cells were fixed in 4% paraformaldehyde in PBS, permeabilized with 0.4% Triton in PBS, blocked for 30 min with 10% bovine serum albumin, and incubated with anti-MAP2 (1:5000, Sigma) overnight at 4 °C. The following day, the cells were washed in 0.4% Triton/PBS three times and incubated with anti-mouse horseradish peroxidase antibody (Amplex Elisa Development Kit, Molecular Probes). Cells were washed three times and incubated with Amplex UltraRed substrate for 30 min. Absorption at 565 nm was measured and data were normalized to control samples.
Statistical Analysis—Statistical analysis of the data were completed using the Prism GraphPad program. All population data were expressed as mean ± S.E. Significance level was set at p < 0.05. Data for colocalization were analyzed by the Mann-Whitney test and Kruskal-Wallis non-parametric tests and data for biotinylation and Western blots were analyzed by Student's t test or analysis of variance.
Animals—All animal experiments were performed in agreement with the guidelines of the policies on the Use of Animals at the University of Toronto.
RESULTS
PDGFβ receptors are activated by dimers of the isoform PDGF-B and heterodimers of PDGF-B and PDGF-A (14). Application of PDGF-BB to isolated or cultured hippocampal neurons caused a long-lasting decrease in peak NMDA-evoked currents (17, 18). To test if the inhibition of NMDA receptor currents involved NR2B over NR2A receptors we applied NR2 subunit-selective antagonists to the bath (27, 28). In isolated CA1 neurons, PDGF-BB application decreased peak NMDA-evoked currents with a maximal inhibition occurring between 8 and 10 min after PDGF-BB treatment (Fig. 1A). However, in the presence of the highly NR2B-selective antagonist, Ro25-6981 (27), PDGF-BB failed to alter NMDA-evoked currents (Fig. 1B). In contrast, when NR2A-containing NMDA receptors were inhibited with NVPAAM077 (28), PDGF-BB decreased peak NMDA-evoked currents (Fig. 1C). NVPAAM077 is a competitive antagonist with limited selectivity between NR2A and NR2B receptors so the concentration of NVPAAM077 was chosen to reduce NR2A currents by about 80%, whereas reducing NR2B only by about 10% (29). The percentage inhibition of both peak and steady-state currents in control cells, in cells treated with NVPAAM077 or Ro25-6981, are presented in Table 1. Taken together these experiments suggest that the inhibition of NMDA currents by PDGF-BB is largely dependent on NR2B- and not NR2A-containing NMDA receptors.
FIGURE 1.
A, vehicle (0.0002% bovine serum albumin, 8 μm HCl) or 10 ng/ml PDGF-BB was applied to isolated CA1 hippocampal neurons (as indicated by the black bar). NMDA-evoked currents were recorded every 60 s by application of 1 μm NMDA, 0.5 μm glycine for 3 s. Sample traces from the same cell are shown with vehicle application (black trace) or with 10 ng/ml PDGF-BB application (gray trace). Data are representative of six experiments. B, NMDA-evoked currents were recorded every 60 s by application of 1 μm NMDA, 0.5 μm glycine for 3 s. Vehicle or PDGF-BB was applied to isolated CA1 hippocampal neurons in the presence of 500 nm Ro25-6981 (NR2B-selective antagonist). Sample traces from the same cell are shown with vehicle application (black trace) or with 10 ng/ml PDGF-BB application (gray trace). Data are representative of six experiments. C, NMDA-evoked currents were recorded every 60 s by application of 1 μm NMDA, 0.5 μm glycine for 3 s. Vehicle or PDGF-BB was applied to isolated CA1 hippocampal neurons in the presence of 50 nm NVP-AAM077 (NR2A-selective antagonist). Sample traces from the same cell are shown with vehicle application (black trace) or with 10 ng/ml PDGF-BB application (gray trace). Data are representative of seven experiments.
TABLE 1.
PDGF-BB inhibition of NMDA-evoked currents in isolated hippocampal neurons in the presence of NR2A- or NR2B-specific antagonists
The t = 14 min time point was chosen for the analysis. Data shown are mean ± S.E. of the mean of five to seven experiments.
|
NMDAR antagonist
|
Peak current
|
Steady-state current
|
||
|---|---|---|---|---|
| Vehicle | PDGF-BB | Vehicle | PDGF-BB | |
| None | 104 ± 5% | 79 ± 5%a | 83 ± 5% | 65 ± 13% |
| Ro25-6981 | 98 ± 2% | 99 ± 5% | 74 ± 6% | 86 ± 4% |
| NVPAAM077 | 100 ± 6% | 75 ± 4%a | 89 ± 7% | 71 ± 16% |
p <0.01 compared to vehicle (Student's t test).
In light of a report that PDGF receptors decreased α-amino-3-hydroxy-5-methylisoxazole-4-proprionic acid receptor (AMPAR)-mediated currents in neurons of the tractus solitarius, we examined if AMPARs were similarly reduced by PDGF-BB in isolated CA1 pyramidal neurons (30). Kainate was used to activate AMPA-mediated currents because this agonist results in a much larger and consistent steady-state current (31, 32). We did not observe any effect of PDGF-BB application on kainate-evoked currents although we observed some characteristic time-dependent decrease in the amplitude of these currents (33, 34) (supplemental Fig. S1).
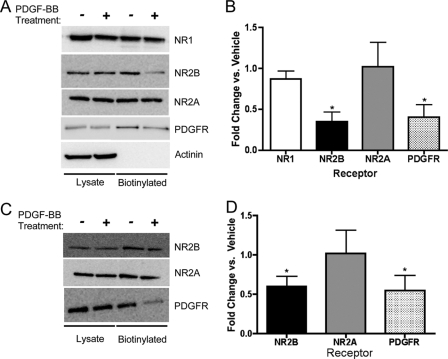
We previously reported that PDGF-BB treatment of hippocampal slices does not significantly alter the surface expression of the NR1 subunit in neurons from the CA1 region of the hippocampus or in cortical neurons (19). To investigate the effects of PDGF-BB treatment on surface expression of NR2 subunits, we treated cultured hippocampal neurons and hippocampal slices with PDGF-BB (10 ng/ml) for 10 min. In hippocampal slices PDGF-BB treatment robustly decreased the surface expression of the NR2B subunit but not the NR2A subunit of the NDMA receptor (Fig. 2, A and B). Surface expression was also reduced for the PDGFβ receptor (Fig. 2, A and B). PDGF-BB treatment decreased surface NR2B expression to 35 ± 12% of control, NR2A surface expression was unchanged at 100 ± 30%, and as previously reported in Beazely et al. (19), NR1 surface expression decreased to 87 ± 11%. The changes in surface expression of NR2A and NR1 were not statistically different from control. As a control, we monitored the biotinylation of the intracellular protein α-actinin (Fig. 2A). Given the slight, but not statistically significant, decrease in NR1, and the variability in the changes of NR2A surface expression, it remains unclear if a portion of the NR1 subunits are internalized with NR2B, or if NR2A replaces NR2B in cell surface NMDA receptors. In cultured hippocampal neurons PDGF-BB application also internalized the NR2B subunit and PDGFβ receptors, but not NR2A (Fig. 2, C and D).
FIGURE 2.
PDGF-BB application results in the selective reduction of the surface expression of NR2B. CA1 hippocampal slices were treated with vehicle or 10 μg/ml PDGF-BB for 10 min. The slices were biotinylated and lysates were incubated overnight with streptavidin beads. A, totallysates or biotinylated samples were blotted using antibodies against NR1 (n = 5), NR2B (n = 7), NR2A (n = 7, C), PDGFβ receptor (n = 4), or α-actinin (n = 4). B, PDGF-BB treatment decreases the surface localization of NR2B and PDGFβ receptors, but not NR1 or NR2A (*, p < 0.05 versus vehicle, Student's unpaired t tests). C and D, PDGF-BB treatment also decreased NR2B (n = 4) and PDGFβ receptor (n = 4) surface localization in hippocampal cultures (*, p < 0.05 versus vehicle, Student's unpaired t tests).
PDGF-BB treatment of hippocampal neurons inhibits NMDA-evoked currents and reduces surface expression of NR2B subunits. To investigate if endocytosis is the mechanism whereby PDGF-BB inhibits NMDA currents we applied PDGF-BB in the absence or presence of two peptides that inhibit clathrin-mediated endocytosis. Neither the amphiphysin-SH3 peptide (35-37) nor the dynamin inhibitory peptide (36) attenuated the inhibition of NMDA currents by PDGF-BB (supplemental Fig. S2, A and B). To control for the ability of the amphiphysin-SH3 peptide to attenuate internalization we demonstrated a complete attenuation of D1 dopamine receptor-mediated increases in NMDA currents in isolated hippocampal neurons (supplemental Fig. S2C). These results demonstrate that the inhibition of NMDA currents by PDGF-BB is not dependent on clathrin-dependent NMDA receptor internalization.
NMDA receptor trafficking, specifically the trafficking of the NR2B subunit, is highly dependent upon the phosphorylation of NR2B (38-40). We examined the total tyrosine phosphorylation of the NR2B and NR2A subunits, as well as the site-specific phosphorylation of tyrosine 1472 on the NR2B subunit, and serines 896 and 897 on the NR1 subunit after treatment of hippocampal slices with PDGF-BB. PDGF-BB significantly increased the phosphorylation of tyrosine 1472 on the NR2B subunit (191 ± 17% of control, n = 5, p < 0.05, unpaired Student's t test, Fig. 3). We did not observe any changes in the total tyrosine phosphorylation state of NR2A or NR2B (81 ± 22 and 117 ± 10% versus control, respectively), nor any changes in the phosphorylation of serines 896 and 897 on the NR1 subunit (107 ± 16 and 110 ± 29% versus control, respectively).
FIGURE 3.
Acutely dissected hippocampal slices were incubated for 10 min with vehicle (control, -) or PDGF-BB 10 ng/ml (+). Lysates were resolved on SDS gels, transferred to nitrocellulose membranes, and probed with antibodies directed against phosphorylated tyrosine 1472. Membranes were stripped and reprobed for total NR2B (n = 4).
Although PDGFβ receptors are expressed in hippocampal neurons (15, 41) very little is known about their localization. To investigate the localization of PDGFβ receptors in hippocampal neurons, as well as their potential colocalization with NMDA receptors, we imaged PDGFβ receptors and NMDA receptor subunits in 2-3-week-old cultured hippocampal neurons. When an antibody directed against the synaptic marker PSD-95 was compared with that recognizing PDGFβ receptor it was apparent that PDGFβ receptors did not show a strong synaptic localization (Fig. 4A). PDGFβ receptor localization was subsequently compared with NR2A and NR2B subunits. PDGFβ receptors colocalized to a much higher degree with the NR2B versus the NR2A subunit (Fig. 4, B and C). Quantification using Manders' overlap coefficient (42, 43) demonstrated that the colocalization of PGDFβ receptors with NR2B was significantly greater than with either PSD-95 or NR2A (Fig. 4D). To ensure that the differences in the Manders' overlap coefficient analysis between PDGFβ receptors localization with NR2A and NR2B was not due to differences in staining intensity, we counted the total number of pixels for PDGFβ receptors (Fig. 4E) and NR2A and -2B (Fig. 4F). These images illustrate that PDGFβ receptors are primarily extrasynaptic and colocalize to a greater degree with NR2B compared with the NR2A subunit.
FIGURE 4.
PDGFβ receptors are primarily extrasynaptic and colocalize with NR2B. A, merged image of PDGFβ receptor staining (green) and PSD-95 staining (red), including a high magnification image of one of the processes. Boxes represent regions of interest that were analyzed for Manders' overlap coefficient. The color scatter plot shows green, red, and yellow (overlap) pixels. The average Manders' overlap coefficient was 0.400. B, merged image of PDGFβ receptor staining (red) and NR2B staining (green), including a high magnification image of one of the processes. Boxes represent regions of interest that were analyzed for Manders' overlap coefficient. The color scatter plot shows green, red, and yellow (overlap) pixels. The average Manders' overlap coefficient was 0.808. C, merged image of PDGFβ receptor staining (red) and NR2A staining (green), including a high magnification image of one of the processes. Boxes represent regions of interest that were analyzed for Manders' overlap coefficient. The color scatter plot shows green, red, and yellow (overlap) pixels. The average Manders' overlap coefficient was 0.438. D, PDGFβ receptors show significantly higher colocalization with NR2B compared with NR2A or PSD-95. *, p < 0.05. Comparison of total pixel count between PDGFβ receptor images and between NR2A and NR2B were not significantly different (data not shown). E and F, comparison of total pixel count between PDGFβ receptor images and between NR2A and NR2B, non-parametric statistical comparison. PDGFβ receptor pixel numbers are not significantly different (p > 0.05) and NR2A and NR2B pixel numbers are not significantly different (p > 0.05).
PDGFβ receptors colocalize with NR2B subunits and both are internalized after PDGF-BB treatment. To determine whether PDGFβ receptors and NR2B subunits share a physical interaction we treated hippocampal CA1 slices with vehicle or 10 ng/ml PDGF-BB. Lysates were subsequently incubated with anti-PDGFβ receptor antibodies pre-conjugated to agarose beads. Although the agarose-PDGFβ receptor antibodies pulled down PDGFβ receptors, Abl and Src kinases (data not shown), we did not detect NR2B in either of the immunoprecipitated samples (supplemental Fig. S3). This result suggests that although the PDGFβ receptor and NR2B are colocalized and both are internalized upon PDGF-BB treatment, they do not appear to physically interact with each other.
We previously reported that PDGF-BB depressed isolated NMDA receptor-mediated synaptic currents at CA3-CA1 synapses without altering the amplitude of LTP of CA1 EPSPs (17). The contribution of NR2A versus NR2B receptors to either LTP or LTD has proven to be highly controversial. It was suggested that NR2B receptors play a more dominant role in LTD (44) but a number of laboratories report that there is only a partial or no subunit dependence to these forms of CA1 synaptic plasticity (29, 45). Furthermore, there are several reports that the NR2B antagonists, ifenprodil and Ro28-6981, enhance rather than block LTD (45, 46). In the visual cortex it is the ratio of expression of NR2A/NR2B receptors that determines the direction of synaptic plasticity, LTD/LTP rather than the actual participation of a given subtype of NMDAR (47). Our results suggest that PDGF-BB selectively reduces the contribution of NR2B receptors therefore we examined the effect of PDGF-BB on LTD. We used a frequency (1 Hz) and amplitude near the threshold that reliably induced LTD. Consistent with several previous reports (45, 46) LTD was substantially enhanced by treatment with Ro25-6981 (% depression at 90 to 100 min; control 13 ± 1.9%; Ro25-6981, 37% ± 2.3, p < 0.01, one-way analysis of variance). Applications of PDGF-BB similarly enhanced the degree of LTD induced by stimulation at 1 Hz frequency (Fig. 5A). This increase in the amplitude of LTD was entirely occluded when Ro25-6981 was included in the bathing solution during the PDGF-BB application (Fig. 5B), confirming the dependence of the action of PDGF-BB on the NR2B-containing NMDA receptor subtype. We did not attempt to use NVPAAM077 as we could not adequately ensure its precise concentration at the receptors in the slice.
FIGURE 5.
PDGF-BB enhanced long-term depression in CA1 region of hippocampal slice. A, vehicle (0.0002% bovine serum albumin, 8 μm HCl) or PDGF-BB (10 ng/ml) was applied for 10 min after stable baseline recording as indicated by the open bar. Long-term depression was evoked by low-frequency stimulation after the vehicle treatment but the magnitude of LTD was increased after PDGF-BB treatment. B, PDGF-BB (10 ng/ml) or PDGF plus Ro25-6981 (0.5 μm) was applied for 10 min after stable baseline recording as indicated by the open bar. Ro25-6981 treatment did not block LTD, but PDGF-BB failed to facilitate LTD in the presence of Ro25-6981. Insets, representative traces recorded at the indicated time (numbers). Calibration bars, vertical = 0.25 mV; horizontal = 5 ms.
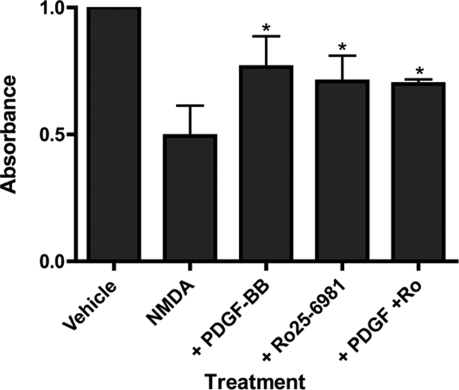
Several reports have suggested that excessive stimulation of NR2B-containing NMDA receptors with a relatively higher degree of extrasynaptic localization may mediate neurotoxicity (7-10). Consistent with reports of the neuroprotective properties of PDGF-BB, we demonstrate that PDGF-BB selectively inhibits NR2B-containing NMDA receptors. However, previous reports used longer incubations of cells with PDGF-BB (1-24 h) (22) compared with the rapid (10 min) effects of PDGF-BB on NMDA currents and NR2B surface expression. To determine whether 10-min PDGF-BB treatment could protect cells against NMDA toxicity, we used a MAP2 immunoreactivity assay similar to that used by Tseng and Dichter (23, 26, 48, 49). NMDA treatment reduced MAP2 immunoreactivity (a measure of the remaining live neuronal cells) by half and a 10-min PDGF-BB pretreatment significantly attenuated this cell death (Fig. 6). Pretreatment with Ro25-6981 also reduced the NMDA-induced cell death and occluded further protection by PDGF-BB (Fig. 7), further suggesting a crucial role for the NR2B-containing receptors in mediating the neuroprotective effects of PDGF-BB.
FIGURE 6.
PDGF-BB is neuroprotective. Day 18-21 cultured hippocampal neurons were pretreated for 10 min with vehicle (ECF), 10 ng/ml PDGF-BB, 2.5 μm Ro25-6981, or both. Cells were subsequently incubated with vehicle (ECF) or 100 μm NMDA, 1 μm glycine for 3 min. ECF was aspirated and cells were returned to media for 24 h before determining cell viability as determined by enzyme-linked immunosorbent assay. Data represent the mean ± S.E. of four experiments (*, p < 0.05, analysis of variance with Bonferroni's post-test compared with NMDA-treated cells alone, second bar).
FIGURE 7.
PDGF-BB treatment alters NMDA signaling to transcription factors in hippocampal neurons. For “total” NMDA treatment (A-C), 14-21-day-old hippocampal cultures were treated for 10 min in warm ECF with vehicle (-) or PDGF-BB (+), followed by treatment with vehicle or 100 μm NMDA and 1 μm glycine. All treatments were in the presence of 1 μm tetrodotoxin, 40 μm 6-cyano-7-nitroquinoxaline-2,3-dione, and 5 μm nifedipine. For “extrasynaptic” NMDA treatment (D-F), cultures were treated for 10 min with vehicle or PDGF-BB. During the last 3 min of the PDGF-BB treatment, 50 μm MK-801, 10 μm bicuculline, 10 μm glycine, and 5 μm nifedipine were included to block synaptic NMDA receptors. After either treatment, cells were washed briefly and incubated with vehicle or 100 μm NMDA, 10 μm glycine for 3 min in the presence of 6-cyano-7-nitroquinoxaline-2,3-dione and nifedipine. A, after total NMDA treatment, membranes were probed with anti-phospho-ERK, phospho-CREB, and ERK. Blots are representative of five to seven experiments and immunoreactivity was quantified versus control (vehicle, -)(B, C). D, after extrasynaptic NMDA treatment, membranes were probed with anti-phospho-ERK, ERK, and phospho-CREB. Blots are representative of five to seven experiments and immunoreactivity was quantified versus control (vehicle, -)(E, F). Phospho-ERK immunoreactivity was normalized to total ERK (both bands were pooled) and phospho-CREB immunoreactivity was normalized to total CREB (not shown).
The differences between NR2B/extrasynaptic and NR2A/synaptic NMDA receptors with regards to cell survival may involve signaling to CREB and MAPK (8, 10, 50-52). PDGFβ receptor activation enhances ERK1/2 phosphorylation in several systems via the recruitment of Grb2 scaffold proteins, via phosphatidylinositol 3-kinase, via activation of Gαi, or after being transactivated by G-protein coupled receptors such as the D4 dopamine receptor (14, 53). To investigate the potential of PDGFβ receptors to alter NMDA signaling to ERK1/2 and CREB, we incubated 14-21-day-old cultured hippocampal neurons with vehicle or PDGF-BB for 10 min, followed by stimulation of the total population of NMDA receptors or alternatively just the extrasynaptic population of NMDA receptors (25, 53). In control hippocampal cultures, activation of all NMDA receptors did not alter CREB phosphorylation, however, after PDGF-BB treatment, NMDA treatment robustly enhanced phospho-CREB levels (Fig. 7, A and B). Conversely, extrasynaptic NMDA receptor activation inhibited both CREB and ERK phosphorylation, but not in cultures pretreated with PDGF-BB (Fig. 7, D-F). These results suggest that PDGF-BB treatment promotes NMDA signaling to ERK and CREB, perhaps by diminishing the contribution of NR2B-containing NMDA receptors to the net response to NMDA.
DISCUSSION
Several growth factors important for neuronal development also play a role in NMDA receptor signaling and subunit trafficking in mature neurons. For example, receptor tyrosine kinase receptors including TrkB and its ligand brain-derived neurotropic factor have been extensively implicated in the modulation of synaptic transmission and plasticity (54-56). Brain-derived neurotropic factor signaling has also been associated with NMDA-induced neuroprotection via preconditioning (57) and the induction of brain-derived neurotropic factor gene expression is enhanced in response to NR2B receptor activation (52). PDGF receptors, which are also receptor tyrosine kinases critical for development of the central nervous system, have received much less attention particularly with respect to their potential function as regulators of synaptic transmission and cell survival (13). The presence of PDGF-B isoform chains in neurons and their processes was initially described by Sasahara et al. (58) who hypothesized that PDGF-BB dimers might be released following injury resulting in potential glial cell chemotaxis, proliferation, and enhanced neuronal survival (59). However, PDGF-B is also located in axons and presumptive synaptic terminals presenting the possibility that it is released during synaptic activity to serve as a modulator of synaptic transmission, a concept that has received some substantial support (13). In addition, staining PDGFβ receptors the hippocampus is strong in pyramidal neurons (15). Therefore, both the receptor and the ligand for PDGFβ receptor signaling are associated with hippocampal synapses, supporting a neuromodulatory role for this growth factor.
NR2B subunits predominate at extrasynaptic sites in hippocampal and other neurons. Both extrasynaptic NMDA receptors and NR2B-containing NMDA receptors, regardless of subcellular location, have been implicated in neuronal cell death (11, 12, 60). Several lines of evidence have shown that PDGFβ receptor signaling is neuroprotective in neuronal cultures and in vivo (21, 23, 61). In this context, our results provide a potential mechanism for the neuroprotective properties of PDGFβ receptor signaling. However, recently published evidence suggest that NR2A- or NR2B-containing NMDA receptors can contribute to NMDA-induced excitotoxicity, and that the neuroprotective effects of NR2A-containing NMDA receptors are only observed at submaximal concentrations of NMDA (12). Several recent studies indicate that in cultured neurons NR2B containing receptors are preferentially linked to excitotoxic pathways, whereas NR2A receptors are actually neuroprotective (11, 12). The selective block of neuronal damage by NR2B versus exacerbation by NR2A antagonists was shown in the MCAO model of stroke in adult animals (11). However, whether or not this relationship holds up for adult animals is somewhat controversial as it has been reported that NR2A containing receptors may become sources of excitotoxicity in older cultures (12). PDGF receptors can signal to ERK directly, therefore the neuroprotective effects of PDGF-BB may involve both a direct enhancement of phospho-ERK levels as well as a decrease in the ability of NR2B receptors to promote ERK dephosphorylation.
In addition to understanding the potential mechanisms of PDGF-BB-mediated neuroprotection, we also demonstrate PDGF-BB modulation of LTD. Both NMDAR and AMPAR-mediated excitatory post-synaptic currents at CA1 synapses undergo LTD but they differ in mechanism (62). The induction of stimulus-activated LTD at CA1 synapses requires a clathrin-dependent internalization of AMPARs (62). In contrast, LTD of the NMDAR component at CA1 synapses is independent of clathrin and instead requires an intact actin cytoskeleton. The inhibition of NMDA-mediated currents in CA1 pyramidal neurons by PDGF-BB is also very long lasting and depends upon an intact actin cytoskeleton (17, 18), however, it is not clathrin-dependent but is also blocked by inhibitors of serine-threonine phosphatases (17). Therefore it closely resembles LTD of NMDA synaptic currents. Although NMDAR receptors can be internalized via clathrin-mediated endocytosis (63), the application of two peptides that interfere with clathrin-mediated endocytosis, applied intracellularly via the patch pipette, did not suppress PDGF-BB-induced inhibition. This suggests that acute inhibition of NR2B subunits is not the result of a clathrin-dependent internalization of the NR2B subunit but may either be the result of functional inhibition of NR2B activity or alternatively it suggests that the internalization of NR2B receptors is via a clathrin-independent mechanism. We previously reported that PDGF-BB had no effect on the amplitude of high frequency LTP (17), however, here we showed that it enhanced the LTD of the CA1 field EPSPs via an NR2B-dependent mechanism. This result is consistent with the suggestion that increasing the ratio of NR2A/NR2B leads to a shift toward LTD over LTP (45, 48). Our previous observation of a lack of change in the surface expression of NR1 subunits implies that NR2A subunits might rapidly replace NR2B subunits. This concept would be consistent with a report that NR2A versus NR2B subunit composition of synaptic NMDA receptors can be rapidly altered by the high frequency stimulation induction of LTP (64). The change in this intracellular Ca2+ signal resulting from a PDGF-BB-induced reduction in NR2B receptors is yet to be determined. Also, the location (synaptic versus extrasynaptic) of the NMDA receptors likely contributes to determination of the direction of synaptic plasticity (48, 65, 66). We demonstrate that PDGFβ receptors colocalize and selectively inhibit and internalize NR2B-containing NMDA receptors, thereby altering the signaling of NMDA receptor populations to CREB and ERK1/2 (Fig. 8). Cell surface receptor expression may be altered by an increase in receptor internalization (as shown in Fig. 8), a decrease in the insertion of NR2B to the membrane, or both. Taken together, these results suggest the selective targeting of NR2B-containing NMDA receptors by PDGFβ receptors likely contributes to the neuroprotective effects of PDGF-BB. PDGFβ receptor signaling is enhanced following ischemic damage and provides some degree of neuroprotection. Therefore, PDGF likely serves to compensate for neuronal damage following stroke perhaps by both minimizing cell damage and by enhancing cell proliferation in animals during development. It remains to be determined if the mechanisms that lead to modulation of excitatory synaptic transmission also provide neuroprotection under ischemic conditions.
FIGURE 8.
The proposed mechanisms of PDGFβR inhibition of NMDA receptors and neuroprotection. PDGFβ receptor activation inhibits NR2B-containing NMDA receptors and selectively removes NR2B subunits from the cell membrane. However, the relationship between these two observations remains unclear. The removal of NR2B may cause, or be the result of, PDGFβ receptor inhibition of NMDA currents. PDGFβ receptor activation is also neuroprotective. PDGF-BB treatment led to an increase in ERK1/2 phosphorylation and prevented extrasynaptic NMDA receptor inhibition of ERK1/2 and CREB phosphorylation.
Supplementary Material
This work was supported by Canadian Institutes of Health Research Grant 15514 (to J. F. M.) and a postdoctoral award (to M. A. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S3.
Footnotes
The abbreviations used are: NMDA, N-methyl-d-aspartate; PDGF, plateletderived growth factor; ERK, extracellular signal-regulated kinase; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid; LTD, long-term depression; LTP, long-term potentiation; AMPARP, α-amino-3-hydroxy-5-methylisoxazole-4-proprionic acid receptor; CREB, cAMP-response element-binding protein; PBS, phosphate-buffered saline; MAP, mitogen-activated protein; ECF, extracellular fluid; EPSP, excitatory post-synaptic potential.
References
- 1.Paoletti, P., and Neyton, J. (2007) Curr. Opin. Pharmacol. 7 39-47 [DOI] [PubMed] [Google Scholar]
- 2.Groc, L., Heine, M., Cousins, S. L., Stephenson, F. A., Lounis, B., Cognet, L., and Choquet, D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 18769-18774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Hallaq, R. A., Conrads, T. P., Veenstra, T. D., and Wenthold, R. J. (2007) J. Neurosci. 27 8334-8343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tovar, K. R., and Westbrook, G. L. (2002) Neuron 34 255-264 [DOI] [PubMed] [Google Scholar]
- 5.Groc, L., Choquet, D., Stephenson, F. A., Verrier, D., Manzoni, O. J., and Chavis, P. (2007) J. Neurosci. 27 10165-10175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris, A. Z., and Pettit, D. L. (2007) J. Physiol. 584 509-519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardingham, G. E., and Bading, H. (2002) Biochim. Biophys. Acta 1600 148-153 [DOI] [PubMed] [Google Scholar]
- 8.Hardingham, G. E., and Bading, H. (2003) Trends Neurosci. 26 81-89 [DOI] [PubMed] [Google Scholar]
- 9.Hardingham, G. E. (2006) Biochem. Soc. Trans. 34 936-938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang, S. J., Steijaert, M. N., Lau, D., Schutz, G., Delucinge-Vivier, C., Descombes, P., and Bading, H. (2007) Neuron 53 549-562 [DOI] [PubMed] [Google Scholar]
- 11.Liu, Y., Wong, T. P., Aarts, M., Rooyakkers, A., Liu, L., Lai, T. W., Wu, D. C., Lu, J., Tymianski, M., Craig, A. M., and Wang, Y. T. (2007) J. Neurosci. 27 2846-2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Engelhardt, J., Coserea, I., Pawlak, V., Fuchs, E. C., Kohr, G., Seeburg, P. H., and Monyer, H. (2007) Neuropharmacology 53 10-17 [DOI] [PubMed] [Google Scholar]
- 13.Valenzuela, C. F., Kazlauskas, A., and Weiner, J. L. (1997) Brain Res. Brain Res. Rev. 24 77-89 [DOI] [PubMed] [Google Scholar]
- 14.Heldin, C. H., Ostman, A., and Ronnstrand, L. (1998) Biochim. Biophys. Acta 1378 F79-F113 [DOI] [PubMed] [Google Scholar]
- 15.Smits, A., Kato, M., Westermark, B., Nister, M., Heldin, C. H., and Funa, K. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 8159-8163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh, H. J., Silos-Santiago, I., Wang, Y. X., George, R. J., Snider, W. D., and Deuel, T. F. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 1952-1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valenzuela, C. F., Xiong, Z., MacDonald, J. F., Weiner, J. L., Frazier, C. J., Dunwiddie, T. V., Kazlauskas, A., Whiting, P. J., and Harris, R. A. (1996) J. Biol. Chem. 271 16151-16159 [DOI] [PubMed] [Google Scholar]
- 18.Lei, S., Lu, W. Y., Xiong, Z. G., Orser, B. A., Valenzuela, C. F., and MacDonald, J. F. (1999) J. Biol. Chem. 274 30617-30623 [DOI] [PubMed] [Google Scholar]
- 19.Beazely, M. A., Tong, A., Wei, W. L., Van Tol, H., Sidhu, B., and MacDonald, J. F. (2006) J. Neurochem. 98 1657-1663 [DOI] [PubMed] [Google Scholar]
- 20.Egawa-Tsuzuki, T., Ohno, M., Tanaka, N., Takeuchi, Y., Uramoto, H., Faigle, R., Funa, K., Ishii, Y., and Sasahara, M. (2004) Exp. Neurol. 186 89-98 [DOI] [PubMed] [Google Scholar]
- 21.Iihara, K., Sasahara, M., Hashimoto, N., Uemura, Y., Kikuchi, H., and Hazama, F. (1994) J. Cereb. Blood Flow Metab. 14 818-824 [DOI] [PubMed] [Google Scholar]
- 22.Ohno, M., Sasahara, M., Narumiya, S., Tanaka, N., Yamano, T., Shimada, M., and Hazama, F. (1999) Neuroscience 90 643-651 [DOI] [PubMed] [Google Scholar]
- 23.Tseng, H. C., and Dichter, M. A. (2005) Neurobiol. Dis. 19 77-83 [DOI] [PubMed] [Google Scholar]
- 24.Wang, L. Y., and MacDonald, J. F. (1995) J. Physiol. 486 83-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov, A., Pellegrino, C., Rama, S., Dumalska, I., Salyha, Y., Ben-Ari, Y., and Medina, I. (2006) J. Physiol. 572 789-798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrier, R. L., Ma, T. C., Obrietan, K., and Hoyt, K. R. (2006) J. Neurosci. Methods 154 239-244 [DOI] [PubMed] [Google Scholar]
- 27.Fischer, G., Mutel, V., Trube, G., Malherbe, P., Kew, J. N., Mohacsi, E., Heitz, M. P., and Kemp, J. A. (1997) J. Pharmacol. Exp. Ther. 283 1285-1292 [PubMed] [Google Scholar]
- 28.Auberson, Y. P., Allgeier, H., Bischoff, S., Lingenhoehl, K., Moretti, R., and Schmutz, M. (2002) Bioorg. Med. Chem. Lett. 12 1099-1102 [DOI] [PubMed] [Google Scholar]
- 29.Bartlett, T. E., Bannister, N. J., Collett, V. J., Dargan, S. L., Massey, P. V., Bortolotto, Z. A., Fitzjohn, S. M., Bashir, Z. I., Collingridge, G. L., and Lodge, D. (2007) Neuropharmacology 52 60-70 [DOI] [PubMed] [Google Scholar]
- 30.Ohi, Y., Ishii, Y., Haji, A., Noguchi, S., Sasaoka, T., Fujimori, T., Nabeshima, Y., Sasahara, M., and Hattori, Y. (2007) Brain Res. 1159 77-85 [DOI] [PubMed] [Google Scholar]
- 31.Wang, L. Y., Salter, M. W., and MacDonald, J. F. (1991) Science 253 1132-1135 [DOI] [PubMed] [Google Scholar]
- 32.Jackson, M. F., Joo, D. T., Al-Mahrouki, A. A., Orser, B. A., and Macdonald, J. F. (2003) Mol. Pharmacol. 64 395-406 [DOI] [PubMed] [Google Scholar]
- 33.Wang, L. Y., Taverna, F. A., Huang, X. P., MacDonald, J. F., and Hampson, D. R. (1993) Science 259 1173-1175 [DOI] [PubMed] [Google Scholar]
- 34.Wang, L. Y., Dudek, E. M., Browning, M. D., and MacDonald, J. F. (1994) J. Physiol. 475 431-437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grabs, D., Slepnev, V. I., Songyang, Z., David, C., Lynch, M., Cantley, L. C., and De Camilli, P. (1997) J. Biol. Chem. 272 13419-13425 [DOI] [PubMed] [Google Scholar]
- 36.Man, H. Y., Lin, J. W., Ju, W. H., Ahmadian, G., Liu, L., Becker, L. E., Sheng, M., and Wang, Y. T. (2000) Neuron 25 649-662 [DOI] [PubMed] [Google Scholar]
- 37.Nong, Y., Huang, Y. Q., Ju, W., Kalia, L. V., Ahmadian, G., Wang, Y. T., and Salter, M. W. (2003) Nature 422 302-307 [DOI] [PubMed] [Google Scholar]
- 38.Zhang, S., Edelmann, L., Liu, J., Crandall, J. E., and Morabito, M. A. (2008) J. Neurosci. 28 415-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, H. Y., Hsu, F. C., Gleichman, A. J., Baconguis, I., Coulter, D. A., and Lynch, D. R. (2007) J. Biol. Chem. 282 20075-20087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, F., Plummer, M. R., Len, G. W., Nakazawa, T., Yamamoto, T., Black, I. B., and Wu, K. (2006) Brain Res. 1121 22-34 [DOI] [PubMed] [Google Scholar]
- 41.Kotecha, S. A., Oak, J. N., Jackson, M. F., Perez, Y., Orser, B. A., Van Tol, H. H., and MacDonald, J. F. (2002) Neuron 35 1111-1122 [DOI] [PubMed] [Google Scholar]
- 42.Manders, E. M., Stap, J., Brakenhoff, G. J., van Driel, R., and Aten, J. A. (1992) J. Cell Sci. 103 857-862 [DOI] [PubMed] [Google Scholar]
- 43.Li, Q., Lau, A., Morris, T. J., Guo, L., Fordyce, C. B., and Stanley, E. F. (2004) J. Neurosci. 24 4070-4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, L., Wong, T. P., Pozza, M. F., Lingenhoehl, K., Wang, Y., Sheng, M., Auberson, Y. P., and Wang, Y. T. (2004) Science 304 1021-1024 [DOI] [PubMed] [Google Scholar]
- 45.Morishita, W., Lu, W., Smith, G. B., Nicoll, R. A., Bear, M. F., and Malenka, R. C. (2007) Neuropharmacology 52 71-76 [DOI] [PubMed] [Google Scholar]
- 46.Hendricson, A. W., Miao, C. L., Lippmann, M. J., and Morrisett, R. A. (2002) J. Pharmacol. Exp. Ther. 301 938-944 [DOI] [PubMed] [Google Scholar]
- 47.Zhou, M., Diwu, Z., Panchuk-Voloshina, N., and Haugland, R. P. (1997) Anal. Biochem. 253 162-168 [DOI] [PubMed] [Google Scholar]
- 48.Chen, W. S., and Bear, M. F. (2007) Neuropharmacology 52 200-214 [DOI] [PubMed] [Google Scholar]
- 49.Zhou, M., and Panchuk-Voloshina, N. (1997) Anal. Biochem. 253 169-174 [DOI] [PubMed] [Google Scholar]
- 50.Hardingham, G. E., Fukunaga, Y., and Bading, H. (2002) Nat. Neurosci. 5 405-414 [DOI] [PubMed] [Google Scholar]
- 51.Pokorska, A., Vanhoutte, P., Arnold, F. J., Silvagno, F., Hardingham, G. E., and Bading, H. (2003) J. Neurochem. 84 447-452 [DOI] [PubMed] [Google Scholar]
- 52.Vanhoutte, P., and Bading, H. (2003) Curr. Opin. Neurobiol. 13 366-371 [DOI] [PubMed] [Google Scholar]
- 53.Lu, W., Man, H., Ju, W., Trimble, W. S., MacDonald, J. F., and Wang, Y. T. (2001) Neuron 29 243-254 [DOI] [PubMed] [Google Scholar]
- 54.Arancio, O., and Chao, M. V. (2007) Curr. Opin. Neurobiol. 17 325-330 [DOI] [PubMed] [Google Scholar]
- 55.Lauterborn, J. C., Rex, C. S., Kramar, E., Chen, L. Y., Pandyarajan, V., Lynch, G., and Gall, C. M. (2007) J. Neurosci. 27 10685-10694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turrigiano, G. (2007) Curr. Opin. Neurobiol. 17 318-324 [DOI] [PubMed] [Google Scholar]
- 57.Marini, A. M., Jiang, X., Wu, X., Pan, H., Guo, Z., Mattson, M. P., Blondeau, N., Novelli, A., and Lipsky, R. H. (2007) Amino Acids 32 299-304 [DOI] [PubMed] [Google Scholar]
- 58.Sasahara, M., Fries, J. W., Raines, E. W., Gown, A. M., Westrum, L. E., Frosch, M. P., Bonthron, D. T., Ross, R., and Collins, T. (1991) Cell 64 217-227 [DOI] [PubMed] [Google Scholar]
- 59.Barres, B. A., Hart, I. K., Coles, H. S., Burne, J. F., Voyvodic, J. T., Richardson, W. D., and Raff, M. C. (1992) Cell 70 31-46 [DOI] [PubMed] [Google Scholar]
- 60.Kohr, G. (2006) Cell Tissue Res. 326 439-446 [DOI] [PubMed] [Google Scholar]
- 61.Ishii, Y., Oya, T., Zheng, L., Gao, Z., Kawaguchi, M., Sabit, H., Matsushima, T., Tokunaga, A., Ishizawa, S., Hori, E., Nabeshima, Y., Sasaoka, T., Fujimori, T., Mori, H., and Sasahara, M. (2006) J. Neurochem. 98 588-600 [DOI] [PubMed] [Google Scholar]
- 62.Morishita, W., Marie, H., and Malenka, R. C. (2005) Nat. Neurosci. 8 1043-1050 [DOI] [PubMed] [Google Scholar]
- 63.Lavezzari, G., McCallum, J., Lee, R., and Roche, K. W. (2003) Neuropharmacology 45 729-737 [DOI] [PubMed] [Google Scholar]
- 64.Bellone, C., and Nicoll, R. A. (2007) Neuron 55 779-785 [DOI] [PubMed] [Google Scholar]
- 65.Bear, M. F. (2003) Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 649-655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Philpot, B. D., Espinosa, J. S., and Bear, M. F. (2003) J. Neurosci. 23 5583-5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.