Abstract
Viral capsids are envisioned as vehicles to deliver the viral genome to the host cell. They are nonetheless dynamic protective shells, as they participate in numerous processes of the virus cycle such as assembly, genome packaging, binding to receptors, and uncoating among others. In so doing, they undergo large scale conformational changes. Capsid proteins with essential enzymatic activities are being described more frequently. Here we show that the precursor (pVP2) of the capsid protein VP2 of the infectious bursal disease virus (IBDV), an avian double-stranded RNA virus, has autoproteolytic activity. The pVP2 C-terminal region is first processed by the viral protease VP4. VP2 Asp-431, lying in a flexible loop preceding the C-terminal most α-helix, is responsible for the endopeptidase activity that cleaves the Ala-441—Phe-442 bond to generate the mature VP2 polypeptide. The D431N substitution abrogates the endopeptidase activity without introducing a significant conformational change, as deduced from the three-dimensional structure of the mutant protein at 3.1 Å resolution. Combinations of VP2 polypeptides containing mutations affecting either the cleavage or the catalytic site revealed that pVP2 proteolytic processing is the result of a monomolecular cis-cleavage reaction. The D431N mutation does not affect the assembly of the VP2 trimers that constitute the capsid building block. Although VP2 D431N trimers are capable of assembling both pentamers and hexamers, expression of a polyprotein gene harboring the D431N mutation does not result in the assembly of IBDV virus-like particles. Reverse genetics analyses demonstrate that pVP2 self-processing is essential for the assembly of an infectious IBDV progeny.
Multiprotein assemblies are the basic cellular entities controlling fundamental biological processes. Viral capsids represent an excellent framework for the analysis of “built-in” features such as structural polymorphism and transient conformations. In addition, complex capsids may require one or more auxiliary proteins (scaffold, accessory, and proteolytic proteins) to trigger structural and functional changes. In this context, assembly of birnavirus capsid has resulted in an appropriate process for studying the coordination of molecular factors involved that, like in many other viruses, rarely leads to aberrant assemblies in vivo.
Birnaviruses are a family of naked icosahedral viruses infecting rotifers, insects, fishes, and birds (1). Their genome, formed by two segments of double-stranded RNA (dsRNA),4 encodes only five mature polypeptides. Although most of the research has been focused on the infectious bursal disease virus (IBDV), an important avian pathogen (2), available information suggests that all members of the family share similar replication strategies.
The IBDV particle possesses a relatively complex icosahedral capsid based on a triangulation number T = 13 levo lattice composed by a single polypeptide known as VP2. Whereas the simplest capsids are built from 60 identical subunits and assembled into pentamers (T = 1), those with more than 60 subunits are assembled into pentamers and hexamers and cannot have identical but “quasi-equivalent” environments (T > 1) (3). The T number also describes the number of different environments occupied by the protein subunit (4). The VP2 protein assembles into 260 trimers that form 12 pentamers and 120 hexamers (5-7). From strict geometric considerations, VP2 trimers have five distinct conformations.
VP2 is synthesized as part of a polyprotein (∼110 kDa) that is cotranslationally self-cleaved, releasing three polypeptides, namely pVP2 (the VP2 precursor), VP4 (the protease), and VP3 (a polypeptide with scaffolding activity among others). pVP2 (512 residues, 54.4 kDa) undergoes subsequent, and much slower, processing events mediated by VP4 at three secondary targets (positions 487, 494, and 501) (8), and the resulting intermediates are further cleaved between residues Ala-441 and Phe-442 to yield VP2 (47 kDa) and several C-terminal fragments (accounting for 7.4 kDa). Both the mechanism and the executor of the last pVP2 → VP2 proteolytic maturation remain unknown and are associated with the pVP2 assembly into a procapsid-like structure (9). The released C-terminal segments remain associated to the capsid (10) and appear to be involved in the entry mechanism by promoting the disruption of host cell membranes (11).
Studies carried out with another birnavirus, infectious pancreatic necrosis virus (IPNV), a pathogen of the aquatic fauna (12), suggest that the pVP2 → VP2 conversion is required for the maturation of quasi-spherical provirions into mature icosahedral virions (13). This maturation mechanism is reminiscent of the post-assembly proteolytic maturation that takes place during the morphogenesis of some ssRNA icosahedral viruses, and in particular to those of noda- and tetraviruses (14, 15). The capsid protein (CP) precursor, the α-protein, associates as a trimer, building a quasi-spherical provirion (16). At low pH, the α-protein is autocatalytically processed, rendering the mature CP, known as β-protein, and the C-terminal γ-peptide (17). This reaction is slow and dependent on a conserved Asp residue located in close proximity of the CP scissile bond (18). Maturation of the β-protein is accompanied by large conformational changes on the CP that irreversibly transforms the quasi-spherical (non-infectious) provirions into icosahedral infective virions (14, 17).
The molecular mechanisms that define the multimeric state of the mature VP2 as hexamers or pentamers have been intensively analyzed (7, 19-21). Expression of VP2 alone results in the spontaneous assembly of icosahedral T = 1 subviral particles (SVP), an all-pentamer capsid ∼23 nm in diameter composed of 20 trimeric clusters of VP2. On the other hand, pVP2 expression leads to the assembly of tubular structures with a hexagonal lattice (7, 19). The pVP2 C-terminal processing events are abolished when this protein is expressed using a baculovirus-based system. Notably, the pVP2 C-terminal region contains an amphipathic α-helix that plays an important role in allowing the formation of multiple VP2 conformations. VP3 also participates in the inherent conformational polymorphism of pVP2 through interaction with the pVP2 C-terminal end, working as a canonical scaffolding protein.
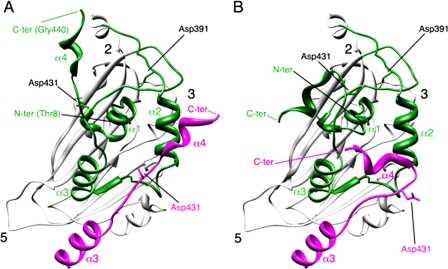
The atomic structure of the VP2 polypeptide has been recently solved from T = 1 SVP (5, 22, 23). The VP2 subunit folds into three domains termed projection (P), shell (S), and base (B). Domains S and P are β barrels with a jelly roll topology, whereas the B domain is formed by α-helices facing the interior of the shell, corresponding to its N and C termini. The helical C-terminal arm of VP2 establishes a domain swapping and mediates interactions between adjacent VP2 trimers increasing their stability. S and B domains share a high degree of structural similarity with noda- and tetravirus α-protein. pVP2 and α-protein cleavage sites are deeply buried inside the capsid shell, thus sheltered from possible attacks by cellular proteases. Fitting the VP2 atomic structure into the three-dimensional cryo-electron microscopy map of IBDV T = 1 SVP allowed us to identify at least two conformations for its C-terminal region (19, 21). These structural considerations tempted us to suggest Asp-391 and/or Asp-431 as candidate catalytic residues capable of triggering a hydrolytic cleavage either on its own scissile bond or on that from a neighboring pVP2 molecule (Fig. 1). The results presented here demonstrate that residue Asp-431 is essential for pVP2 proteolytic maturation, thus leading us to propose that birnavirus pVP2 polypeptides are autocatalytically processed. This finding provides yet another piece of evidence reinforcing the hypothesis about a common ancestor linking icosahedral ssRNA and dsRNA viruses.
FIGURE 1.
Conformational flexibility of the VP2 C-terminal α-helix around its cleavage site. A, bottom view, facing the inner surface of the viral capsid, of the VP2 protein x-ray model (PDB entry 2GSY). α-Helices (1-4) of domain B (green) are indicated for a VP2 chain; the candidate catalytic residues Asp-391 and Asp-431 are also indicated. α3 and α4 helices of the closer neighboring VP2 chain (magenta) that projects toward the 3-fold axis of the trimer are shown, and to simplify the view, only its Asp-391 residue is indicated. The locations of 5-, 3-, and 2-fold axes of icosahedral symmetry are indicated. B, same as panel A, but the C-terminal α4 helices of VP2 chains were remodeled after fitting the x-ray model into an equivalent cryo-electron microscopy map (21).
EXPERIMENTAL PROCEDURES
Cells and Viruses—Recombinant vaccinia virus (rVV) VT7LacOI/POLY and recombinant baculoviruses (rBV) FB/VP2-452 and FB/VP2-501 were previously described (7, 24, 25). Expression experiments were carried out with BSC-1 mammalian cells or QM7 quail muscle cells for rVV infections and Trichoplusia ni (H5) insect cells (Invitrogen) for rBV infections. BSC-1 and QM7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum. H5 cells were grown in TC-100 medium (Invitrogen) containing 10% fetal calf serum. rVVs and rBVs were grown and titrated as previously described (25, 26).
Generation of the Recombinant Viruses VT7/LacOI—Using PCR overlap extension five mutant pVP2 genes were constructed containing the following single and double mutations: A441G, F442G, A441G/F442G, D431N, and D391N. PCR reactions were carried out using the pVP2 gene, cloned in the pFB/POLY plasmid as template, and the set of mutator primers (supplemental Table 1) in combination with primers 5′-GCCATCACAAGCCTCAGCGTTGG and 5′-GTGCACCGCGGAGTACCCCAG for A441G, F442G, and A441G/F442G. Primers 5′-GATGCCATCACAAGCCTCAGC and 5′-CGCAGTCGAGGTTGTGTGCAC were used for D431N and D391N. The resulting DNA fragments were digested with NdeI and SphI and used to replace the original NdeI-SphI fragment of the pVOTE.2/POLY plasmid previously described (25). The plasmids pVOTE/A441G, pVOTE/F442G, pVOTE/A441G/F442G, pVOTE/D391N, and pVOTE/D431N were used to obtain the recombinant vaccinia viruses VT7LacOI/A441G, VT7LacOI/F442G, VT7LacOI/A441G/F442G, VT7LacOI/D391N, and VT7LacOI/D431N. For this, BSC-1 cells were infected with the vaccinia virus recombinant VT7LacOI (27) and transfected with the previous plasmids. Selection and amplification of VT7LacOI/A441G, VT7LacOI/F442G, VT7LacOI/A441G/F442G, VT7LacOI/D391N, and VT7LacOI/D431N were carried out as previously described (28).
Construction of Recombinant Baculoviruses—rBV FB/VP2-452 and FB/VP2-501 were previously described (7, 19). The pVOTE.2/D431N and pVOTE.2/D391N plasmids were used as a template for PCR synthesis to generate rBV/VP2-452D431N, rBV/VP2-452D391N, and rBV/VP2-501D431N. PCR was performed with Vent DNA polymerase (New England Biolabs) with common 5′ and 3′ end primer (5′-GCGCAGATCTATGACAAACCTGTCAGATCAAACCC and 5′-GCGCAAGCTTACCTTATGGCCCGGATTATGTCTTTGAAGC). BglII-HindIII-digested PCR fragments were cloned into FastBac (Invitrogen) BamH1-HindIII polylinker sites for protein expression. Selection of derived bacmids from the DH10Bac Escherichia coli strain and preparation for Lipofectine transfection were performed according to the manufacturer's protocols (Invitrogen). The constructs were expressed in H5 insect cells (29).
Immunoprecipitation of VP2—QM7 cell monolayers were infected with the corresponding rVV at a multiplicity of infection of 2 plaque-forming units/cell and maintained in the presence of isopropyl β-d-thiogalactosidase. At 18 h postinfection (pi), cells were washed twice and metabolically labeled with methionine-free DMEM containing 125 μCi/ml [35S]Met and isopropyl β-d-thiogalactosidase for 1 h. After this period cells were washed 3 times with DMEM containing a 10-fold concentration of cold methionine and then maintained in DMEM supplemented with isopropyl β-d-thiogalactosidase and 2% fetal calf serum. At 0, 24, 48, and 72 h post-labeling, cells were harvested, washed twice with phosphate-buffered saline, and resuspended in lysis buffer (50 mm Tris, pH 7.5, 100 mm NaCl, 5 mm EDTA, 0.5% IGEPAL CA-630). Immunoprecipitations were carried as described elsewhere (26). Immunoprecipitated samples were resuspended in Laemmli sample buffer to a 1× final concentration and subjected to 11% SDS-PAGE followed by autoradiography.
Reverse Genetic Analysis—DNA fragments containing cDNA versions corresponding to the complete positive strand RNAs of to the IBDV Soroa strain segments A and B, fused to the T7 bacteriophage promoter (in 5′ position with respect of the IBDV cDNAs), and a cDNA corresponding to the hepatitis delta virus ribozyme (in 3′ position with respect of the IBDV cDNAs) sequences were generated by in vitro gene synthesis (Genescript Co.) and inserted into the multiple cloning site of the pUC57 cloning plasmid (GenBank™/EMBL). These plasmids were named pT7-SA-Rz and pT7-SB-Rz, containing the cDNA corresponding to segment A and B, respectively.
Plasmid pVOTE/D431N, described above, was digested with SphI and NdeI and ligated to pT7-SA-Rz previously digested with the same enzymes. The resulting plasmid, pT7-SA(D431N)-Rz, was sequenced to determine the correctness of its sequence.
QM7 cells were transfected with a combination of either pT7-SA-Rz and pT7-SB-Rz or pT7-SA(D431N)-Rz and pT7-SB-Rz, respectively, using Lipofectamine (Invitrogen). 6 h after transfection cultures were infected with 1 plaque-forming unit/cell of VT7LacOI, an rVV inducibly expressing the T7 RNA polymerase. After infection, cultures were maintained at 37 °C in DMEM containing 10% fetal calf serum supplemented with 1 mm isopropyl β-d-thiogalactosidase. At 72 h pi, cultures were harvested and subjected to three freeze-thawing cycles. After removing cell debris by low speed centrifugation, supernatants were recovered and passed through 0.1-μm filters (Millipore) to eliminate contaminant rVV particles and used to infect fresh QM7 cell monolayers. Infections were performed in triplicate with undiluted and 10-1-10-5 serial dilutions of the initial stocks. A set of infected cells was used to determine the IBDV titer at 72 h pi. Another set was harvested at 72 h pi, and the corresponding extracts were analyzed for the presence of VP1 and VP2 proteins. The absence of contaminant infecting rVV particles was also monitored by Western blot using the mAbC3. The third set was also harvested at 72 h pi and used to collect cells originally infected with the undiluted stocks. These samples were subjected to three freeze-thawing cycles, and the procedure described above was repeated, thus allowing the analysis of the initial virus stock and two subsequent virus amplification rounds.
Purification of IBDV Polyprotein-derived Structures—QM7 and H5 cells (2-5 × 108 cells) were infected with appropriate rVV or rBV (multiplicity of infection 1-5 plaque-forming units/cell). At 72 h pi in rVV assays and at 48 h pi in rBV assays, cells were harvested, lysed in PES buffer (25 mm piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.2, 150 mm NaCl, and 20 mm CaCl2) plus 1% IGEPAL CA-630 (Sigma) on ice, and processed on a 25% sucrose cushion and a linear 20-50% sucrose gradient (19). The particulate material containing polyprotein-derived structures was concentrated 20-fold by ultracentrifugation and used for SDS-PAGE, Western blot, and electron microscopy analysis.
SDS-PAGE and Western Blot—Infected cell extracts (25 μl) or concentrated sucrose gradient fractions (10 μl) were added to Laemmli' s sample buffer to a 1× final concentration and heated at 100 °C for 3 min. Electrophoresis was performed in 11% polyacrylamide gels. Western blot analyses were carried out using an anti-VP2 serum as described (26).
Electron Microscopy—2-5-μl samples of concentrated sucrose gradient fractions were applied to glow-discharged carbon-coated grids for 2 min and negatively stained with 2% aqueous uranyl acetate. Micrographs were recorded with a JEOL 1200 EXII electron microscope operating at 100 kV at a nominal magnification of 40,000×.
Crystallization and Data Collection—Cubic crystals belonging to space group P213 (a = b = c = 326.72 Å) were obtained by the vapor diffusion method in hanging drops at room temperature by mixing 2 volumes of VP2-452_D431N-derived SVP (1.6 mg/ml) and 1 volume of the reservoir solution containing 14-17% polyethylene glycol 4000, 0.1 m Tris, pH 9.0, and 5% isopropanol. Crystals were transferred to a cryoprotecting solution containing 20% glycerol in the crystallization buffer and incubated for 1 min before they were flash-frozen by immersion in liquid nitrogen. X-ray data were collected from a single crystal using synchrotron radiation at the European Synchrotron Radiation Facility, Grenoble, France (beamline ID14-2) to 3.1 Å resolution. Diffraction images were processed using MOSFLM (30) and internally scaled with SCALA (31).
Structure Refinement—Crystals of D431N-mutated SVP were isomorphous to the previously obtained of native VP2 SVPs (PDB code 2GSY (22)). The initial maps for D431N-mutant were obtained after a rigid body fitting of the coordinates of native SVPs (PDB code 2GSY) to the new unit cell. The D431N mutation and other subtle changes were manually rebuilt using Coot (32), and the structure was then refined with CNS (33) using the 20-fold non crystallographic symmetry as a restraint. The final refinement statistics are summarized in supplemental Table 2. The coordinates and structure factors have been deposited into the Protein Data Bank (code 3FBM).
RESULTS
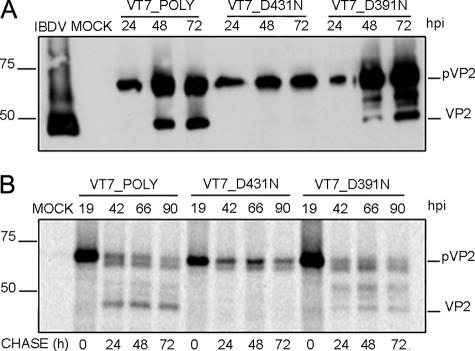
Effect of Mutations on Asp Residues Potentially Involved on pVP2 → VP2 Proteolytic Maturation—(p)VP2 residues Asp-391 and Asp-431 appeared to be likely candidates to trigger a nucleophilic attack on the pVP2 441-442 scissile bond. The IBDV polyprotein expressed by an inducible rVV provided an excellent framework to analyzing the possible role of residues Asp-391 and Asp-431 on pVP2 proteolytic maturation. As described above, the use of this system allows the accumulation of pVP2 and VP2 and the efficient assembly of virus-like particles (VLP), structurally undistinguishable from purified IBDV virions (26, 34). Hence, two single-point mutant versions of the IBDV polyprotein gene were generated and used to construct two rVV, called VT7LacOI/D431N and VT7LacOI/D391N, in which Asp residues under study at positions 431 and 391 were replaced by Asn residues, respectively. Cultures infected with these two rVV were harvested at different times pi (24, 48, and 72 h), and the corresponding extracts were analyzed by Western blot using VP2-specific serum. As a control for this experiment, cultures were also infected with VT7LacOI/POLY, a rVV expressing the wild-type polyprotein gene (26).
Although expression of wild-type and D391N polyproteins led to accumulation of both pVP2 and VP2 polypeptides, samples from cells expressing polyprotein D431N showed equivalent pVP2 levels but did not contain detectable VP2 (Fig. 2A). These results strongly suggested that the D431N substitution specifically abrogated pVP2 proteolytic processing and were further confirmed using an alternative experimental approach. QM7 cells were infected with the three described rVV and, at 19 h pi, the cultures were metabolically labeled with [35S]methionine for 1 h. Thereafter, cultures were extensively washed and incubated with fresh medium supplemented with an excess of cold methionine. At different times post-labeling (0, 24, 48, and 72 h), cells were harvested, and the corresponding extracts were subjected to immune precipitation using anti-VP2 antiserum. The results of this analysis (Fig. 2B) demonstrate that whereas in cells infected with VT7LacOI/POLY and VT7LacOI/D391N, the pVP2 polypeptide is progressively processed to mature VP2, the pVP2 polypeptide encoded by VT7LacOI/D431N remains intact for the duration of the experiment (72 h after its synthesis). Taken together, these results demonstrate that the conservative D431N substitution causes a complete blockade on pVP2 → VP2 proteolytic maturation.
FIGURE 2.
Effect of conservative mutations of residues Asp-431 and Asp-391 on pVP2/VP2 proteolytic maturation. A, Western blot analysis of proteins expressed in cells infected with rVV VT7LacOI/POLY, VT7LacOI/D431N, or VT7LacOI/D391N, respectively. Infected cultures were harvested at 24, 48, and 72 h pi, respectively. The corresponding extracts were subjected to SDS-PAGE and Western blot using anti-VP2 serum. B, QM7 cells infected with rVV VT7LacOI/POLY, VT7LacOI/D431N, or VT7LacOI/D391N were metabolically pulse-labeled with [35S]Met for 1 h at 19 hpi (hpi). The radioactivity was chased with by adding fresh medium containing an excess of cold methionine. Cultures were harvested at 0, 24, 48, and 72 h post-labeling, and the corresponding extracts were immunoprecipitated using anti-VP2 serum. The resulting samples were subjected to SDS-PAGE followed by autoradiography. Molecular weight markers (expressed in kDa, left) and bands corresponding to proteins pVP2 and VP2 (right) are indicated.
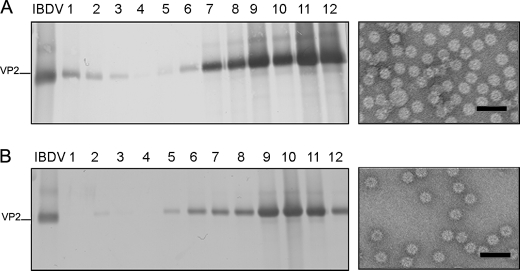
D431N Substitution Allows the Assembly of VP2 Trimers—VP2 trimers constitute the building block of the IBDV-related icosahedral capsids with different T as well as different tubular structures with helical symmetry (19, 21). Therefore, it seemed likely that the results described above might be caused by deficiency on trimer assembly induced by a VP2 conformational change generated in the D431N mutant. Such a conformational change might prevent both the assembly of IBDV provirions and the subsequent pVP2 proteolytic processing event.
Expression of chimerical VP2 genes in insect cells leads to the spontaneous assembly of T = 1 SVP built by 20 VP2 trimers arranged into pentamers. We have exploited this system to determine the effect of the D431N mutation on the assembly of VP2 trimers. A VP2 gene with the N-terminal 452 residues harboring the D431N substitution was generated, VP2-452_D431N, inserted into the pFastBac1 transfer vector, and then used to generate the corresponding rBV, rBV/VP2-452_D431N. Extracts from cells infected with rBV/VP2-452_D431N were used for SVP purification. As shown in Fig. 3, expression of the VP2-452D431N gene results in the assembly of T = 1 SVP that is undistinguishable under electron microscopy observation by negative staining from those produced by wild-type protein and D391N mutant (data not shown).
FIGURE 3.
VP2 with the D431N substitution assembles as T = 1 subviral particle VP2 trimers. A, wild-type VP2-452 SVP were purified by ultracentrifugation on sucrose gradients. Gradients were collected into 12 fractions, concentrated by ultracentrifugation, and analyzed by SDS-PAGE and Western blot using anti-VP2 serum. The direction of sedimentation was right to left, with fraction 12 representing the gradient top. The image shown on the right corresponds to a representative electron microscopy micrograph (negative staining) of wild-type SVP, present at the upper gradient fractions. B, mutant VP2-452_D431N assemblies were analyzed as described above. The image shown on the right corresponds to a representative electron microscopy micrograph (negative staining) of mutant VP2-452_D431N SVP present at the upper gradient fractions. Bars = 100 nm.
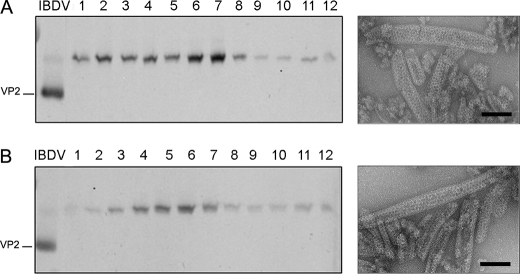
The possibility nevertheless existed that the mutation D431N might somehow affect the formation of VP2 hexamers formed by six VP2 trimers. To analyze this possibility we took advantage of a previous observation. As described above, we have shown that expression of VP2 long forms in a rBV expression system leads to the assembly of tubular structures formed exclusively by hexamers built by pVP2 intermediate precursor trimers (7, 19). Therefore, the D431N gene was subcloned into pFB/VP2-501, and the resulting plasmid, pFB/VP2-501_D431N, was used to generate the corresponding rBV. As shown in Fig. 4, expression of VP2-501 _D431N in insect cells results in the assembly of rigid tubules with an identical morphology to that of tubules purified from cells expressing wild-type VP2-501 gene. The results of this series of experiments demonstrate that the D431N substitution do not prevent either the trimer assembly or the subsequent formation of pentamers and hexamers.
FIGURE 4.
pVP2 variant with the D431N substitution assembles as tubular assemblies with hexagonal symmetry. A, wild-type p VP2-501 assemblies were purified by ultracentrifugation on sucrose gradients. Gradients were collected into 12 fractions, concentrated by ultracentrifugation, and analyzed by SDS-PAGE and Western blot using anti-VP2 serum. The direction of sedimentation was right to left, with fraction 12 representing the gradient top. The image shown on the right corresponds to a representative electron microscopy micrograph (negative staining) of wild-type tubular structures present at the lower fractions. B, mutant pVP2-501 _D431N assemblies were analyzed as described above. The image shown on the right corresponds to a representative electron microscopy micrograph (negative staining) of pVP2-501 _D431N rigid tubules at the lower fractions. Bars = 100 nm.
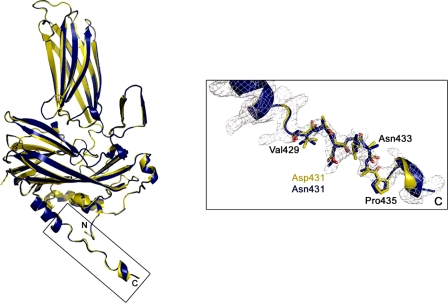
Finally, to rule out the possibility that the D431N substitution might induce conformational changes on the VP2 polypeptide undetectable by electron microscopy, the crystal structure of VP2-452_D431N-derived SVP was determined at 3.1 Å resolution (supplemental Table S1). The root mean square deviation of the superimposition of all 428 equivalent Cα atoms from VP2 in the structures of native and D431N SVP is 0.20 Å, showing that these structures are essentially identical in conformation. This similarity is also maintained within the flexible linker between helices α3 and α4 (residues 428-435), containing the D431N mutation (Fig. 5).
FIGURE 5.
Structure of the VP2 D431N T = 1 SVP monomer. Ribbon diagrams of wild-type (yellow) and mutant D431N (blue) VP2 proteins. The superposition was performed using all the 428 residues of both molecules. Shown on the right is an electron density map of the VP2 C-terminal boxed region around residue Asp-431. Residues from Val-429 to Pro-435 are shown as sticks and explicitly labeled every two residues.
pVP2 Undergoes an Intramolecular Proteolytic Processing—To get a better understanding of the pVP2 maturation mechanism, it was important to determine whether the pVP2 → VP2 cleavage is the result of an intra-(cis-cleavage) or intermolecular (trans-cleavage) event.
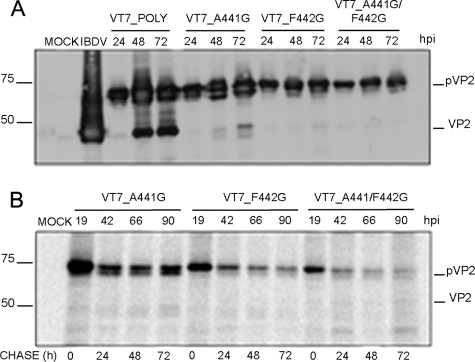
To discriminate between these two possibilities, a polyprotein mutant containing an intact Asp-431 residue but resistant to self-proteolytic processing was required. Such a mutant would be used for co-expression analysis to provide the pVP2 → VP2 proteolytic activity in trans. With this aim, three polyprotein genes containing one or two amino acid substitutions affecting either one or both amino acids at the pVP2 scissile bond (Ala-441—Phe-442) were generated. The mutant genes contain the following substitutions: (i) A441G, (ii) F442G, and (iii) A441G/F442G. Mutant genes were then inserted within the vaccinia virus genome, giving rise to rVV VT7LacOI/A441G, VT7LacOI/F442G, and VT7LacOI/A441G/F442G, respectively. The effect of the described mutations on pVP2 processing was analyzed by Western blot and pulse-chase/immune precipitation analysis. As shown in Fig. 6, whereas single substitutions cause a major but partial pVP2 → VP2 blockade, the double mutation A441G/F442G completely abolishes pVP2 processing. Accordingly, this mutant gene was selected for subsequent experiments.
FIGURE 6.
Analysis of the pVP2 cleavage site Ala-441-Phe-442. A, Western blot analysis of proteins expressed in cells infected with rVV VT7LacOI/POLY, VT7LacOI/A441G, VT7LacOI/F442G, and VT7LacOI/A441G/F442G. Infected cultures were harvested at 24, 48, and 72 h pi (hpi). B, QM7 cells were infected with different recombinants VT7LacOI/A441G, VT7LacOI/F442G, or VT7LacOI/A441G/F442G were pulse-labeled with [35S]Met for 1 h at 19 h pi. The radioactivity was chased with by adding with fresh medium containing an excess of cold methionine. Cultures were harvested at 0, 24, 48, and 72 h post-labeling, and the corresponding extracts immunoprecipitated using anti-VP2 serum. The resulting samples were subjected to SDS-PAGE followed by autoradiography. Molecular weight markers (expressed in kDa, left), and bands corresponding to proteins pVP2 and VP2 (right) are indicated.
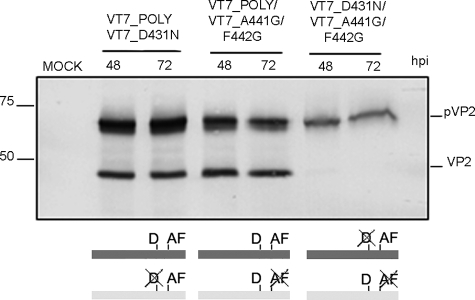
Next, cells were coinfected with VT7LacOI/POLY and VT7LacOI/D431N, VT7LacOI/POLY and VT7LacOI/A441G/F442G or VT7LacOI/D431N and VT7LacOI/A441G/F442G. Cultures were harvested at 48 and 72 h pi, and the corresponding extracts were analyzed by Western blot. As shown in Fig. 7, the presence of mature VP2 was exclusively detected in samples from cells expressing the wild-type polyprotein gene that provides both the catalytic Asp-431 residue and the intact cleavage site. Coinfection with VT7LacOI/D431N and VT7LacOI/A441G/F442G showed that pVP2-D431N remains uncleaved and is not proteolytically processed in trans by the wild-type pVP2, thus strongly suggesting that pVP2 processing is the result of a monomolecular cis-cleavage reaction.
FIGURE 7.
pVP2 intramolecular proteolytic processing. Western blot analysis of proteins expressed in QM7 cells coinfected with VT7LacOI/POLY and VT7LacOI/D431N, VT7LacOI/POLY and VT7LacOI/A441G/F442G, or VT7LacOI/A441G/F442G and VT7LacOI/D431N. Infected cultures were harvested at 48 and 72 h pi (hpi). A scheme indicating the combinations of rVV used in this experiment (mutated positions are crossed out) is shown at the bottom.
D431N Substitution Completely Abolishes IBDV Infectivity—It was important to assess the relevance of the Asp-431 on IBDV infectivity. This analysis was carried out using a reverse genetics approach using plasmids pT7_SA_Rz and B pT7_ SB_Rz containing the cDNA sequences corresponding to IBDV segments A and B, respectively, and plasmid pT7_SA/D431N_Rz, harboring a mutant version of segment A containing the D431N substitution.
The results obtained demonstrate that the D431N substitution completely abolishes the production of an infectious IBDV progeny (Table 1). Western blot analysis indicated that cells transfected with plasmids pT7_SA/D431N_Rz and pT7_SB_Rz failed to trigger the generation of infectious IBDV. This finding, in agreement with the observations described above, highlights the critical importance of the VP2 D431N residue on IBDV infectivity.
TABLE 1.
Virus titers obtained by reverse genetics
|
Coinfections
|
Virus titers
|
||
|---|---|---|---|
| First round | Second round | Third round | |
| pfu/ml | |||
| pT7-SA-Rz/pT7-SB-Rz | 1.3 ± 0.5 × 102 | 9.0 ± 1.3 × 104 | 6.0 ± 1.5 × 107 |
| pT7-SA(D431N)-Rz/ | 0 | 0 | 0 |
| pT7-SB-Rz | |||
pVP2 → VP2 Maturation Blockade Prevents the Assembly of Stable IBDV-derived VLP—IPNV morphogenesis involves the assembly of provirions containing a high proportion of pVP2 molecules. Provirion maturation appears to be associated to pVP2 proteolytic processing (13). Hence, it seemed feasible that blocking pVP2 → VP2 processing might favor the accumulation and isolation of IBDV provirions. To test this possibility, QM7 cultures were infected with VT7LacOI/POLY or VT7LacOI/D431N. Expression of the IBDV polyprotein in avian cells using an inducible rVV results in the accumulation of abundant levels of VLP with similar size and shape to those of authentic IBDV particles (26, 35). Remarkably, the assemblies produced with the rBV system in insect cells were nevertheless structurally varied, and in general, the formation of IBDV-like particles was rather inefficient. Insect cells might, therefore, have an important cellular factor(s) that interferes negatively with VLP assembly (e.g. cellular proteins and/or host environment).
Cultures harvested at 2 different times, 36 and 72 h pi, and the corresponding extracts were then used for the isolation of IBDV-derived assemblies using a sucrose gradient-based purification protocol followed by electron microscopy and Western blot analyses of the collected fractions. As described previously, extracts from cells infected with the rVV expressing the wild-type polyprotein gene contained abundant icosahedral VLPs with a relatively high VP2/pVP2 ratio (26). In contrast, no IBDV-derived assemblies were detected in samples from VT7LacOI/D431N-infected cells. The use of gentler extraction conditions and/or alternative purification protocols yielded similar results. The same results were observed with extracts from cells infected with the three rVVs expressing polyprotein genes containing mutations on the pVP2 cleavage site described above.
DISCUSSION
We analyzed the protease activity of the IBDV CP VP2. The IBDV polyprotein is a fusion of capsid precursor pVP2 with the viral protease (VP4) and the scaffolding protein (VP3). pVP2, a 512-residue precursor, undergoes sequential C-terminal processing events mediated by the viral protease VP4 to render shorter pVP2 polypeptides. These pVP2 intermediates with C-terminal extensions of variable length are further cleaved to render the mature VP2 (441 residues). Based on the VP2 x-ray structure, we examined the role of two likely candidate residues, Asp-391 and Asp-431, for catalyzing the last proteolytic event that occurs during capsid maturation. These residues were selected taking into account the structural analogies of domains S and B of IBDV CP to their counterparts in some positive-sense ssRNA viruses such as noda- and tetraviruses, in which the cleavage is mediated by acid residues, specifically Asp-75 of Flock House virus (FHV) (18) and Glu-103 of Nudaurelia capensis ω virus (36).
Our results indicate that he D431N mutation blocks the pVP2 → VP2 proteolytic trimming and affects the assembly of stable IBDV particles. Assembly of VLPs with the proper size and morphology, i.e. the T = 13 capsid, was not detected in cells expressing the polyprotein gene harboring the D431N mutation. Expression of pVP2 intermediate polypeptides with the point mutation D431N nonetheless can assemble either as allpentamer structures (T = 1 SVP) or tubular structures built of pVP2 hexamers. The isolation of these assembly products excluded a possible misfolding of the polypeptide as they are built by the same structural block, VP2 trimers (19). The crystal structure of the mutant D431N VP2 protein provides additional experimental evidence supporting this interpretation. Similarly, mutations on the scissile bond Ala-441—Phe-442 yielded no production of particles.
We previously reported that the molecular switch controlling VP2 polymorphism resides in the C-terminal 443GFKDII-RAIR452 segment that is organized as an amphipathic α-helix (19). In addition, most of the temporally bound 71-residue C-terminal segment consists of α-helices (11). Our working model assumes that assembly control of the complex IBDV T = 13 capsid requires the electrostatic interaction of the acidic segment of the VP3 C-terminal region, whose importance was previously established (29, 37), with the pVP2 amphipathic α-helix as an initial event in the adoption of different conformational states. In this context it is easily envisioned how the D431N mutation is not only unable to cleavage the Ala-441—Pro-442 scissile bond but also might preclude or alter the interaction of VP3 with the helical region of VP2 at the inner basement of the VP2 S domain. Furthermore, we believe that the interaction of the C-terminal VP3 segment with the pVP2 amphipathic α-helix might be necessary to temporally inhibit the last proteolytic event because, as reported previously, the pVP2 conformational state becomes irreversible (only able to assemble as pentamers) when its C-terminal region has been removed (21). Another fact that remains to be clarified in the sophisticated succession of interactions between VP3 and (p)VP2 refers to the high flexibility exhibited by the VP2 C-terminal end (21). It is likely that the conformational change originates from a rearrangement of some helical regions in the procapsid internal VP2 S domains as described for the Nudaurelia capensis ω virus conformational change (14, 17, 38).
Noda- and tetraviruses undergo a biphasic maturation process. During the first step, triggered by a reduction in pH, the procapsids undergo large scale structural rearrangements, thus quickly evolving to smaller, less porous, capsids; the second step is dependent on the first and consists of a relatively slow auto-proteolytic cleavage of the precursor CP subunits. Maturation of IPNV, a closely related birnavirus, also occurs by pVP2 processing to VP2 in which spherical provirions evolve to relatively smaller icosahedral virions (13). Our attempts to isolate IBDV-derived provirions using a wide variety of approaches have failed at present. This could be due to several factors: (i) a comparatively faster pVP2 → VP2 conversion rate for the IBDV capsid than that of its IPNV counterpart; or (ii) a weaker interaction between the IBDV pVP2 with the scaffolding protein VP3, thus leading to the assembly of more labile provirions.
In addition to the structural similarities between their CP polypeptides described above, birna-, noda-, and tetravirus share many other features at multiple levels (39). These viruses possess bipartite genomes with a similar overall organization (one segment coding for the RNA polymerase, and the other one encoding the capsid precursor protein). Interestingly, birna-, noda-, and tetravirus mRNA lack 3′-terminal poly(A) tails (15, 40). Their RNA-dependent RNA polymerases exhibit a non-canonical organization exclusive of this three-virus group containing a specific sequence permutation of the catalytic palm subdomain (41-43). An additional similarity between birnaviruses and positive-sense ssRNA viruses is the protein priming of RNA synthesis, described for IPNV (44) and originally discovered in picornaviruses (45). A further parallel among noda-, tetra-, and birnaviruses refers to the presence of active capsid-associated α-helical peptides generated during the proteolytic processing of the CP precursor C-terminal region. These peptides form hydrophilic channels that permeabilize biological membranes, allowing genome translocation during the virus entry process (11, 46). Finally, the multifunctional IBDV VP3 protein (involved in virus assembly as an scaffolding protein and RNA synthesis as an RNA-dependent RNA polymerase activator) is mostly found associated to the dsRNA segments forming ribonucleoprotein complexes (47). In this sense, nodavirus also expresses a dsRNA-binding protein that acts as a potent suppressor, inhibiting host RNA silencing (48, 49). Our finding that the IBDV CP precursor is self-cleaved in a cis-processing event mediated by an aspartic acid residue adds further experimental evidence reinforcing the structural and functional relationships among icosahedral ssRNA and dsRNA viruses.
Viral CPs ensure the successful propagation of viral genomes by sheltering them from a hostile environment and guiding them through the interaction with specific cell receptors in their journey to reach an appropriate milieu for a successful replication process. Nevertheless, there is increasing evidence that CPs play other important roles. In addition to the Asp- or Glu-based proteolytic activities associated to birna-, noda-, and tetravirus, other CPs such that of Semliki Forest virus, an enveloped plus strand RNA virus, exhibits a self-cleaving serine protease activity (50). Enzymatic activities associated to CP in most known examples found in fully assembled virus particles are not exclusively restricted to protease activities. The CP of Leishmania RNA virus, a dsRNA virus that persistently infects the parasitic protozoan Leishmania, possesses a site-specific RNA endoribonuclease activity that cleaves viral positive-sense RNA (51, 52). L-A virus, another dsRNA virus that infects the yeast Saccharomyces cerevisiae, has a CP that removes the m7GMP caps from host cellular mRNAs and covalently attaches them to a His residue on the capsid outer surface (53, 54). Unfortunately, the lack of specific sequence consensus motifs complicates the search for CP-associated enzymatic activities. Finally, these studies might settle the basis for the future design of capsid-targeted antiviral compounds trapping IBDV virion particles at an intermediate maturation stage rather than interrupting the assembly of capsid subunits, e.g. binding to trimers.
Supplementary Material
Acknowledgments
We thank J. L. Carrascosa (Consejo Superior de Investigaciones Científicas, Madrid) for stimulating discussions and other helpful input.
The atomic coordinates and structure factors (code 3FBM) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by Spanish Dirección General de Investigación (MEC) Grants BIO2006-09407, BFU2005-02376, and BFU2005-06487. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
Footnotes
The abbreviations used are: dsRNA, double-stranded RNA; ssRNA, single-stranded RNA; IBDV, infectious bursal disease virus; IPNV, infectious pancreatic necrosis virus; CP, capsid protein; rBV, recombinant baculovirus; rVV, recombinant vaccinia virus; SVP, subviral particle; VLP, virus-like particle; DMEM, Dulbecco's modified Eagle's medium; pi, postinfection.
References
- 1.Delmas, B., Kibenge, F. S. B., Leong, J. C., Mundt, E., Vakharia, V. N., and Wu, J. L. (2005) in Virus Taxonomy (Fauquet, C. M., Mayo, M. A., Maniloff, J., Desselberger, U., and Ball, L. A., eds) pp. 561-569, Elsevier Science Publishers B. V., Amsterdam
- 2.van den Berg, T. P., Eterradossi, N., Toquin, D., and Meulemans, G. (2000) Rev. Sci. Tech. 19 509-543 [PubMed] [Google Scholar]
- 3.Caspar, D. L. D., and Klug, A. (1962) Cold Spring Harbor Symp. Quant. Biol. 27 1-24 [DOI] [PubMed] [Google Scholar]
- 4.Baker, T. S., Olson, N. H., and Fuller, S. D. (1999) Microbiol. Mol. Biol. Rev. 63 862-922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulibaly, F., Chevalier, C., Gutsche, I., Pous, J., Navaza, J., Bressanelli, S., Delmas, B., and Rey, F. A. (2005) Cell 120 761-772 [DOI] [PubMed] [Google Scholar]
- 6.Böttcher, B., Kiselev, N. A., Stel'Mashchuk, V. Y., Perevozchikova, N. A., Borisov, A. V., and Crowther, R. A. (1997) J. Virol. 71 325-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castón, J. R., Martínez-Torrecuadrada, J. L., Maraver, A., Lombardo, E., Rodríguez, J. F., Casal, J. I., and Carrascosa, J. L. (2001) J. Virol. 75 10815-10828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sánchez, A. B., and Rodríguez, J. F. (1999) Virology 262 190-199 [DOI] [PubMed] [Google Scholar]
- 9.Chevalier, C., Lepault, J., Erk, I., Da Costa, B., and Delmas, B. (2002) J. Virol. 76 2384-2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Da Costa, B., Chevalier, C., Henry, C., Huet, J. C., Petit, S., Lepault, J., Boot, H., and Delmas, B. (2002) J. Virol. 76 2393-2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galloux, M., Libersou, S., Morellet, N., Bouaziz, S., Da Costa, B., Ouldali, M., Lepault, J., and Delmas, B. (2007) J. Biol. Chem. 282 20774-20784 [DOI] [PubMed] [Google Scholar]
- 12.Dobos, P. (1996) Annu. Rev. Fish Dis. 5 25-54 [Google Scholar]
- 13.Villanueva, R. A., Galaz, J. L., Valdes, J. A., Jashes, M. M., and Sandino, A. M. (2004) J. Virol. 78 13829-13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canady, M. A., Tihova, M., Hanzlik, T. N., Johnson, J. E., and Yeager, M. (2000) J. Mol. Biol. 299 573-584 [DOI] [PubMed] [Google Scholar]
- 15.Schneemann, A., Reddy, V., and Johnson, J. E. (1998) Adv. Virus Res. 50 381-446 [DOI] [PubMed] [Google Scholar]
- 16.Gallagher, T. M., and Rueckert, R. R. (1988) J. Virol. 62 3399-3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor, D. J., Krishna, N. K., Canady, M. A., Schneemann, A., and Johnson, J. E. (2002) J. Virol. 76 9972-9980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zlotnick, A., Reddy, V. S., Dasgupta, R., Schneemann, A., Ray, W. J., Jr., Rueckert, R. R., and Johnson, J. E. (1994) J. Biol. Chem. 269 13680-13684 [PubMed] [Google Scholar]
- 19.Saugar, I., Luque, D., Ona, A., Rodriguez, J. F., Carrascosa, J. L., Trus, B. L., and Caston, J. R. (2005) Structure 13 1007-1017 [DOI] [PubMed] [Google Scholar]
- 20.Oña, A., Luque, D., Abaitua, F., Maraver, A., Castón, J. R., and Rodríguez, J. F. (2004) Virology 322 135-142 [DOI] [PubMed] [Google Scholar]
- 21.Luque, D., Saugar, I., Rodriguez, J. F., Verdaguer, N., Garriga, D., Martin, C. S., Velazquez-Muriel, J. A., Trus, B. L., Carrascosa, J. L., and Caston, J. R. (2007) J. Virol. 81 6869-6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garriga, D., Querol-Audi, J., Abaitua, F., Saugar, I., Pous, J., Verdaguer, N., Castón, J. R., and Rodriguez, J. F. (2006) J. Virol. 80 6895-6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, C. C., Ko, T. P., Chou, C. C., Yoshimura, M., Doong, S. R., Wang, M. Y., and Wang, A. H. (2006) J. Struct. Biol. 155 74-86 [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Arias, A., Martinez, S., and Rodriguez, J. F. (1997) J. Virol. 71 8014-8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Torrecuadrada, J. L., Castón, J. R., Castro, M., Carrascosa, J. L., Rodriguez, J. F., and Casal, J. I. (2000) Virology 278 322-331 [DOI] [PubMed] [Google Scholar]
- 26.Lombardo, E., Maraver, A., Castón, J. R., Rivera, J., Fernandez-Arias, A., Serrano, A., Carrascosa, J. L., and Rodriguez, J. F. (1999) J. Virol. 73 6973-6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward, G. A., Stover, C. K., Moss, B., and Fuerst, T. R. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 6773-6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Earl, P. L., and Moss, B. (1993) in Current protocols in molecular biology (Ausubel, F. M., Brent, R., Kingston, R. E., More, D. D., Seidman, J. G., Smith, J. A., and StruhI, K., eds) pp. 16.17.1-16.18.10, John Wiley & Sons, New York
- 29.Maraver, A., Oña, A., Abaitua, F., Gonzalez, D., Clemente, R., Ruiz-Díaz, J. A., Castón, J. R., Pazos, F., and Rodríguez, J. F. (2003) J. Virol. 77 6438-6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leslie, A. (1991) in Crystallographic computing (Moras, D., Podjarny, A., and Thiery, J., eds) pp. 27-38, Oxford University Press, Oxford
- 31.Evans, P. (1994) Acta Crystallogr. D Biol. Crystallogr. 50 760-763 [DOI] [PubMed] [Google Scholar]
- 32.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 33.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 34.Castón, J. R., Rodríguez, J. F., and Casrrascosa, J. L. (2008) in Segmented double-stranded RNA viruses: Structure and Molecular Biology (Patton, J. T., ed) pp. 133-144, Caister Academic Press, Norfolk
- 35.Fernandez-Arias, A., Risco, C., Martinez, S., Albar, J. P., and Rodriguez, J. F. (1998) J Gen. Virol. 79 1047-1054 [DOI] [PubMed] [Google Scholar]
- 36.Taylor, D. J., and Johnson, J. E. (2005) Protein Sci. 14 401-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chevalier, C., Lepault, J., Da Costa, B., and Delmas, B. (2004) J. Virol. 78 3296-3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bothner, B., Taylor, D., Jun, B., Lee, K. K., Siuzdak, G., Schultz, C. P., and Johnson, J. E. (2005) Virology 334 17-27 [DOI] [PubMed] [Google Scholar]
- 39.Ahlquist, P. (2005) Curr. Biol. 15 465-467 [Google Scholar]
- 40.Hanzlik, T. N., and Gordon, K. H. (1997) Adv. Virus Res. 48 101-168 [PubMed] [Google Scholar]
- 41.Garriga, D., Navarro, A., Querol-Audi, J., Abaitua, F., Rodriguez, J. F., and Verdaguer, N. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20540-20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorbalenya, A. E., Pringle, F. M., Zeddam, J. L., Luke, B. T., Cameron, C. E., Kalmakoff, J., Hanzlik, T. N., Gordon, K. H., and Ward, V. K. (2002) J. Mol. Biol. 324 47-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan, J., Vakharia, V. N., and Tao, Y. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7385-7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, H. T., Si, W. D., and Dobos, P. (2004) Virology 322 199-210 [DOI] [PubMed] [Google Scholar]
- 45.Paul, A. V., van Boom, J. H., Filippov, D., and Wimmer, E. (1998) Nature 393 280-284 [DOI] [PubMed] [Google Scholar]
- 46.Cheng, R. H., Reddy, V. S., Olson, N. H., Fisher, A. J., Baker, T. S., and Johnson, J. E. (1994) Structure 2 271-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hjalmarsson, A., Carlemalm, E., and Everitt, E. (1999) J. Virol. 73 3484-3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chao, J. A., Lee, J. H., Chapados, B. R., Debler, E. W., Schneemann, A., and Williamson, J. R. (2005) Nat. Struct. Mol. Biol. 12 952-957 [DOI] [PubMed] [Google Scholar]
- 49.Chen, H. Y., Yang, J., Lin, C., and Yuan, Y. A. (2008) EMBO Rep. 9 754-760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morillas, M., Eberl, H., Allain, F. H., Glockshuber, R., and Kuennemann, E. (2008) J. Mol. Biol. 376 721-735 [DOI] [PubMed] [Google Scholar]
- 51.MacBeth, K. J., and Patterson, J. L. (1995) J. Virol. 69 3458-3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacBeth, K. J., and Patterson, J. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 8994-8998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanc, A., Ribas, J. C., Wickner, R. B., and Sonenberg, N. (1994) Mol. Cell. Biol. 14 2664-2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang, J., Naitow, H., Gardner, N. A., Kolesar, A., Tang, L., Wickner, R. B., and Johnson, J. E. (2005) J. Mol. Recognit. 18 158-168 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.