Abstract
The nuclear receptor vitamin D receptor (VDR) is known to associate with three vitamin D response element (VDREs)-containing regions within the CDKN1A (p21) gene region. Here we show in MDA-MB453 breast cancer cells that the natural VDR ligand 1α,25-dihydroxyvitamin D3 causes cyclical transcription factor binding and chromatin looping of distal VDREs to the transcription start site (TSS) of the p21 gene, leading to cyclical accumulation of the p21 mRNA. At the chromatin level, association of the mediator protein MED1 precedes both the peaks of VDR binding to VDREs and phosphorylated RNA polymerase (p-Pol II) to the TSS. The loss of co-repressor NCoR1-histone deacetylase (HDAC) 3 complex and inhibitory chromatin looping from VDREs to the TSS are also initial events followed by increased acetylation of histone 3 at lysine 9 at the TSS prior to initiation of transcription. Simultaneous to VDR and p-Pol II peaks, chromatin loops between VDREs and the TSS are formed, and the lysine demethylase LSD1 and the histone acetyltransferase CBP are enriched in both regions. This is followed by a moderate peak in p21 transcript accumulation, repeated in cycles of 45-60 min. The transcript accumulation pattern is disturbed by siRNA inhibition of the mediator protein MED1, LSD1, NCoR1, or various HDACs, whereas CBP appears unnecessary for the response. Inhibition of MED1, HDAC4, or LSD1 by siRNA also attenuates ligand-induced chromatin looping. In conclusion, 1α,25-dihydroxyvitamin D3 regulates p21 transcription by inducing cyclical chromatin looping that depends on both histone deacetylation and demethylation.
As a member of the nuclear receptor (NR)2 superfamily the vitamin D receptor (VDR) acts as a transcription factor that binds to specific response elements (VDREs) within the regulatory regions of its primary target genes (1). The natural VDR ligand 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3) has an important role in the regulation of calcium and phosphate homeostasis and bone mineralization (2). In addition to this classical role, there is both epidemiological and preclinical evidence that 1α,25(OH)2D3 is an anti-proliferative agent (3). Vitamin D deficiency increases the risk of certain cancers, whereas the administration of 1α,25(OH)2D3 in cell culture or in animal models for cancer inhibits angiogenesis and induces G0/G1 arrest, differentiation, and apoptosis (4). These anti-proliferative effects are exerted by various 1α,25(OH)2D3 target genes, among which the product of cyclin-dependent kinase inhibitor 1A (CDKN1A, also called p21) gene induces G0/G1 cell cycle arrest and differentiation (5). We have previously characterized three functional VDREs residing 2-7-kb upstream of the TSS of p21 gene (6).
Non-liganded VDR is associated with primary co-repressors, such as the nuclear receptor co-repressor (NCoR1), that attenuate transcription via interaction with histone deacetylases (HDACs) that pose the chromatin modifications unfavorable for transcription or, at least in the case of NCoR1, are also able to recruit a H2A ubiquitin ligase that inhibits the elongation by RNA polymerase II (Pol II) beyond the first nucleosome of the transcribed region (7).
Introduction of a ligand results in a conformational change in the ligand-binding domain of the VDR, leading to enhanced binding to its heterodimerization partner retinoid X receptor (1) and an exchange of co-repressors to primary co-activators. Those include the members of the p160 family that recruit secondary co-activators, such as the histone-modifying enzymes, the histone acetyltransferase CBP, and the lysine demethylase LSD1, and the mediator complex subunit MED1 that enables the contact to transcriptional machinery.
The histone modifications serve as specific signals for chromatin-binding proteins, affecting cofactors that remodel the chromatin to permit transcription. For example, dimethylation of histone 3 at lysine 4 (H3K4me2) is linked to active core promoter and enhancer regions (8) and is recognized by the ATP-dependent chromatin-remodeling factor CHD1 that can alter the accessibility of DNA for transcription (9). Acetylated histone 3 at lysine 9 (H3K9ac) has a strong association to positive regulation of transcription (10) and is recognized by CBP that acetylates H3K14 and TAF1, which a subunit of the general transcription factor TFIID (11, 12).
In addition to providing a contact between sequence-specific transcription factors and the basal transcription machinery, the Mediator complex promotes the formation of the preinitiation complex and the phosphorylation of Pol II at serine 5 (13). As VDREs are not restricted to proximal promoters of VDR target genes, chromatin looping provides the physical proximity between the response element and the TSS. Stimulus-dependent changes in chromatin looping have been studied extensively in the β-globin locus at different stages of development, but also in response to NR ligands, such as estradiol and 1α,25(OH)2D3 (14-17).
Recently, cyclical models have been proposed for the activation of transcription by NRs, including those for estrogen receptor α on the trefoil factor-1 gene (18) and for VDR on the 24-hydroxylase (19), GADD45, and VDUP1 genes (20). In these models the ligand-dependent transcription is seen as a cyclical process, where alternating activating and repressive actions on chromatin are required, providing means to stringently regulate the endurance and strength of the transcriptional response.
To investigate the interactive transcriptional role of NRs and chromatin looping, we studied protein-chromatin associations on both the VDREs and the TSS, the association frequency between the TSS and the VDREs as well as the outcome in the form of p21 transcript accumulation. The role of histone-modifying cofactors along with the mediator complex subunit MED1 in the latter two processes was also investigated by siRNA knock-down. After synchronization by 1α,25(OH)2D3 alone, we observed that the ligand induces cyclical mRNA accumulation, chromatin looping and association of p-Pol II, MED1, and VDR with the VDRE containing regions of the p21 promoter. Furthermore, MED1, HDAC4, and LSD1 were found to be essential for ligand-dependent looping from distal regions to the TSS and the cyclic induction of p21 transcription. In conclusion, both histone deacetylation and demethylation are essential for the ligand-dependent dynamic looping of chromatin and the increased transcription of p21 in response to 1α,25(OH)2D3.
EXPERIMENTAL PROCEDURES
Cell Culture—MDA-MB453 human mammary epithelial metastatic carcinoma cells were grown in α-MEM supplemented with 10% fetal bovine serum in a humidified 95% air/5% CO2 incubator. Before mRNA extraction, 650,000 cells for each well of a 6-well plate were seeded ∼24 h prior to collection in phenol red-free Dulbecco's modified Eagle's medium with 5% charcoal-stripped fetal bovine serum. For chromatin immunoprecipitation (ChIP) assay or chromosome conformation capture (3C) assay, cells were grown in the same medium overnight to reach 50 to 60% confluency. For all experiments, cells were treated for indicated time periods with 10 nm 1α,25(OH)2D3 (kindly provided by Dr. Milan Uskokovic, BioXell Inc, Nutley, NJ) or ethanol (0.001%).
RNA Extraction and Real-time Quantitative PCR—Total RNA extraction and cDNA synthesis were performed as described previously (17). Real-time quantitative PCR for cDNA was performed using a LightCycler® 480 System (Roche Applied Science) and FastStart SYBR Green Master mix (Roche Applied Science). The PCR cycling conditions were: preincubation for 10 min at 95 °C, 38 cycles of 20 s at 95 °C, 15 s at 60 °C and 15 s at 72 °C. The sequences of the specific primer pairs for the genes CBP, HDAC3, HDAC4, HDAC5, HDAC7, LSD1, MED1, NCoR1, p21, SMRT, and the control gene acidic riboprotein P0 (RPLP0) are listed in supplemental Table S1. Fold inductions were calculated using the formula 2-(ΔΔCt), where ΔΔCt is the ΔCt(ligand)-ΔCt(vehicle), ΔCt is Ct(p21) - Ct(RPLP0) and Ct is the cycle at which the threshold is crossed. Quality of the PCR product was monitored using post-PCR melt curve analysis.
ChIP Assay—ChIP and re-ChIP were performed as described previously (6) except for the following changes: (i) formaldehyde cross-linking time was reduced to 5 min, (ii) chromatin was lysed in 0.6 ml (instead of 1 ml) and sonicated using a Bioruptor UCD-200 (Diagenode, Liege, Belgium) with 10 × 30 s pulses, (iii) preincubation with salmon sperm beads was left out and instead protein A-agarose beads (Upstate Biotechnology, Lake Placid, NY) were blocked overnight with 10 mg/ml of bovine serum albumin and 0.1 mg/ml of salmon sperm DNA at 4 °C, and (iv) chromatin bound to agarose beads were eluted with a different elution buffer (25 mm Tris-HCl pH 7.5, 10 mm EDTA, 0.5% SDS) for 30 min at 65 °C followed by second elution for 2 min at room temperature. Antibodies against VDR (sc-1008), HDAC3 (sc-11417), HDAC4 (sc-11418), CBP (sc-369), NCoR1 (sc-8994), and p53 (sc-6243) were obtained from Santa Cruz Biotechnologies (Santa Cruz, CA). The antibody against p53 was used as a negative control because in MDA-MB453 cells the DNA binding domain of the p53 protein is mutated (21). Antibodies against H3K4me2 (07-030) and H3K9ac (07-352) were obtained from Upstate, while antibodies against the phosphorylated Pol II (p-Pol II; ab5131), LSD1 (ab17721) and T1457-phosphorylated MED1 (ab60950) were obtained from Abcam (Cambridge, UK).
3C Assay—Chromatin from two cells of a 6-well plate (750,000 cells plated per well) was collected, cross-linked, and lysed as for ChIP assays, but the sonication was reduced to one 15-s pulse. After removal of cellular debris by centrifugation, 25 μl of chromatin was diluted in 75 μl of ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm NaCl, protease inhibitors, and 16.7 mm Tris-HCl, pH 8.1) supplemented with protease inhibitors (Complete protease inhibitor mixture, Roche Applied Science), Red or Tango buffer (Fermentas, Vilnius, Lithuania) and digested overnight at 37 °C with 25 units of the restriction enzymes MvaI or Hpy8I (Fermentas), respectively. Digested chromatin was diluted 1:6 to T4 ligation buffer supplemented with 0.5 mm ATP and ligated with 15 units of T4 DNA ligase (Fermentas) for 4 h at room temperature. Samples were reverse cross-linked and DNA was recovered as described previously for ChIP assay (6). As positive controls, plasmids carrying the p21 promoter regions-7930 to -6072 or -4968 to +37 were digested and ligated with plasmid covering the TSS region (-676 to +535).
PCR of Chromatin Templates—For each of the three VDRE-carrying regions and the TSS of the p21 gene, genomic primers were designed (supplemental Table S2) and for their quantification 6-carboxyfluorescein (FAM)-modified probes were used (supplemental Table S3). Similarly, primers were designed for the detection of C3 ligation products (supplemental Table S4) and their TaqMan quantification (supplemental Table S5). To achieve quantifiable product specificity, the probes used for 3C ligation products were targeted against the ligation site, as described previously for quantification of chromatin looping (16). All oligonucleotides used in this study were obtained from Eurogentec (Liege, Belgium). Real-time quantitative PCR was performed with the Maxima Probe qPCR master mix (Fermentas) on a LightCycler® 480 System (Roche). The PCR cycling conditions were: preincubation for 10 min at 95 °C, 50 cycles of 20 s at 95 °C, 60 s at 60 °C. For the PCR on the TSS, preincubation for 10 min at 95 °C, 50 cycles of 20 s at 95 °C, 60 s at 61 °C was used with GC-rich solution (Roche Applied Science) in addition to the Maxima Probe qPCR master mix. The PCR products were also resolved on 2% agarose gels to control correct product size. Relative association of chromatin-bound proteins or histone modifications were calculated using the formula 2-(ΔCt), where ΔCt is Ct(output) - Ct(input), output is the immunoprecipitated DNA and input is the purified genomic DNA from starting material of the ChIP assay. For the 3C assay relative chromatin looping was calculated using the formula 2-(ΔΔCt), where ΔΔCt is the ΔCt(target) -ΔCt(non-treated p21-2 for Hpy8Irestricted template or p21-3 for MvaI restricted template), ΔCt is Ct(3C ligation product) - Ct(positive control plasmid).
siRNA Inhibition—MDA-MB453 cells were reverse transfected with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions using a mixture of three double-stranded siRNA oligonucleotides per gene (Eurogentec, 200 pmol of each siRNA, supplemental Table S6). For cDNA synthesis 650,000 cells, for 3C assays 750,000 cells, and for ChIP assays 4,500,000 cells were used. Cell treatments were started 24 h after plating and RNA extraction, real-time quantitative PCR, ChIP assays, and 3C assays were carried out as described above.
RESULTS
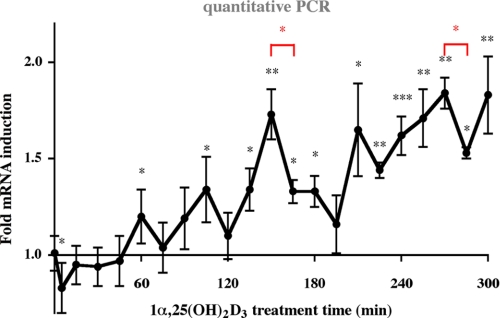
Cyclical Induction of p21 mRNA Expression by 1α,25(OH)2D3—The p21 gene has been shown to respond to 1α,25(OH)2D3 in several mammary cell lines, including MDA-MB453 cells (17). To further elucidate the dynamics of p21 induction, we performed real-time quantitative PCR analysis of p21 mRNA expression in response to 1α,25(OH)2D3 in a detailed time course of 300 min with 15-min intervals (Fig. 1). The first peak of p21 mRNA expression appeared after 60 min, the second at 105 min followed by a third at 150 min. A fourth peak appeared at 210 min and a fifth peak at 255 to 270 min. After each of these peaks, the accumulation of p21 mRNA ceased resulting in a decrease of p21 mRNA levels. The peaks of p21 mRNA levels appeared in cycles of 45-60 min with the longest lag time after the major peak at 150 min, where a 1.7-fold induction decreased within 45 min to levels close to that of untreated cells. Please note that the cells had not been synchronized. Thus, 1α,25(OH)2D3 in itself seems to be sufficient for the induction of cyclicity in p21 transcription.
FIGURE 1.
Cyclical induction of p21 transcription by 1α,25,(OH)2D3. Real-time quantitative PCR was performed to measure the time-dependent mRNA expression of the p21 gene in MDA-MB453 cells after treatment with 10 nm 1α,25(OH)2D3. The data were normalized to the expression of the housekeeping gene RPLP0, and fold inductions were calculated in reference to vehicle control. Data points indicate the means of at least three independent cell treatments, and the bars represent standard deviations. A two-tailed Student's t test was performed to determine the significance of the stimulation in reference to vehicle-treated control (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Taken together, these observations suggest that 1α,25(OH)2D3 induces p21 transcription only for short periods at selected time points. Moreover, the drastic decreases of the transcript accumulation level between the phases of mRNA induction indicate that the gene is actively repressed at these time points.
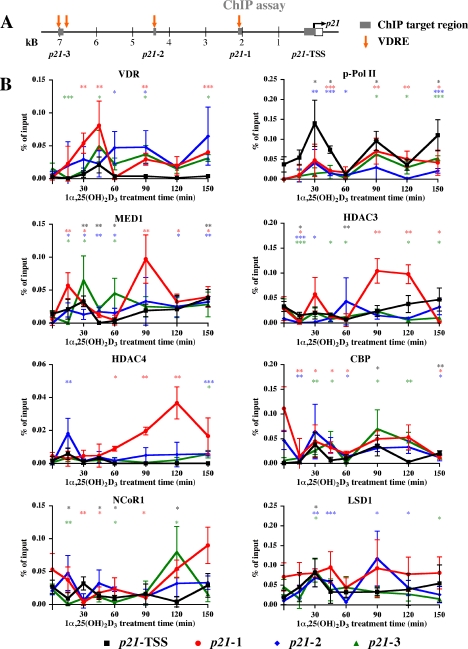
Cyclical Enrichment of VDR, p-Pol II, and MED1 on 1α,25(OH)2D3-responsive Regions—To study whether the cyclical induction of the p21 mRNA in response to 1α,25(OH)2D3 is based on parallel cyclical association of VDR and its partner proteins to the regulatory regions of the p21 gene, we performed in 1α,25(OH)2D3-treated MDA-MB453 cells ChIP assays with antibodies against VDR, p-Pol II, MED1, HDAC3, HDAC4, CBP, NCoR1, H3K4me2, H3K9ac, and LSD1 (Fig. 2). We analyzed the time period 0 to 150 min, in which the p21 mRNA peaked three times (Fig. 1). On the chromatin templates we determined by real-time quantitative PCR the 1α,25(OH)2D3-induced enrichment of the three previously identified VDR-associated regions (6, 17) and the p21 TSS in comparison to that in untreated cells (Fig. 2A).
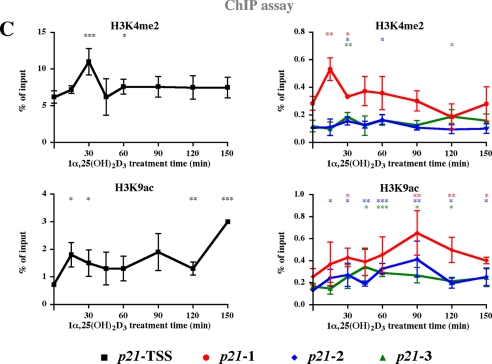
FIGURE 2.
Dynamic association of VDR, cofactors, and histone modifications to VDRE containing regions and the TSS of the p21 gene. Schematic overview on the human p21 promoter indicating the location of VDREs described previously (6) and of targeted genomic regions (A). On each of these four regions ChIP assays with indicated antibodies were performed on chromatin extracted from MDA-MB453 cells that had been treated with 10 nm 1α,25(OH)2D3 for the indicated times (B and C). Analysis was performed by real-time quantitative PCR using FAM-labeled probes, and association was calculated relative to input samples. Association of p53 was subtracted from relative association levels to control for nonspecific binding. Data points indicate the means of at least three independent cell treatments, and the bars represent standard deviations. A two-tailed Student's t test was performed to determine the significance of the stimulation in reference to vehicle-treated control (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Because the central events of 1α,25(OH)2D3-dependent gene transcription are the direct binding of VDR and the induction of transcription by Pol II, these two proteins were analyzed first (Fig. 2B). The association of VDR peaked at all three VDRE containing genomic regions (p21-1, -2, and -3) at 30-45, 90, and 150 min after the onset of ligand treatment. Similar pattern with the same maxima was observed also for p-Pol II. However, already at 15 min, i.e. before VDR and p-Pol II peaks, acetylation of H3K9 was induced 2-fold on the TSS. Similarly, MED1 peaked at 15 min on regions 1 and 2, HDAC3 decreased on all regions, whereas NCoR1 was significantly decreased only on the TSS and on region 3 (Fig. 2, B and C). It should be noted that the basal levels of H3K4me2 and H3K9ac were 20- and 3-times higher, respectively, on the TSS than on the VDRE-containing regions. Therefore, they are presented in Fig. 2C in separated graphs with different scales. At 30 min, i.e. together with VDR and p-Pol II, MED1 peaked at the TSS and on region 3, CBP on the TSS and H3K9ac on regions 1 and 2. Simultaneously, H3K4me2 was induced on all regions, and LSD1 on regions 2, 3, and the TSS. After 45 min of treatment, the enrichment of VDR still peaked on regions 1 and 3 and levels of p-Pol II remained higher than in untreated cells, whereas MED1 escaped from the TSS and region p21-1. Concurrently, the enrichment of H3K4me2 and H3K9ac were reduced on the TSS. At the time point 60 min, most proteins displayed low associations, while MED1 showed a minor peak on region 3.
At 90 min after onset of ligand treatment, i.e. concurrently with the second peak of VDR and p-Pol II association, MED1 and HDAC3 peaked on region 1, H3K9ac on both regions 1 and 2, CBP on region 3, and LSD1 on region 2. In contrast, NCoR1 stayed low on all regions, while HDAC4 increased only on region 1. At 120 min the rather low association of VDR and p-Pol II with all regions was well reflected by MED1, while HDAC3 and HDAC4 were still high on region 1, CBP on region 3, and NCoR1 on all regions. In addition, H3K4me2 was high on region 3, whereas H3K9ac was reduced from the TSS. Finally, the third peak of VDR and p-Pol II at 150 min coincided with increased binding of MED1 to the TSS, H3K9ac of the TSS and NCoR1 binding to region 1.
In summary, VDR and p-Pol II binding patterns showed similarity to each other on all regions studied, as well as to the patterns of MED1 and LSD1 association. H3K9ac displayed analogy to VDR, p-Pol II and MED1 association. Unlike other enrichments studied, those of H3K4me2 and HDAC4 did not show cyclicity. Generally, the effects on histone modifications and protein association were modest but significant, reflecting the modest effects of 1α,25(OH)2D3 on the transcript levels of the p21 gene. All observations are summarized in Table 1.
TABLE 1.
Summary of changes on the p21 gene promoter
1α,25(OH)2D3-triggered associations of proteins with four regions of the p21 promoter are summarized and compared with histone modifications, looping to the TSS and mRNA accumulation. “+” indicates a region showing significantly increased association with the mentioned protein/looping to the TSS compared to untreated cells. At the time 0 point, this indicates that the region shows significantly higher association with the mentioned protein/looping to the TSS compared to the lowest point in treated cells, “−” indicates regions showing significantly decreased association with the mentioned protein/looping to the TSS compared to untreated cells, whereas blank entries represent no significant change.
| 1α,25(OH)2D3 treatment time | Region | VDR | p-Pol II | MED1 | HDAC3 | HDAC4 | CBP | NCoR | LSD1 | H3K4me2 | H3K9ac | Looping to the TSS | p21 mRNA accumulation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | p21-TSS | + | + | + | + | 1.00 | |||||||
| p21-1 | + | + | + | + | |||||||||
| p21-2 | + | + | |||||||||||
| p21-3 | + | + | + | + | + | + | |||||||
| 15 | p21-TSS | + | 0.95 | ||||||||||
| p21-1 | + | + | |||||||||||
| p21-2 | + | + | + | ||||||||||
| p21-3 | |||||||||||||
| 30 | p21-TSS | + | + | + | + | 0.94 | |||||||
| p21-1 | + | + | + | + | |||||||||
| p21-2 | + | + | + | + | + | + | |||||||
| p21-3 | + | + | + | + | |||||||||
| 45 | p21-TSS | + | - | - | 0.96 | ||||||||
| p21-1 | + | + | - | - | |||||||||
| p21-2 | + | + | + | + | |||||||||
| p21-3 | + | - | + | + | |||||||||
| 60 | p21-TSS | - | - | + | 1,20 | ||||||||
| p21-1 | + | - | |||||||||||
| p21-2 | + | + | + | - | + | + | |||||||
| p21-3 | + | - | - | + | |||||||||
| 90 | p21-TSS | + | + | 1,19 | |||||||||
| p21-1 | + | + | + | + | - | + | + | ||||||
| p21-2 | + | + | + | + | + | ||||||||
| p21-3 | + | + | + | + | + | ||||||||
| 120 | p21-TSS | - | + | 1,11 | |||||||||
| p21-1 | + | + | + | + | |||||||||
| p21-2 | + | + | + | + | |||||||||
| p21-3 | + | + | + | + | + | + | |||||||
| 150 | p21-TSS | + | + | + | + | 1,73 | |||||||
| p21-1 | + | + | + | - | - | + | |||||||
| p21-2 | + | + | + | - | + | ||||||||
| p21-3 | + | + | - | + | - | - |
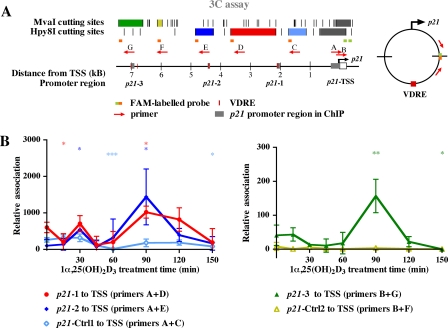
Dynamic Chromatin Looping Provides a Contact between VDRE Regions and the TSS—To unravel the spatial requirements as to how the VDRE carrying regions up to 7-kb upstream of the TSS harvest p-Pol II and hence contribute to enhanced p21 transcription, we performed 3C assays (Fig. 3). In this assay chromatin is cross-linked in living cells as in ChIP assays, but instead of shearing and antibody-based selection, chromatin is restriction digested and then ligated in circumstances favoring intramolecular interactions. Hence the probability that two distant chromatin regions separated by multiple restriction sites will form a ligation product increases, if they are spatially close via either random interactions or specific chromatin looping. We studied the TSS association of five upstream chromatin fragments located 1.5-7 kb from the p21 TSS, where the fragments with location nearest to the TSS should show the highest association based on random interactions (22). Association of the more proximal regions to the TSS was studied using the restriction enzyme Hpy8I, whereas for the more distal regions MvaI was used. To quantify specifically the ligation products of VDRE-containing regions to the TSS of p21, we used FAM-labeled oligonucleotides in quantitative PCR (Fig. 3A).
FIGURE 3.
1α,25(OH)2D3 induces dynamic looping of VDR binding regions to the TSS of the p21 gene. Schematic overview on the human p21 promoter indicating previously described VDREs (6), MvaI and Hpy81I restriction enzyme recognition sites and location of primers along with FAM-labeled probes used for quantification of the 3C products (A). 3C assays were performed with chromatin from MDA-MB453 cells that were treated for the indicated times with 10 nm 1α,25(OH)2D3 (B). Chromatin was extracted, cross-linked, and digested with either MvaI or Hpy8I. After ligation, the DNA was extracted and analyzed by PCR with primer A in combination with primers C, D, or E for Hpy8I-digested chromatin (left figure) or with primers F or G for MvaI-digested chromatin (right figure). Re-ligated digestions of subcloned p21 promoter fragments served as positive controls. Analysis was performed by real-time quantitative PCR using a FAM-labeled probe targeting the ligation site specific for the product. PCR efficacy was normalized to positive controls. Values indicate looping as relative values compared with basal looping of region p21-2 to the TSS. Data points indicate the means of at least three independent cell treatments, and the bars represent standard deviations. A two-tailed Student's t test was performed to determine the significance of the stimulation in reference to vehicle-treated control (*, p < 0.05; **, p < 0.01; ***, p < 0.001). Targeted restriction fragments in A and quantitative results in B are shown in the same color.
MDA-MB453 cells were treated identically as for the ChIP assay (Fig. 2), i.e. the same time points were chosen for chromatin extraction after 1α,25(OH)2D3 treatment (Fig. 3B). While the basal looping of the VDRE containing region 2 to the TSS was found to be lower than that of control region 1, as expected based on its location, regions 1 and 3 showed higher association to the TSS than the unresponsive control fragments at -1.5 and -6 kb, respectively. This could indicate active repression of p21 transcription via these regions. Consistently, both NCoR1 and HDAC3 were initially present on these regions and their enrichment decreased ligand-dependently within 15 to 30 min, although VDR was not initially present on region p21-1 (Fig. 2B). For region p21-1, the association with the TSS first decreased at 15 min and was re-introduced at 30 min of ligand treatment. For region 2, the first, albeit minor peak of looping to the TSS was found already at 30 min (Fig. 3B). Interestingly, in this time period the looping of region 3 to the TSS was at its minimum. These observations also fit with the p-Pol II binding at this time point, which showed a maximum for regions 1 and 2, but not for region 3 (Fig. 2B). On all three VDRE-containing regions, 1α,25(OH)2D3 maximally induced their association with the TSS by 2-15-fold at 90 min after onset of treatment (Fig. 3B), concomitantly with p-Pol II enrichment on these regions (Fig. 2B).
Taken together, basal looping of VDRE containing regions to the TSS correlates with the enrichment of NCoR1 and HDAC3, while the looping after 30 and 90 min of 1α,25(OH)2D3 treatment coincides with p-Pol II associations to these regions, suggesting simultaneous looping and transcription initiation. However, at 150 min, the association of the VDRE-containing regions to the TSS is low, although p-Pol II is significantly associated with these regions. This could in principle result from association with TSSs of other genes than of p21. Alternatively, it may suggest that upon loss of ligand-responsive looping, the transcriptional complexes are partitioned to both the TSS and the ligand-responsive regions, explaining enrichment of Pol II on distal regions without association with the TSS.
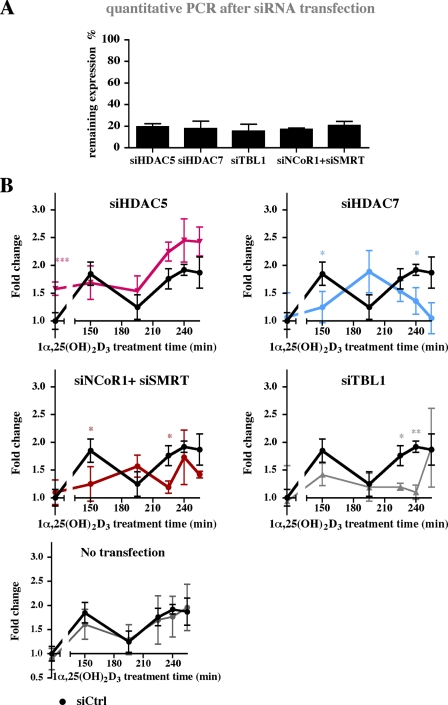
MED1, LSD1, and HDAC4 Are Essential for the Cycling of p21 Transcription—Our ChIP results on the ligand-responsive association of co-activators, co-repressors and histone modifying enzymes with the genomic regions of the p21 gene (Fig. 2, B and C) suggest that these proteins have a role in the transcriptional response of the gene to 1α,25(OH)2D3. To test this assumption, we individually diminished the expression of MED1, HDAC3, HDAC4, CBP, NCoR1, and LSD1 by siRNA oligonucleotide transfection of MDA-MB453 cells (Fig. 4). Western blot was used to monitor the efficacy of siRNA on the protein level (supplemental Fig. S2). In addition to the factors studied in ChIP assays, we studied the effect of knock-down of VDR-linked cofactors, such as HDAC5 and HDAC7 (17), and the NCoR1 related co-repressor Silencing Mediator of Retinoid and Thyroid Receptors (SMRT, also called NCoR2) in combination with NCoR1 (Fig. S2). The inhibition by specific siRNAs resulted in 15-25% remaining expression of the targeted mRNAs, when compared with cells transfected with non-targeted siRNA (supplemental Fig. S2A). After a 24-h transfection period with siRNA oligonucleotides, the MDA-MB453 cells were stimulated with 1α,25(OH)2D3 and RNA was extracted. We focused on the period of 150 to 255 min after the onset of stimulation, which showed the most drastic effects on mRNA accumulation (Fig. 1).
FIGURE 4.
Effect of cofactor silencing on the p21 mRNA response to 1α,25,(OH)2D3. MDA-MB453 cells were transfected for 24 h with siRNA oligonucleotides against the genes MED1, LSD1, HDAC4, NCoR1, HDAC3, CBP, or with a non-targeted siRNA (siCtrl), and subsequently either not stimulated (A) or stimulated (B) for the indicated times with 10 nm 1α,25(OH)2D3. Real-time quantitative PCR was used to determine the mRNA expression of indicated siRNA target genes (A). Remaining expressions were calculated in reference to samples transfected with a non-targeted siRNA (siCtrl). Real-time quantitative PCR was performed to measure the time-dependent mRNA expression of the p21 gene relative to the control gene RPLP0 (B). Fold inductions were calculated in reference to vehicle control that had been transfected with non-targeted siRNA (siCtrl). Data points indicate the means of at least three independent cell treatments, and the bars represent standard deviations. For B, a two-tailed Student's t test was performed to determine the significance of the effects of the specific siRNAs in reference to control siRNA (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
We found that knock-down of MED1 totally abolished the transcriptional cycles, validating the essential role of this member of the mediator complex in transmitting the transcriptional response of NRs (Fig. 4). Knock-down of LSD1 decreased the basal p21 mRNA expression and disturbed the cyclical induction of p21 transcript, but did not completely abolish the ligand induced rise in the transcript levels. Knock-down of the other secondary co-activator CBP did not affect either basal or 1α,25(OH)2D3 induced p21 gene transcription. This suggests that CBP, despite the ligand-dependent enrichment on the regulatory regions of the p21 gene (Fig. 2B), is unnecessary for the ligand response.
Contrary to the co-activator CBP, the reduction of co-repressor expression by siRNA affected both the basal expression of p21 and its responsiveness to 1α,25(OH)2D3. The silencing of NCoR1 induced the basal expression of p21 and resulted in shifted timing of the cyclical fluctuation of p21 mRNA upon ligand treatment. Simultaneous silencing of both NCoR1 and SMRT had similar effects on ligand response but lacked effects on basal expression, as did the silencing of TBL1, a member of the NCoR/SMRT silencing complexes. The knock-down of HDACs 3, 4, 5, and 7 (based on our previous studies on their importance in the 1α,25(OH)2D3 response of cyclin-dependent kinase inhibitor genes (17)) showed that the roles of individual HDACs in the response were non-redundant and distinct. Knock-down of either HDACs 3, 4, or 5 induced basal expression of p21 and inhibited the ligand-induced rise in p21 transcript level at 150 min. The silencing of HDAC4 lead to disturbed p21 transcript pattern at later time points, whereas the transcript patterns in HDAC3 silenced cells were after 150 min similar to patterns of cells transfected with non-targeted siRNA.
In summary, HDACs and NCoR1 repress the basal expression of the p21 transcript, whereas LSD1 induces it. MED1 is non-redundant and essential to the ligand response, while knock-down of LSD1, HDAC3, HDAC4, or NCoR1 attenuates it. In addition to MED1, silencing of HDAC4 and LSD1 severely disturb the cyclical pattern of p21 transcript accumulation in response to 1α,25(OH)2D3 treatment.
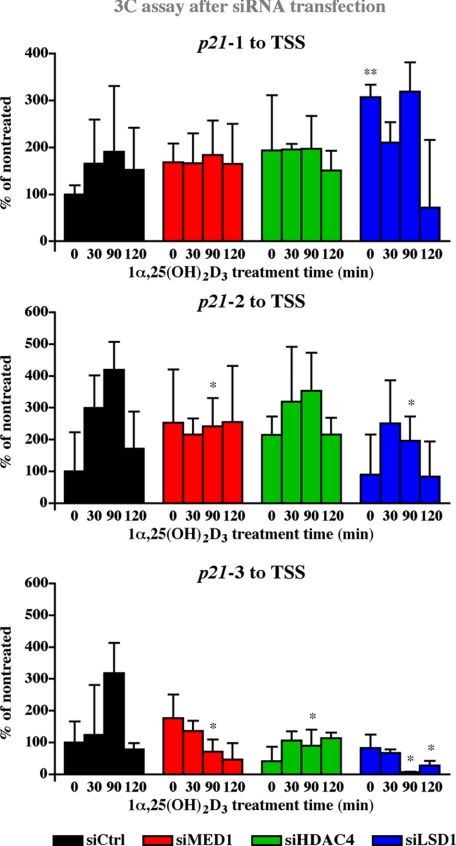
Histone Dimethylation and Deacetylation Are Indispensable for Chromatin Looping—Because the proteins MED1, HDAC4, and LSD1 seem to have a major impact on the 1α,25(OH)2D3-dependent cyclical accumulation of p21 transcript levels (Fig. 4) and are ligand-dependently enriched on the 1α,25(OH)2D3-responsive regions and the TSS of the p21 gene (Fig. 2B), they might also affect the looping of these regions to the TSS. Therefore, we individually knocked down the expression of MED1, HDAC4, and LSD1 by siRNA in MDA-MB453 cells, stimulated the cells with 1α,25(OH)2D3 and performed 3C assays for the association of regions 1, 2 and 3 with the TSS (Fig. 5). On the proximal region 1 1α,25(OH)2D3 only mildly induced association to the TSS and the effects of a knock-down of MED1, HDAC4, or LSD1 were not significant. In contrast, the expressions of MED1 and LSD1 were found to be essential for the induction of maximal ligand-induced association between regions 2 and 3 and the TSS after 90 min of 1α,25(OH)2D3 treatment. For the ligand-dependent looping between region 3 and the TSS, also HDAC4 showed to be essential. As a reference, the behavior of either of the two control regions was not significantly affected by a knock-down of MED1, HDAC4, or LSD1 (supplemental Fig. S3).
FIGURE 5.
Effect of cofactor silencing on chromatin looping VDRE containing regions to the TSS of the p21 gene. MDA-MB453 cells were transfected for 24 h with siRNA oligonucleotides against HDAC4, MED1, or LSD1 and subsequently stimulated with 10 nm 1α,25(OH)2D3 for the indicated times. Chromatin was extracted, cross-linked, and digested with either Hpy8I (upper and center graphs representing regions p21-1 and p21-2, respectively) or MvaI (lower graph representing region p21-3). After ligation, the DNA was extracted and analyzed by PCR with primers and FAM-labeled probes as indicated in Fig. 3. Values indicate looping as percentage to that in untreated cells transfected with non-targeting siRNA from indicated region to the p21 TSS. Data points indicate the means of at least three independent cell treatments, and the bars represent standard deviations. A two-tailed Student's t test was performed to determine the significance of the effects of the specific siRNAs in reference to control siRNA (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
In summary, the 3C results after siRNA knock-down suggest that for the bridging of two distal VDR binding chromatin regions to the basal transcriptional machinery on the TSS of the p21 gene, MED1 is indispensable. For p21-3 also HDAC4 is essential for maximal looping to TSS. Moreover, specific demethylation by LSD1 is essential for chromatin looping of even two most distal VDR binding regions.
DISCUSSION
In this study, we used the well-known gene p21 as a model to describe how a simple signal, such as the stimulation of MBA-MD453 cells with the VDR ligand 1α,25(OH)2D3, can result in cyclicity at the level of transcription factor binding, chromatin looping and eventually transcription (summarized in Table 1). We observed the cyclic responses at high resolution over a time frame of up to 300 min and distinguished an early phase of minor transcriptional induction and a later phase of more pronounced activation, but still modest effects on mRNA induction. Following the peaks, p21 mRNA level decreased rather fast, consistently with previously measured half-life of ∼1 h in human cancer cell lines (23, 24). In the early phase we found two concurrent peaks in VDR and p-Pol II enrichment. The first peak at 30-45 min was preceded by association of MED1 with the VDRE containing regions of the p21 gene. Simultaneous to the second peak in VDR and p-Pol II association at 90 min, looping from the three VDR binding regions to the TSS is 2-15-fold induced allowing physical interaction between the sites. The partially concurrent looping and association of p-Pol II on the distal VDRE containing regions suggests that looping and transcription initiation within a single piece of chromatin are simultaneous events. Whether looping occurs from only one VDRE to the TSS at a time or whether all three VDREs and the TSS connect simultaneously, still remains to be elucidated. These data do not reveal either, whether the elongation of the transcript occurs concurrently with the looping. Consistent with the ChIP results, the p21 transcript accumulation was induced on time points following the peaks in VDR and p-Pol II enrichment.
Our observations for the transcriptional regulation of the p21 gene in response to 1α,25(OH)2D3 activation are summarized in Table 1. Increase in H3K9ac appears to be a very initial effect of the ligand on the TSS, whereas increased MED1 is seen on the ligand-responsive regions and loss of repressors is observed on both the responsive regions and on the TSS. Simultaneously, the association of the proximal ligand-responsive region, p21-1, to the TSS is reduced. These results imply that increased acetylation of the TSS is not resulting from transcription, as it occurs prior to increased p-Pol II association, nor from NR-associated co-activators, as it happens during the lowest association of TSS and the VDRE-containing regions, but from decreased association of co-repressors via loss of the initial repressive looping to VDRE-containing regions.
H3K9ac is recognized by the TAF1 subunit of the basal transcriptional machinery, enabling the proper positioning of the polymerase to the core promoter (11). The Mediator complex promotes both the recruitment and the catalytic activity of TFIIH, leading to increased serine 5-phosphorylated Pol II. This form of Pol II is not able to bind the Mediator complex and eventually phosphorylation leads to the dissociation of the Mediator from the complex (13). Hence both the increased H3K9ac on the TSS and MED1 on the VDREs prepare the association of the p-Pol II complex, which is also supported by ChIP data, where H3K9ac and MED1 association both precede p-Pol II association.
Following this, the association of TSS to two of the responding regions increases, concurrently with induced association of VDR, p-Pol II, and secondary histone-modifying cofactors, such as CBP and LSD1. CBP is able to recognize H3K9ac through its bromodomain and acetylates H3K14 and accordingly CBP peaks at 30 min on the TSS after the increase in H3K9ac (12). The binding patterns of CBP on the regulatory regions of the p21 gene resemble those of p-Pol II and show cyclicity, but CBP appears not to be essential to the ligand response or the basal expression of p21. This suggests that it can be replaced by another histone acetyltransferase, such as the related p300 protein.
Recently, LSD1 has been shown to be essential in the activation of androgen receptor target genes and in the estrogen-induced chromatin looping via demethylation of H3K9 (25, 26). We show here that LSD1 is enriched to the TSS simultaneously with MED1 and H3K4me2, i.e. 30 min after the onset of ligand treatment, and also associates with VDRE-containing regions in a ligand-responsive manner. Perillo et al. (26) have proposed a model, where H3K9 demethylation by LSD1 causes oxidative damage on DNA, alluring a base-excision repair complex with a topoisomerase that catalyzes the transport of one DNA double helix through another to enable chromatin looping. In contrast to this report, where LSD1 constantly bound to both the TSS and the estrogen receptor α enhancer and did not respond to ligand, we observed ligand-dependent changes in LSD1 association. Consistent with previous data on steroid NRs, siRNA knock-down of LSD1 inhibits chromatin looping from distal 1α,25(OH)2D3-responsive regions to the TSS of the p21 gene and its siRNA knock-down produces a similar p21 transcript pattern than loss the of MED1 in response to the ligand.
Loss of HDAC3, HDAC4, HDAC5, or NCoR1 increases the basal expression of p21 and inhibits the transcriptional response to the ligand at 150 min. The p21 transcript does not reach higher levels when 1α,25(OH)2D3 is combined with siRNA inhibition of NCoR1 or both NCoR1 and SMRT compared with ligand treatment and untargeted siRNA, whereas the maximum transcript level is accomplished by the combination of the ligand and loss of HDAC4. Loss of HDAC4 or HDAC7 also severely disturbed the p21 mRNA accumulation pattern upon ligand treatment, unlike the loss of HDAC3 or HDAC5, which only raised the basal expression level. This implies that first of all, individual HDACs have distinct roles in regulation of the ligand response. Secondly, the disunited effects of HDAC removal on the ligand responsive p21 mRNA accumulation pattern suggest that the disturbed transcript accumulation patterns associated with loss of HDAC4 or HDAC7 do not result only from induced basal expression. It remains to be elucidated, whether these distinct roles arise from differential preferences for histone lysine residues or for non-histone targets. Surprisingly, HDAC4 is also essential for optimal chromatin looping from the distal region to the TSS in response to the ligand. The binding pattern of HDAC4 is more aberrant than that of HDAC3 or NCoR1, as it appears relatively late and incoherently on distinct regions. Unlike for LSD1, where exact histone residue targets are defined and a molecular mechanism in looping suggested, the mechanism by which HDAC4 affects looping remains undefined. As the binding and substrate specificity as well as protein associations of HDAC4 are still largely unknown, the reason for its crucial importance in the 1α,25(OH)2D3 response of the p21 gene remains unresolved.
In conclusion, 1α,25(OH)2D3 induces a dynamic and orchestrated response of the p21 gene, where cyclical binding of VDR and p-Pol II in concert with chromatin looping from enhancer regions to the TSS, leads to repeated induction of p21 mRNA production. This response is initiated by increased MED1 association with the VDRE-containing regions and loss of co-repressor complex, both inducing p-Pol II association and initiation of transcription. In the transcriptional response, the demethylation and deacetylation of lysine residues on histones are essential, suggesting a role for LSD1 and HDAC4 in setting the rhythm of histone modifications that enable dynamic chromatin looping, association of transcription factors on regulatory regions and eventually, transcription of the p21 gene.
Acknowledgments
We thank Dr. Milan Uskokovic for 1α,25(OH)2D3 and Maija Hiltunen for skilled technical assistance and Dr. Sami Heikkinen for careful reading of the manuscript. The authors declare they have no conflict of interest.
This work was supported by grants from the Academy of Finland, the Finnish Cancer Organization, and the Juselius Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1-S6 and Figs. S1-S3.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: NR, nuclear receptor; 1α,25(OH)2D3,1α,25-dihydroxyvitamin D3; 3C, chromosome conformation capture; CBP, CREB-binding protein; CDKN1A, cyclin-dependent kinase inhibitor 1A, also called p21; ChIP, chromatin immunoprecipitation; FAM, 6-carboxyfluorescein; H3K4me2, dimethylated histone 3 at lysine 4; H3K9ac, acetylated histone 3 at lysine 9; HDAC, histone deacetylase; LSD1, lysine demethylase 1; MED1, mediator complex subunit 1; NCoR1, nuclear co-repressor 1; p-Pol II, serine 5-phosphorylated RNA polymerase II; RPLP0, acidic riboprotein P0; siRNA, small inhibitory RNA; TBL1, transducin β-like protein 1; TSS, transcription start site; VDR, vitamin D receptor; VDRE, vitamin D response element.
References
- 1.Carlberg, C., and Polly, P. (1998) Crit. Rev. Eukaryot. Gene Expr. 8 19-42 [DOI] [PubMed] [Google Scholar]
- 2.DeLuca, H. F. (2004) Am. J. Clin. Nutr. 80 1689S-1696S [DOI] [PubMed] [Google Scholar]
- 3.Ingraham, B. A., Bragdon, B., and Nohe, A. (2008) Curr. Med. Res. Opin. 24 139-149 [DOI] [PubMed] [Google Scholar]
- 4.Deeb, K. K., Trump, D. L., and Johnson, C. S. (2007) Nat. Rev. Cancer 7 684-700 [DOI] [PubMed] [Google Scholar]
- 5.Liu, M., Lee, M.-H., Cohen, M., Bommakanti, M., and Freedman, L. P. (1996) Genes Dev. 10 142-153 [DOI] [PubMed] [Google Scholar]
- 6.Saramäki, A., Banwell, C. M., Campbell, M. J., and Carlberg, C. (2006) Nucleic Acids Res. 34 543-554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou, W., Zhu, P., Wang, J., Pascual, G., Ohgi, K. A., Lozach, J., Glass, C. K., and Rosenfeld, M. G. (2008) Mol. Cell 29 69-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heintzman, N. D., Stuart, R. K., Hon, G., Fu, Y., Ching, C. W., Hawkins, R. D., Barrera, L. O., Van Calcar, S., Qu, C., Ching, K. A., Wang, W., Weng, Z., Green, R. D., Crawford, G. E., and Ren, B. (2007) Nat. Genet. 39 311-318 [DOI] [PubMed] [Google Scholar]
- 9.Pray-Grant, M. G., Daniel, J. A., Schieltz, D., Yates, J. R., 3rd, and Grant, P. A. (2005) Nature 433 434-438 [DOI] [PubMed] [Google Scholar]
- 10.Jenuwein, T., and Allis, C. D. (2001) Science 293 1074-1080 [DOI] [PubMed] [Google Scholar]
- 11.Agalioti, T., Chen, G., and Thanos, D. (2002) Cell 111 381-392 [DOI] [PubMed] [Google Scholar]
- 12.Zeng, L., Zhang, Q., Gerona-Navarro, G., Moshkina, N., and Zhou, M. M. (2008) Structure 16 643-652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esnault, C., Ghavi-Helm, Y., Brun, S., Soutourina, J., Van Berkum, N., Boschiero, C., Holstege, F., and Werner, M. (2008) Mol. Cell 31 337-346 [DOI] [PubMed] [Google Scholar]
- 14.Barnett, D. H., Sheng, S., Charn, T. H., Waheed, A., Sly, W. S., Lin, C. Y., Liu, E. T., and Katzenellenbogen, B. S. (2008) Cancer Res. 68 3505-3515 [DOI] [PubMed] [Google Scholar]
- 15.Turunen, M. M., Dunlop, T. W., Carlberg, C., and Väisänen, S. (2007) Nucleic Acids Res. 35 2734-2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jing, H., Vakoc, C. R., Ying, L., Mandat, S., Wang, H., Zheng, X., and Blobel, G. A. (2008) Mol. Cell 29 232-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malinen, M., Saramäki, A., Ropponen, A., Degenhardt, T., Väisänen, S., and Carlberg, C. (2008) Nucleic Acids Res. 36 121-132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metivier, R., Penot, G., Hubner, M. R., Reid, G., Brand, H., Kos, M., and Gannon, F. (2003) Cell 115 751-763 [DOI] [PubMed] [Google Scholar]
- 19.Kim, S., Shevde, N. K., and Pike, J. W. (2005) J. Bone Miner. Res. 20 305-317 [DOI] [PubMed] [Google Scholar]
- 20.Banwell, C. M., MacCartney, D. P., Guy, M., Miles, A. E., Uskokovic, M. R., Mansi, J., Stewart, P. M., O'Neill, L. P., Turner, B. M., Colston, K. W., and Campbell, M. J. (2006) Clin. Cancer Res. 12 2004-2013 [DOI] [PubMed] [Google Scholar]
- 21.Lacroix, M., Toillon, R. A., and Leclercq, G. (2006) Endocr. Relat. Cancer 13 293-325 [DOI] [PubMed] [Google Scholar]
- 22.Dekker, J. (2006) Nat. Methods 3 17-21 [DOI] [PubMed] [Google Scholar]
- 23.Liu, J., Shen, X., Nguyen, V. A., Kunos, G., and Gao, B. (2000) J. Biol. Chem. 275 11846-11851 [DOI] [PubMed] [Google Scholar]
- 24.Esposito, F., Cuccovillo, F., Vanoni, M., Cimino, F., Anderson, C. W., Appella, E., and Russo, T. (1997) Eur. J. Biochem. 245 730-737 [DOI] [PubMed] [Google Scholar]
- 25.Metzger, E., Wissmann, M., Yin, N., Muller, J. M., Schneider, R., Peters, A. H., Gunther, T., Buettner, R., and Schule, R. (2005) Nature 437 436-439 [DOI] [PubMed] [Google Scholar]
- 26.Perillo, B., Ombra, M. N., Bertoni, A., Cuozzo, C., Sacchetti, S., Sasso, A., Chiariotti, L., Malorni, A., Abbondanza, C., and Avvedimento, E. V. (2008) Science 319 202-206 [DOI] [PubMed] [Google Scholar]