Abstract
Lhcb5 is an antenna protein that is highly conserved in plants and green algae. It is part of the inner layer of photosystem II antenna system retained in high light acclimated plants. To study the structure-function relation and the role of individual pigments in this complex, we (i) “knocked out” each of the chromophores bound to multiple (nine total) chlorophyll sites and (ii) exchanged the xanthophylls bound to the three xanthophyll sites. The occupancy and associated energy of the pigment binding sites were determined. The role of the individual pigments in protein folding, stability, energy transfer, and dissipation was studied in vitro. The results indicate that lutein has a primary role in the folding and stability of the complex, whereas violaxanthin and zeaxanthin have a negative effect on folding yield and stability, respectively. The data showed a distinct function for the L1 and L2 carotenoid binding sites, the former preferentially involved in gathering the excitation energy to chlorophyll a (Chl a), whereas the latter modulates the concentration of chlorophyll singlet excited states dependent on the xanthophylls bound to it, likely via an interaction with Chl-603. Our results also underscored the role of zeaxanthin and lutein in quenching the excitation energy, whereas violaxanthin was shown to be very effective in energy transfer. The characteristics of the isolated proteins were consistent with the observed role of Lhcb5 in vivo in catalyzing fluorescence quenching upon zeaxanthin binding.
Sunlight energy is absorbed and converted into chemical energy by pigments bound to photosystems I and II (PSI2 and PSII). Photosystems are composed of two moieties: (i) a reaction center devoted to the conversion of light energy into chemical energy and (ii) an antenna system that increases the capacity of light absorption and contributes to photoprotection (1). In photosynthetic eukaryotes, the antenna system is composed of members of a multigenic family called light-harvesting complexes (Lhc). In higher plants, 10 different subunits are associated with the photosystems, Lhca1-4 with PSI and Lhcb1-6 with PSII (2). All of these proteins share the same evolutionary origin and a common structural organization (3). Only the structure of LHCII, a trimer composed mainly of Lhcb1 (4), has been resolved at the atomic level, revealing the presence of three transmembrane and two amphipathic helices named, respectively, A-C and D-E (5). Each Lhcb1 polypeptide has been shown to coordinate four carotenoids and 14 Chl molecules. Two carotenoid binding sites, L1 and L2,3 are close to helices A and B, respectively (6), and a third site, N1, specific for neoxanthin, is located in proximity to helix C (7). Finally, a peripheral and less stable binding site, V1, has been shown to accommodate violaxanthin and zeaxanthin (5, 8, 9).
Other members of the Lhc protein family coordinate a variable number of pigments (8-14 Chl and 2-4 carotenoid molecules, depending on the polypeptide (9-14)) implying that some of the binding sites identified in Lhcb1 might be absent or empty in other Lhc complexes. The L1 site is conserved and binds lutein in all Lhc analyzed thus far. Site L2 is also present in all complexes, but its selectivity for xanthophyll ligands is more variable; in Lhcb1-3 it is occupied mainly by lutein, whereas lutein, violaxanthin, zeaxanthin, and neoxanthin have been proposed to be bound to L2 in Lhcb4, Lhcb5 (15-17) and, with the exclusion of neoxanthin, Lhcb6 and Lhca1-4 (18-19). Neoxanthin is bound to the N1 site in Lhcb1-5 (9, 20).
Most Chl are coordinated to the proteins by nucleophylic amino acid residues via their central Mg2+ (5, 6). Eight putative Chl binding residues are conserved in all Lhc proteins as suggested by sequence analysis (2) and confirmed for several complexes by mutation analysis (15, 21-24). Although the binding sites are conserved, their specificity for Chl a or Chl b can vary between different complexes. Conserved Chl a binding sites are located for the most part near helices A and B in the center of the structure, whereas the more peripheral sites differ in their affinity depending on the gene product (25).
Despite their similarity in sequence and three-dimensional organization, distinct Lhc isoforms were conserved during evolution of the green lineage (26), and mutants lacking individual Lhc complexes show fitness reduction in a natural environment (27), suggesting a specific function for each of them. For example, knock-out of Lhcb6 significantly affects non-photochemical quenching (NPQ), a mechanism of heat dissipation of excitation energy absorbed in excess (28, 29), whereas depletion of the other Lhc subunits does not (30, 31). Within this picture, the properties and specificity of the binding sites and the ability of polypeptide chains to tune the spectral properties of pigments have a fundamental role in determining the different functions of the Lhc proteins.
Lhcb5 is a PSII antenna protein involved in photoprotection by quenching of Chl singlets (32, 33) and triplets (34). Consistently, biochemical (32) and genetic (31) analyses have shown that Lhcb5 is involved in the slowly activated NPQ component qI. Furthermore, together with Lhcb4 and Lhcb6, Lhcb5 has been found capable of generating radical carotenoid cations involved in the qE mechanism, the energy-dependent quenching of excitonic energy (33, 35). In this work we analyzed the properties of each pigment located in the binding sites of Lhcb5, to clarify the molecular basis of the physiological function of this protein.
EXPERIMENTAL PROCEDURES
DNA Cloning, Mutations, and Isolation of Overexpressed Lhcb5 Apoprotein—cDNA from Arabidopsis thaliana encoding Lhcb5 (GenBank™ accession number AF134129) was supplied by the Arabidopsis Biological Resource Center at Ohio State University. Mature Lhcb5 was amplified and cloned in pQE-50 expression vector as described previously (32). Mutations were obtained as described (15) using the QuikChange™site-directed mutagenesis kit (Stratagene). Lhcb5 WT and mutant apoproteins were overexpressed in the SG13009 strain of Escherichia coli and purified following a protocol described previously (36, 37).
Reconstitution in Vitro of Recombinant Lhcb5—Reconstitution and purification of recombinant Lhcb5 pigment-protein complexes were performed as described previously (13) with the following modifications: 83.3 μm carotenoids and 231.5 μm chlorophylls with a Chl a/b ratio of 3 were added to 13.9 μm apoprotein. Pigments were purified from spinach as described previously (38). For reconstitution with modified carotenoid composition the mixture was obtained from pure pigments either purchased from Sigma-Aldrich (Chl a and Chl b) or purified by HPLC (xanthophylls). When more than one carotenoid was present in the pigment mix, all of the species were added in equal amounts.
Pigment Analysis—HPLC analysis was performed according to Gilmore and Yamamoto (39). The chlorophyll to carotenoid ratio and Chl a/b ratio were measured independently by fitting the spectrum of acetone extracts with the spectra of individual purified pigments (17).
Spectroscopy—Room temperature absorption spectra were recorded using an SLM-Aminco DK2000 spectrophotometer in 10 mm HEPES, pH 7.5, 0.2 m sucrose, and 0.03% n-dodecyl-β-d-maltopyranoside. The wavelength sampling step was 0.4 nm. Fluorescence emission spectra were measured using a Jasco FP-777 spectrofluorimeter and corrected for the instrumental response. Differences between mutant absorption spectra were calculated after normalization to the chlorophyll stoi-chiometries as described by Bassi et al. (15). Fluorescence quantum yields were calculated as the ratio between the emission spectra area (650-800 nm) and the absorption at 625 nm wavelength (5 nm bandwidth). This is the same wavelength (625 nm, 5 bandwidth) we employed for the excitation and was chosen because Chl a absorbs in this region, and thus the yield calculation was less affected by the different Chl a/b ratios in the samples. CD spectra were measured at 10 °C on a Jasco 600 spectropolarimeter using an R7400U-20 photomultiplier tube; samples were placed in the same solution described for absorption, with an OD of 1 at the maximum absorption peak in the Qy transition. The measurements were performed in a 1-cm cuvette. Denaturation temperature measurements were performed by following the decay of the CD signal at 682 nm when increasing the temperature from 20 to 80 °C with a time slope of 1 °C/min and a resolution of 0.2 °C. The thermal stability of the samples was determined by finding the t½ of the signal decay.
RESULTS
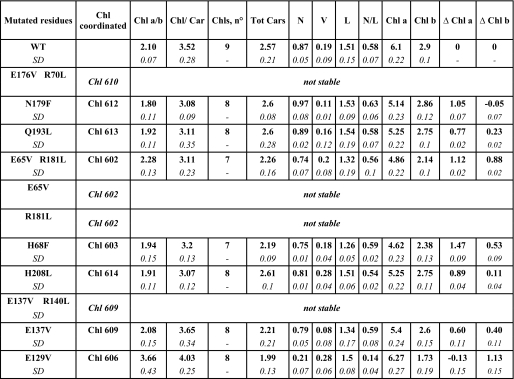
Mutational Analysis of Chl Binding Sites—Chl binding residues were identified in the Lhcb5 sequence by comparison (2) with other Lhc polypeptides, namely Lhcb1, Lhcb4, Lhca1, and Lhca3. Information was made available by crystallography (5, 40) or mutation analysis (15, 21, 23). The cDNA encoding Lhcb5 from A. thaliana was mutated at sequences encoding the putative Chl binding residues (Table 1) by substituting the nucleophilic residues with apolar amino acids unable to coordinate Mg2+. WT and mutant apoproteins were overexpressed in E. coli and reconstituted in vitro upon the addition of pigments. WT-Lhcb5 and seven mutants yielded stable reconstituted products and showed a monomeric aggregation state when analyzed by sucrose gradient ultracentrifugation. In the case of four mutants, it was not possible to obtain stable products (see Table 1).
TABLE 1.
Pigment binding properties of mutants in individual Chl binding residues
Pigment binding properties of WT and mutant-Lhcb5 were analyzed through a combined approach of fitting analysis of acetonic pigment extracts absorption spectra and HPLC. Data are normalized to the number of chlorophylls present in each mutant. Chl a/b, molar ratio between Chl a and Chl b; Chl/car, molar ratio between chlorophylls and carotenoid; Chls, n°, number of chlorophylls bound by each Lchb5 complex; Tot Cars; number of carotenoids bound by each Lhcb5 complex; N/L, molar ratio between neoxanthin and lutein; Chl a/b, number of Chl a and Chl b bound by each Lhcb5 complex. Variations of Chl a and Chl b (Δ Chla; Δ Chlb) with respect to WT are also shown; these values were calculated by subtracting the number of Chl a(b) bound by each mutant to the number of Chl a(b) bound by WT. S.D. is indicated in italics.
To determine the nature and the spectroscopic properties of the pigments coordinated to the individual sites, the pigment content (Table 1) and the absorption spectra (Fig. 1) of the reconstituted mutants were measured and compared with those of the WT. The Chl a/b ratio of Lhcb5 WT from Arabidopsis is 2.1 ± 0.1, corresponding to the binding of six Chl a and three Chl b molecules for the proposed stoichiometry of nine Chl-protein molecules (10, 17). Most of the mutants (with the exception of E65V/R181L and E129V) show a decrease in Chl a/b ratio, implying that Chl a is the ligand for the majority of the sites. Moreover, for all mutants but E129V, the ratio between lutein and neoxanthin is identical to that of the WT, suggesting that the mutations do not induce in first approximation a specific loss of xanthophylls, thus allowing normalizing of the chlorophyll content to the carotenoid content. Detailed results of the individual mutants are presented below.
FIGURE 1.
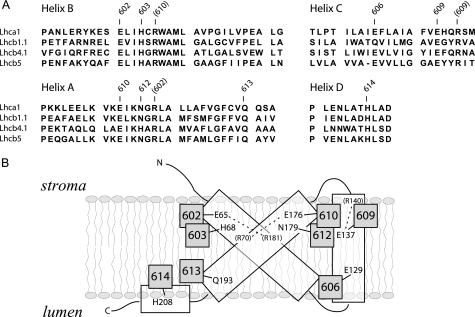
Identification of putative Chl binding residues in Lhcb5. A, sequence alignment of Lhcb5 with Lhca1, Lhcb1.1, and Lhcb4.1 from A. thaliana. Only transmembrane helices are shown. Conserved Chl binding residues are indicated as identified by structural and mutational analyses (5). Basic residues involved indirectly in Chl binding are indicated in parentheses. Chl-602, Chl-603, Chl-613, Chl-614, Chl-610, Chl-612, Chl-609, and Chl-606 are indicated as 602, 603, 613, 614, 610, 612, 609, and 606, respectively. B, schematic model of Lhcb5 tridimensional organization. The amino acid residue responsible for Chl binding is indicated.
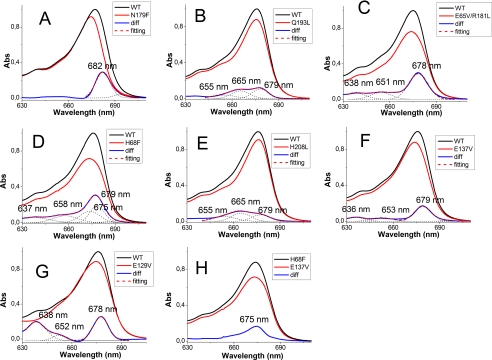
Mutant N179F (Chl-612)—The change in Chl a/b and Chl/Car ratios (Table 1) induced by the mutation are fully consistent with the loss of a single Chl a molecule, whereas the xanthophyll content is not affected by the mutation. The WT minus N179F difference absorption spectrum (Fig. 2A) indicates that Chl-612, which has a maximum at 682 nm at room temperature, is the red-most form of Lhcb5. As shown previously, similar absorption maxima were observed for Chl-612 in all Lhc complexes analyzed thus far (22-24, 64), indicating that this site is fully conserved across the family.
FIGURE 2.
Difference absorption spectra of mutants in individual Chl binding residues. Differences between WT and mutants absorption spectra were calculated after normalization to the Chl stoichiometry as done in Bassi et al. (15). Fitting with Gaussian distributions of the difference spectrum in the red region is indicated to help identify different contributions. Difference absorption spectra (blue traces) between Lhcb5-WT and N179F (A), Q193L (B), E65V/R181L (C), H68F (D), H208L (E), E137V (F), and E129V (H) mutants and between the E137V mutant and the H68F mutant (G) are shown.
Mutants Q193L and H208L (Chl-613 and -614)—The pigment analysis (Table 1) suggests that both mutants lost one Chl molecule. Sites 613 and 614 are occupied mainly by Chl a, although the mutations induce a loss of a small amount of Chl b, suggesting that they are able to accommodate Chl b as well. In the difference spectra (Fig. 2, B and E) of both mutants, two major bands are visible at 665 and 678 nm; these can be attributed to the loss of interaction between these two Chl, which are indeed located close in the structure (5). An additional band at 655 nm accounts for the loss of Chl b. In both mutants we would expect to lose the excitonic interaction and gain a new absorption of the monomeric Chl still present in the complex (Chl-614 in the Q193L mutant and Chl-613 in the H208L mutant). This is consistent with the experimental data, where the same bands are present in both spectra. Moreover, the ratio between the bands in the two mutants is different, with mutant Q197L loosing more of its signal at 678 nm and mutant H208L losing more at 665 nm. The absence of a negative contribution in the difference spectra is expected based on a difference in band broadening between interacting and noninteracting Chl and on a similar distribution of the oscillator strength over the two bands. Moreover, the presence of Chl b in both sites strongly complicates the picture, because Chl a-Chl b interaction (and vice versa) is expected. These observations also imply that the shift in the site energy of the Chl induced by the interaction is rather small and that the site energy of these two chromophores differs substantially, with Chl-613 absorbing around 678 nm and Chl-614 at 665 nm. A very similar picture was found in Lhca1, Lhcb1, and Lhcb4 (15, 21, 22).
Mutant E65V/R181L (Chl-602)—Normalization to the carotenoid content indicates the loss of one Chl molecule (Table 1). However, this would mean that site 602 has a mixed occupancy (50:50 Chl a:Chl b), at variance compared with the other Lhc complexes, where this chlorophyll binding site was suggested to bind Chl a and the corresponding mutation led to a loss of carotenoids (15, 21-24). The specificity for Chl a of this site is likely to be essential for the mechanism of Lhc protein folding, which is initiated by Chl a and xanthophylls (41, 42). It was proposed that during Lhc assembly in vivo, the first polypeptide-Chl a interaction involves a conserved motif, Glu-X-X-(His/Asn)-X-Arg, which corresponds to Chl binding sites 610, 612, 602, and 603 (43, 44). It is thus likely that mutant E65V/R181L loses one Chl a and one Chl b (Table 1), with the Chl a accommodated in site 602. The second Chl affected by the mutation needs to have a high affinity for Chl b. In the difference absorption spectrum (Fig. 2C), in addition to the main contribution at 678 nm, two bands in the Chl b region were detected at 638 and 651 nm. Loss of Chl b upon mutation on the Chl-602 binding site was obtained also for Lhcb1 (21) and Lhca1 (22). In those cases it was suggested that the Chl b lost upon the mutation was Chl-611, which in the structure is the nearest Chl lacking a protein ligand (5). A similar interpretation also holds for Lhcb5, thus suggesting that Chl-602 and -611 in Lhcb5 are Chl a and b molecules, respectively.
The presence of a second Chl b band in the difference spectrum is probably a consequence of an alteration on protein structure due to mutation on the ionic couple Glu-65/Arg-181. Because this ionic bridge is important for protein folding and stability, its absence might affect a second Chl b binding site. With the present data, we were not able to assess whether this secondary effect was only a perturbation in the absorption spectrum or the effective loss of pigment.
Mutant H68F (Chl-603)—Normalization to the carotenoid content indicates the loss of one Chl molecule (0.8 Chl a and 0.2 Chl b). However, similar to the case for Chl-602, Chl-603 also belongs to the conserved “Chl a core,” suggesting that this mutation leads instead to the loss of two Chl molecules (1.4 Chl a and 0.6 Chl b). The nearest neighbor to Chl-603 is Chl-609, which is thus the best candidate for the second Chl lost in this mutant. The WT minus H68F absorption difference spectrum peaks at 678 nm. The peak is broad and cannot be fitted with a single Gaussian distribution but requires two of them, with maxima at 675 and 679 nm, respectively (Fig. 2D). The presence of two Chl a forms supports the hypothesis that more than one Chl a is affected by the mutation. A shoulder around 656 nm and a small contribution at 638 nm account for the loss of Chl b. Based on the hypothesis that this mutant is losing Chl-603 and -609, the absorption spectrum of Chl-603 can be obtained by the difference between the absorption of mutant E173V, which loses only Chl-609, and that of mutant H68F. The result indicates that Chl-603 has an absorption maximum at 675 nm, as is the case in Lhcb1 and Lhcb4 (21).
Mutant E137V (Chl-609)—Pigment analysis suggests that this mutant loses one Chl and that the binding site has mixed occupancy (Table 1). A small loss of neoxanthin and lutein, probably from the N1 and L2 sites, was also observed. The difference absorption spectrum shows the Chl a component at 679 nm (Fig. 2F) and two contributions in the Chl b absorption region at 636 and 653 nm, indicating that the absence of Chl-609 also has an effect on a neighboring chlorophyll, probably Chl-606, which indeed absorbs at 638 nm (see below).
Mutant E129V (Chl-606)—Pigment analysis showed a strong reduction of the neoxanthin content, an indication that the mutation affected the binding of this xanthophyll to the N1 site. Normalization of the Chl content to lutein suggested that this mutant lost 1.13 Chl b and gained 0.13 Chl a. In LHCII, which has a Gln in this position, the carboxyl group was involved in the binding of Chl-606 via a water molecule, whereas the two hydrogens of the amide form H-bonds with the formyl groups of Chl b 609 and 607. In the case of Lhcb5, the Gln is substituted by a Glu, which can form only one H-bond, likely with Chl-609. Thus, the substitution of the Glu with an apolar residue not only disturbs the binding of Chl-606 but also induces a change in affinity of Chl binding site 609 because of the loss of the H-bond, which favors the binding of Chl b in the WT complex and explains the results. This hypothesis was confirmed by the presence of a high degree of disconnected Chl b in E129V mutant (supplemental Fig. S1), probably due to destabilization of Chl b in the Chl-609 binding site. However, the possibility that the mutation induced a loss of lutein from the L2 site and of two Chl cannot be ruled out. From the difference spectrum the contribution of Chl b 606 can be observed at 638 nm (Fig. 2F). A second positive contribution at 652 nm may be associated with the change of affinity/loss of Chl in site 609 (Fig. 2F). A positive component at 678 nm (Fig. 2F) may indicate a Chl a loss at Chl-609 (Fig. 2G) but could also be attributed to a change in the oscillator strength of some Chl because of the large perturbation induced by the mutation to the hydrogen network.
Chl-610—The biochemical and spectroscopic properties of Chl-610 cannot be studied through a mutational analysis because the E176V/R70L mutant is not stable. However, Chl-610 is suggested to be a Chl a in Lhcb5, as shown in Lhcb1, where the mutant missing this Chl is partially stable (21). Moreover, Chl-610 is part of the inner Chl a cluster, conserved among Lhc proteins (15, 21). This hypothesis of a Chl a bound in site 610 is also in full agreement with the proposed stoichiometry of six Chl a and three Chl b molecules bound to Lhcb5.
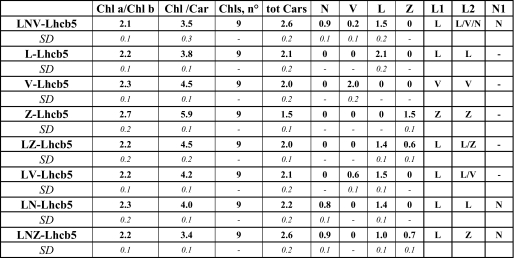
Carotenoid Binding Sites—For most of the carotenoid binding sites the amino acid residues involved in the binding are not known, making it impossible to produce mutants lacking individual xanthophylls. To gain information on the biochemical and functional properties of carotenoids in Arabidopsis-Lhcb5, we reconstituted Lhcb5-WT in the presence of different xanthophyll species. Xanthophyll substitution in specific sites actually occurs in vivo during the operation of the xanthophyll cycle(s) (45-48). Thus, the characterization of Lhc proteins with altered xanthophyll composition yields information on the functional roles of the carotenoids involved in xanthophyll cycles. Lhcb5 was reconstituted in the presence of lutein (L), violaxanthin (V), zeaxanthin (Z), lutein + violaxanthin (LV), lutein + zeaxanthin (LZ), lutein + neoxanthin (LN), and lutein + neoxanthin + zeaxanthin (LNZ). A complex with neoxanthin only could not be obtained, in agreement with previous results (17).
The pigment composition of reconstituted samples are reported in Table 2; these results are very similar to those found previously for Lhcb5 from Zea mays (17), although in Arabidopsis-Lhcb5 the number of carotenoids bound is generally higher and the amount of violaxanthin lower (32). The data presented here provide indications of the presence of three carotenoid binding sites as suggested previously (9, 20). All samples reconstituted in the presence of neoxanthin bind more carotenoids than the corresponding samples without neoxanthin, confirming the presence of a neoxanthin-specific binding site, identified as site N1 (9, 20). The presence of a second neoxanthin binding site is suggested by the observation that reconstitution without neoxanthin (Lhcb5-V, -LV, -L) increases the content of lutein and violaxanthin compared with Lhcb5-LNV, partially compensating for the absence of neoxanthin to the level of two xanthophylls per monomer. The occupancy of the carotenoid binding sites in the different complexes is summarized in Table 2.
TABLE 2.
Pigment binding properties of Lhcb5 reconstituted with different carotenoid species
The samples were named after the carotenoid species used in the reconstitution mix (e.g., Lhcb5-L, complex reconstituted in the presence of only lutein as a xanthophyll). Pigment binding properties of different samples were analyzed through a combined approach of fitting analysis of acetonic pigment extract absorption spectra and HPLC. Data are normalized to the number of chlorophylls bound by Lhcb5. The occupancy of the different carotenoid binding sites (L1, L2, N1) is reported for each sample. Chl a/b; molar ratio between Chl a and Chl b, Chl/Car, molar ratio between chlorophylls and carotenoid; Chls, n°, number of chlorophylls bound by each Lchb5 complex; Tot Cars, number of carotenoids bound by each Lhcb5 complex. S.D. is indicated in italics.
The absorption maximum of the xanthophylls in Arabidopsis-Lhcb5 was determined by a second derivative analysis of the absorption spectra of the different complexes (not shown). The results obtained are consistent with previous data from maize-Lhcb5, indicating that the environment of the xanthophylls is very similar in the two proteins (17, 49).
It can thus be concluded that, as in the case of maize-Lhcb5, site L1 accommodates lutein, site L2 accommodates lutein, violaxanthin, and possibly neoxanthin under normal conditions, and zeaxanthin under a stress condition, whereas site N1 is specific for neoxanthin. The L2 binding flexibility is a common feature of all Lhc proteins, even if the relative affinities vary in different proteins (9, 16, 50). For the N1 site we show that the Lhcb5-E129V mutant, missing Chl-606, undergoes selective loss of neoxanthin. Similar results were obtained with Lhcb1, confirming that the N1 site is structurally conserved in Lhcb5 and LHCII (20).
Role of Pigments in Protein Folding and Stability—To evaluate the role of the different chromophores on the folding and stability of Lhcb5 two parameters were analyzed: (i) the yield of refolding in vitro and (ii) the stability during thermal denaturation (Table 3). All mutations induced a decrease of refolding yield, suggesting that each chlorophyll binding site has a role in the cooperative process of Lhcb5 folding. The stronger effect was observed for mutations E176V/R70L (affecting Chl-610 binding site), E65V and R181L (both affecting the Chl-602 binding site), and E137V/R140L (Chl-609), which inhibit the folding of the complex, and mutation E129V (Chl-606), which lowers the reconstitution yield by 75% compared with the WT. The other mutations lowered the yield by about 40% as compared with the WT, with the exception of N179F (Chl-612), for which only a 15% decrease was observed (Tables 1 and 4). Upon refolding with an altered xanthophyll complement, changes in yield were greater; only 10% of Lhcb5 was refolded in the presence of violaxanthin as the only xanthophyll and 63% in the presence of zeaxanthin, whereas refolding with lutein increased the yield to 120%. The presence of violaxanthin or zeaxanthin in the L2 binding site, instead, is substantially equivalent, leading to a yield decreased by 20% compared with lutein, whereas filling the N1 site with neoxanthin did not significantly modify the yield. These data thus clearly suggest that in Lhcb5 lutein promotes folding more efficiently than other xanthophylls.
TABLE 3.
Protein refolding yield and thermal stability
Protein refolding yield relative to WT Lhcb5 reconstituted with lutein, neoxanthin, and violaxanthin and the thermal stability of chlorophyll binding site mutants expressed as t½ during thermal denaturation are reported. The percentage of thermal stability is reported in comparison with control (WT-LNV). S.D. is less than 15% for protein folding yield and 2% for thermal stability.
| Mutated residues | Protein folding yield with respect to WT | Thermal stability |
|---|---|---|
| % | °C (% control) | |
| WT | 100 | 67.5 (100) |
| E176V/R70L | Not stable | |
| N179F | 85 | 67.9 (100) |
| Q193L | 56 | 51.5 (76) |
| E65V/R181L | 62 | 58.4 (86) |
| E65V | Not stable | |
| R181L | Not stable | |
| H68F | 61 | 59.0 (87) |
| H208L | 57 | 59.5 (88) |
| E137V/R140L | Not stable | |
| E137V | 56 | 50.0 (74) |
| E129V | 26 | 61.7 (91) |
| Different carotenoids binding Lhcb5 | Protein folding yield with respect to LNV-Lhcb5a | Thermal stabilityb |
|---|---|---|
| % | °C | |
| LNV-Lhcb5 | 100 | 67.5 (100) |
| LN-Lhcb5 | 125 | 66.1 (98) |
| LNZ-Lhcb5 | 103 | 63.0 (93) |
| LV-Lhcb5 | 99 | 60.7 (90) |
| LZ-Lhcb5 | 99 | 58.0 (86) |
| V-Lhcb5 | 11 | 58.1 (86) |
| L-Lhcb5 | 119 | 61.4 (91) |
| Z-Lhcb5 | 63 | 55.8 (83) |
S.D. < 15%.
S.D. < 2 °C.
TABLE 4.
Energy transfer efficiencies
Energy transfer (ET) Chl b > Chl a and Car > Chl a efficiencies in different mutants and WT reconstituted with different carotenoids was determined by comparing absorption and fluorescence excitation spectra (see “Results” for details). Data are normalized to 100% efficiency of Chl a - Chl a energy transfer. S.D. < 6% for each sample.
| Mutated residues | Chl b > Chl a ET | Car > Chl a ET |
|---|---|---|
| WT-LNV | 87 | 76 |
| N179F | 81 | 68 |
| Q193L | 70 | 76 |
| E65V/R181L | 84 | 76 |
| H68F | 87 | 76 |
| H208L | 84 | 72 |
| E137V | 74 | 76 |
| E129V | 63 | 62 |
| Different carotenoids binding Lhcb5 | ||
|---|---|---|
| LV-Lhcb5 | 80 | 58 |
| L-Lhcb5 | 86 | 60 |
| V-Lhcb5 | 85 | 75 |
| LN-Lhcb5 | 86 | 82 |
| LNZ-Lhcb5 | 86 | 74 |
| LZ-Lhcb5 | 88 | 65 |
| Z-Lhcb5 | 59 | 55 |
The stability of the reconstituted complex during heat denaturation (Table 3) shows that deletion of one Chl ligand decreased the stability by 6-8 °C with the exception, once more, of mutant N179F, which was as stable as the WT, and Q193L (Chl-613) and E137V (Chl-609), in which a decrease in stability of 15-20 °C was observed. The occupancy of the carotenoid binding sites by different xanthophylls had a similar effect on the stability of the complex, with the exception of neoxanthin in site N1, where it increased the protein stability by 5 °C, similar to that observed for Lhcb1 (7). Surprisingly, despite a very strong effect on the refolding yield, the sample reconstituted with violaxanthin only was almost as stable as the sample with lutein, suggesting that the selection between these two xanthophylls occurs at the level of folding. The presence of zeaxanthin in the L2 site decreased the stability of the samples by 4 °C, and its binding to L1 induced an additional destabilization, suggesting a change in the structure of the complex.
Energy Transfer Efficiency—The efficiency of energy transfer from xanthophylls to Chl a can be evaluated by comparing the 1-T spectra to the fluorescence excitation spectra. A detailed analysis can be obtained by fitting the spectra of the complexes with the spectra of individual pigments as shown previously (51). The blue absorption region of the WT 1-T spectrum was described with two absorption forms for lutein, one for violaxanthin, one for neoxanthin, and three and two for chlorophyll a and b, respectively. The fluorescence excitation spectra were fitted with the same spectral forms allowing for amplitude changes. The energy transfer efficiencies (Table 4) were calculated assuming 100% Chl a to Chl a transfer, which is justified by the observation that in any mutant we observed a significant Chl a disconnection (see supplemental data).
Among the mutants of the Chl binding sites, we observed a clear decrease of Chl b > Chl a energy transfer on Q193L, E137V, and E129V mutants, losing, respectively, Chl-613, Chl-609, and Chl-606. This result was due partially to the presence of some disconnected Chl, as evident from fluorescence emission spectra (supplemental Fig. S1), especially in the case of mutant E129V. The Car > Chl a energy transfer was partially affected only in N179F and E129V mutants, the former by losing Chl-612 located close to L1 (5), the latter by partially destabilizing the N1 site. Analysis of complexes with altered carotenoid composition showed that the absence of neoxanthin induced a large reduction of Car > Chl a energy transfer, indicating that neoxanthin is more efficient than the average of the carotenoids in light harvesting. The low values observed for the sample reconstituted with only lutein or LV, which accommodates the xanthophylls in the central sites, suggests that at least one of the carotenoids is not very efficient in energy transfer. The presence of violaxanthin also in the L1 site (Lhcb5-V) led to an increase of the energy transfer, indicating that violaxanthin is more efficient than lutein in transferring energy from L1. The presence of zeaxanthin together with other xanthophylls did not significantly affect the transfer efficiency from carotenoids to Chl a. However, when zeaxanthin was the only xanthophyll bound, a strong decrease of energy transfer to Chl a from both Chl b and carotenoids was detected, as described previously (49), suggesting a change in the structure of the complex in agreement with the thermal stability data.
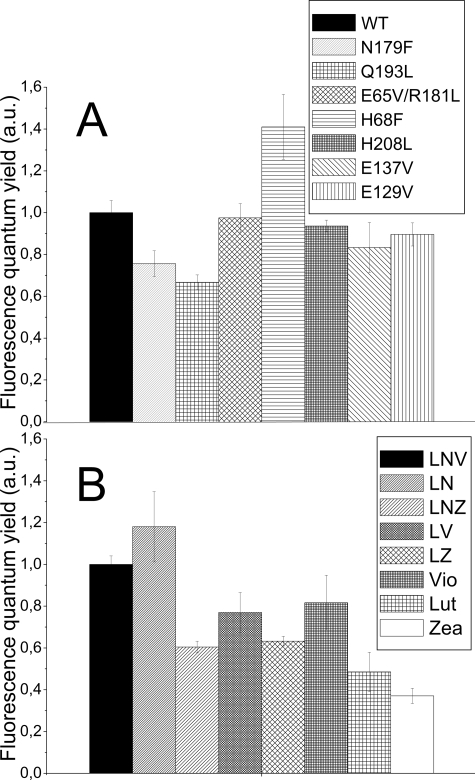
Pigment Effect on Chlorophyll Fluorescence Quantum Yield—Xanthophylls are involved in photoprotection (28, 52, 53) by dissipating the Chl excitation energy as heat. To check the role of the individual chromophores in energy dissipation, the fluorescence yield of all samples was measured. Fig. 3A shows the relative fluorescence quantum yield of the mutants of the Chl binding sites. Mutants N197F and Q193L, losing Chl-612 and -613, have lower fluorescence quantum yield than the WT, whereas mutation at site H68F, affecting Chl-603, increases the yield. Mutations at the other binding sites, instead, do not induce significant differences compared with WT.
FIGURE 3.
Chl and carotenoid effect on Lhcb5 fluorescence yield. A, fluorescence quantum yield of Lhcb5 chlorophyll binding site mutants compared with WT-Lhcb5. B, fluorescence yield of Lhcb5 recombinant samples reconstituted with different carotenoid species. Values are normalized to 100% for WT/LNV-Lhcb5.
In the complexes with altered xanthophyll composition (Fig. 3B), a specific effect of neoxanthin in increasing the fluorescence yield of Lhcb5 could be observed (compare LNV versus LV and LN versus lutein samples). On the contrary, V-Lhcb5 and LV-Lhcb5 showed very similar yields, whereas this value was significantly lower for L-Lhcb5 (compare LV versus L-Lhcb5). These results indicate that lutein binding to the L2 site induces a decrease of fluorescence quantum yield compared with violaxanthin, whereas in the case of the L1 site the opposite occurs.
A strong decrease of fluorescence yield was obtained upon zeaxanthin binding to both sites. The comparison between the LNV and LNZ samples indicates that when this xanthophyll is bound to site L2, a reduction of 40% of fluorescence yield is obtained, which is not altered by the absence of neoxanthin (compare LNZ versus LZ samples). A further reduction of fluorescence yield is evident when zeaxanthin is bound also to L1 site (compare zeaxanthin versus LZ, LNZ-Lhcb5).
DISCUSSION
In this work we coupled optical spectroscopy to mutation analysis to obtain information on the properties of individual chromophores in pigment-protein complexes reconstituted in vitro. Moreover, a comparison of the effect of the same mutation in different members of the Lhc family also yields structural information (15, 21-24). We have applied this approach to Lhcb5 in the present work.
Lhcb5 Apoprotein: Identification of Residues Involved in Protein Stability—Several mutations were shown to inhibit the folding of the complex. Interestingly, they are all mutations that target charged residues in the membrane and that clearly have an important role in the stability of the complex.
The absence of folding upon mutation E176V/R70L is a common feature of Lhc proteins, and it indicates that, as is the case for LHCII (5, 6), these residues form an ionic pair, stabilizing the interaction between helices A and B in all members of the Lhc family. According to the structure of LHCII, a second ionic pair between the same two helices is formed by residues Glu-65 and Arg-181, with the former coordinating Chl-602. However, in Lhcb5 these mutations do not impair the folding, provided that both residues are mutated into non-charged species, which implies that the contribution of the Glu-176/Arg-70 ionic pair to protein stability is higher compared with that of Glu-65/Arg-181 as was the case for other Lhcb complexes. A third ionic pair is involved in the coordination of Chl-609; in this case both residues are located in helix C (Glu-137 and Arg-140). In Lhcb5 only the single E137V mutant formed a stable pigment-protein complex, whereas the double mutant did not survive purification. This is similar to what was reported previously for Lhcb4, Lhca1, and Lhcb1 (although in the latter case the double mutant was obtained in a highly unstable form) (15, 22) but is at variance with data for Lhca3 and Lhca4, where only the double mutant was stable (23, 24). These results indicate that the organization of helix C in the Lhc complexes is not conserved. Comparing the available structures of Lhc proteins, we observed that in Lhca3 and Lhca4 the arginine is located in the middle of the helix (40) and, in the absence of the compensating negative charge, represents a factor of destabilization, while in LHCII and Lhca1 this residue is located at the C-terminus (5, 40), being thus a factor of stabilization. The results of the mutants of Lhcb5 indicate that the organization of the C helix in Lhcb5 is very similar to that of Lhcb1 and Lhca1 with the Arg at the C terminus.
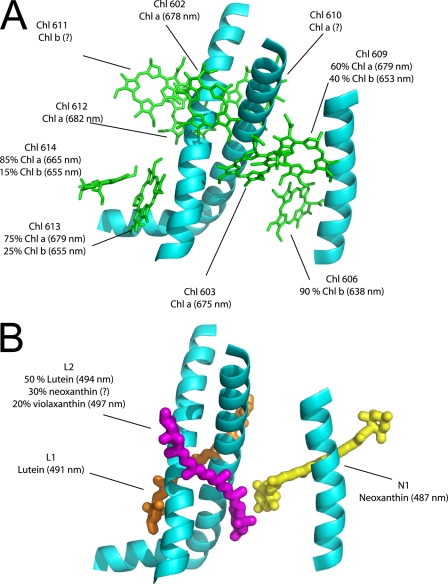
Chlorophyll Binding Sites in the Different Lhc Proteins—The occupancy of chlorophyll binding sites and the absorption of individual chlorophylls in Lhcb5 are summarized in Fig. 4. A comparison of the pigment properties in the same binding site of different Lhc complexes allows underscoring common features and specificities that can be correlated with the functional specialization of each Lhc gene product (Table 5). At least three different domains are conserved through the family. 1) In all complexes, site 612 accommodates a Chl a with absorption at 681-682 nm, which in Lhcb complexes represents the lowest energy state of the system. 2) Sites 602 and 603 also accommodate Chl a in all Lhc complexes (although in the case of Lhcb5 we cannot exclude a limited affinity for Chl b). Chl-603 absorbs around 675 nm. 3) Sites 613 and 614 retain the ability to accommodate both Chl a and b in all members of the family, and with the exception of Lhcb4, they have similar energy in the different complexes.
FIGURE 4.
Map of Lhcb5 chromophores. Data from mutational analysis were employed to build an Lhcb5 chromophore map. Chl, carotenoids, and α helix positions are from LHCII (Liu et al. (5); Protein Data Bank code 1rwt). A, different chlorophylls are shown in green. The binding site selectivity and absorption maxima of each chlorophyll are also reported. B, carotenoid bound by Lhcb5 is shown together with the binding site occupancy and absorption values.
TABLE 5.
Chlorophyll binding site occupancy in Lhca1, Lhcb1, Lhcb4, and Lhcb5 antenna proteins
The nature of the chlorophylls in the different chlorophyll binding sites and the absorption (Abs.) maxima in Qy are shown in Lhca1, Lhcb1, Lhcb4, and Lhcb5. Where no or uncertain information are available about chlorophyll nature and absorption maxima, a question mark (?) is indicated.
|
Pigment binding site
|
Lhca1a
|
Lhcb1b
|
Lhcb4c(CP29)
|
Lhcb5d(CP26)
|
||||
|---|---|---|---|---|---|---|---|---|
| Chl | Abs. maxima | Chl | Abs. maxima | Chl | Abs. maxima | Chl | Abs. maxima | |
| nm | nm | nm | nm | |||||
| Chl-610 | Chl a | ? | Chl a | 679 | Chl a | 669 (?) | Chl a (?) | ? |
| Chl-612 | Chl a | 682 | Chl a | 681 | Chl a | 680 | Chl a | 682 |
| Chl-613 | Chl a/Chl b (80/20) | 681/642 | Chl a/Chl b (50/50) | 662/650 | Chl a/Chl b (70/30) | 668/638 | Chl a/Chl b (75/25) | 679/655 |
| Chl-602 | Chl a | 683 | Chl a | 674 | Chl a | 676 | Chl a | 678 |
| Chl-603 | Chl a | 681/685 | Chl a | 675 | Chl a | 675 | Chl a | 675 |
| Chl-604 | Chl a/Chl b (50/50) | 678/652 | ||||||
| Chl-607 | Chl b | 652 | ||||||
| Chl 608 | Chl a | ? | Chl a | 679 | ||||
| Chl-611 | Chl a/Chl b (50/50) | 682/649 | Chl b | 646/660 | Chl b (?) | ? | ||
| Chl-614 | Chl a/Chl b (90/10) | 660/681 | Chl a/Chl b (50/50) | 665/650 | Chl a/Chl b (30/70) | 679/639 | Chl a/Chl b (85/15) | 665/655 |
| Chl-609 | Chl a/Chl b (20/80) | ?/642 | Chl b | 652 | Chl a/Chl b (60/40) | 678/650 | Chl a/Chl b (60/40) | 679/653 |
| Chl-606 | Chl a/Chl b (30/70) | 685/649 | Chl b | 652 | Chl a/Chl b (40/60) | 678/652 | Chl b | 638 |
| Car L1 | L | 495 | L | 489 | L | 494.5 | L | 491 |
| Car L2 | V | 499 | L/V | 495/492 | V/N | 492/? | L/V/N | 494/497/? |
| Car N1 | L/V/N | ?/488/? | N | 486 | N | 486 | N | 487 |
| Car V1 | V | 484.8 | ||||||
The domain of the C helix is less conserved both with respect to the affinity for Chl a or b and to the energy of the Chl. The difference depends largely on the extension of the H-bond network that in Lhcb1 involves most of the Chl in this domain. The key difference is the presence of a Gln in Lhcb1 as a ligand for Chl-606 and of a Glu in most of the other complexes. Because this residue is involved in a complex network of H-bonds that stabilize the binding of Chl b in this region, the substitution has a large effect on Chl a versus Chl b affinity. Indeed, Chl-609 is a Chl b in Lhcb1 and a site with mixed occupancy in all other complexes. Most of the pigments present in this region are also involved in excitonic interactions. Thus a small change in the structure can lead to a large change in the energy of the pigments, allowing for fine-tuning of the light harvesting properties of the individual complexes. This is clearly the case for Chl-606, which absorbs at 638 nm in Lhcb4 and Lhcb5 and shifts to the red in Lhcb1.
Light Harvesting—The efficiency in energy transfer from Chl b > Chl a and Car > Chl a allows the acquisition of information on the light harvesting function of the individual pigments coordinated to Lhcb5. Chl b to Chl a energy transfer efficiency decreases upon mutations at the ligands of Chl-613, -606, and -609, as in the case of Lhcb1 (54), suggesting that energy equilibration in Lhcb5 takes a very similar path to that in LHCII (55).
Analysis of the complexes reconstituted with different carotenoid composition indicates that neoxanthin transfers energy to Chl with high efficiency. This was confirmed by the analysis of the E129V mutant, which lost neoxanthin and showed a strong decrease of the energy transfer efficiency.
The data also show that violaxanthin is very efficient in energy transfer, clearly more so than lutein and zeaxanthin, which lower the transfer efficiency to 60 and 55%, respectively, versus 75% for Lhcb5-V and for the WT complex. This is in agreement with an earlier proposal, which suggested that violaxanthin bound to Lhcb protein is involved in light harvesting, whereas zeaxanthin can act as a quencher of the Chl a excitation energy (56).
Photoprotection—In addition to its role in light harvesting, it has been proposed that Lhcb5 is involved in the photoprotective mechanisms that under conditions of high light protect plants from photoinhibition by dissipating excess energy as heat. The efficiency of the dissipative channel is proportional to the quenching of fluorescence and thus can be measured. The excited state lifetime of Chl associated with Lhcb complexes is shorter than that of Chl in solution, 3.6-3.9 ns versus 5 ns (57, 58). It has been suggested that this may be due to a mixing of the excited states of Chl and xanthophylls, which have a very short S1 lifetime (59). Analysis of the mutants showed that the fluorescence quantum yield of Lhcb5-H68F is strongly increased as compared with that of the WT, whereas all other mutants showed a yield at the WT level or lower. This indicates that Chl-603 is involved in a mechanism that shortens the lifetime of the excited states of the complex. Interestingly, Chl-603 is located in close proximity to the carotenoid binding site L2, suggesting that a mixing of the excited states between Chl-603 and carotenoid in L2 possibly occurs. Conversely, Lhcb5-N179F and Lhcb5-Q193L mutants, affecting Chl molecules located nearby lutein L1, are characterized by a significant decrease of florescence quantum yield, indicating that Chl-612 and Chl-613 are involved in light harvesting rather than quenching, at least at the level of complexes in solution.
Although the maximum reduction of the fluorescence yield observed in the Chl mutants is 25% with respect to WT, the effect of the exchange of xanthophylls is far more pronounced. Moreover, analysis of the complexes reconstituted with different carotenoids showed that the modulation of the fluorescence quantum yield depends not only on the particular binding site in which the xanthophyll is located but also on the xanthophyll species associated with it. The presence of zeaxanthin or lutein as the only xanthophyll decreases the yield to 50 and 40%, respectively, as compared with the WT, whereas the effect of violaxanthin is far weaker, in agreement with the proposed role of quenchers for zeaxanthin and lutein (56, 60). Interestingly, most of the samples coordinating neoxanthin showed a fluorescence yield higher than that of the complexes without this species but with similar xanthophyll composition (e.g. Lhcb5 LN versus lutein). The only exception is Lhcb5-LNZ, which is far more quenched than the WT. It has been suggested that during the mechanism of non-photochemical quenching a conformational change occurs, switching the complex from a light harvesting to a photoprotective function. This conformational change has been monitored in vivo following the Raman signal of the neoxanthin, which changes configuration in a stress condition (60). In the case of Lhcb5 it has been shown that this conformational change is driven by the binding of zeaxanthin to side L2, leading to a decrease in fluorescence yield (32). It can thus be suggested that the presence of neoxanthin stabilizes the light-harvesting conformation, whereas the presence of the zeaxanthin stabilizes the quenched conformation. This conformational change could lead to a new functional organization of the pigments bound to Lhcb5, possibly inducing quenching also on the L1 site, as proposed recently for LHCII (60), or enhancing the interactions between Chl-603 and zeaxanthin in L2, which lead to the formation of a zeaxanthin radical cation (33).
Physiological Role of Lhcb5 in NPQ—The above results open the question of the physiological relevance in vivo of these in vitro observations. The amount of energy dissipated via NPQ in vivo is up to 80% of the energy harvested by the system. This indicates that fluorescence quenching in vivo is stronger than that observed in vitro. The fluorescence yield of leaves is known to be modulated by a process called non-photochemical quenching. It might be asked whether the effect of zeaxanthin on Lhcb5 fluorescence yield has a role in NPQ. Indeed, it has been shown that Lhcb5 plays a major role in the slower component of NPQ, qI, which is reduced in Lhcb5-depleted plants (31, 61).
Lhcb5 has been shown to catalyze charge transfer quenching upon zeaxanthin binding (33, 35), inducing formation of both lutein and zeaxanthin radical cation formation (62). Because the formation of carotenoid radical cations in Lhc proteins is one of the mechanisms suggested to be responsible for the fastest component of NPQ, qE (35), we hypothesized that Lhcb5 also has a role in this process. The discrepancy of genetic investigations showing no phenotype in the absence of Lhcb5 (31) can be explained by a compensatory role of Lhcb4 and Lhcb6. Moreover, in Lhcb4 Chl-603, which we suggest has a role in fluorescence quantum yield reduction through interaction with the carotenoid on the L2 site in Lhcb5, was shown to be involved directly in energy dissipation through charge transfer quenching; the interaction between carotenoids in L2 and Chl-603 seems to be a determinant for quenching in the minor complexes.
Conclusion—We have performed a detailed analysis of the monomeric antenna protein Lhcb5 by mutation analysis of Chl binding sites and exchange of xanthophyll species in the carotenoid binding sites. Biochemical and spectroscopic analysis allows the investigation of the nature of and energy levels associated with the different Chl bound to Lhcb5. The occupancy of Chl sites is distinct as compared with that of LHCII (5, 21), Lhcb4 (15), and Lhca proteins (22) (Table 2), especially in the helix C domain, providing a base for the different physiological roles of these homologous proteins.
In regard to carotenoids, Lhcb5 has three xanthophyll binding sites, L1, L2, and N1. Site N1 is specific for neoxanthin and is characterized by a high Car > Chl a energy transfer. Sites L1 and L2 are promiscuous and can bind all xanthophyll species; the L1 and L2 sites appear to have distinct properties, the former being specialized in harvesting light and transferring excitation energy to Chl a, the latter, instead, in controlling the fluorescence quantum yield of the system by quenching Chl a excited states. The L2 site, moreover, is particularly effective in exchanging violaxanthin with zeaxanthin, which further enhances the level of quenching. These properties, which in vivo are likely to be enhanced by the presence of other factors such as PsbS (63), are consistent with the observed function of Lhcb5 in both the fast and slowly reversible components of NPQ as observed in vivo (32, 61) and in vitro (33).
Supplementary Material
This work was supported by Grants FIRB RBLA03455F-002 and RBIP06CPBR-006 from the Italian Ministry of Research Special Fund for Basic Research and by a Vidi grant (to R. C.) from The Netherlands Organization for Scientific Research (NWO), Earth and Life Science. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: PS, photosystem; Car, carotenoid; Chl, chlorophyll; Lhca, light-harvesting complex a (photosystem I); Lhcb, light-harvesting complex b (photosystem II); LHCII, major light-harvesting complex of photosystem II; NPQ, non-photochemical quenching; WT, wild type; L, lutein; V, violaxanthin; N, neoxanthin; Z, zeaxanthin; HPLC, high pressure liquid chromatography.
Carotenoid binding sites L1, L2, N1, and V1 are named, respectively, Lut620, Lut621, Neo, and Xant in the work of Liu et al. (5).
References
- 1.Albertsson, P.-A., Andreasson, E., and Svensson, P. (1990) FEBS Lett. 273 36-40 [DOI] [PubMed] [Google Scholar]
- 2.Jansson, S. (1999) Trends Plant Sci. 4 236-240 [DOI] [PubMed] [Google Scholar]
- 3.Green, B. R., and Durnford, D. G. (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 685-714 [DOI] [PubMed] [Google Scholar]
- 4.Caffarri, S., Croce, R., Cattivelli, L., and Bassi, R. (2004) Biochemistry 43 9467-9476 [DOI] [PubMed] [Google Scholar]
- 5.Liu, Z., Yan, H., Wang, K., Kuang, T., Zhang, J., Gui, L., An, X., and Chang, W. (2004) Nature 428 287-292 [DOI] [PubMed] [Google Scholar]
- 6.Kühlbrandt, W., Wang, D. N., and Fujiyoshi, Y. (1994) Nature 367 614-621 [DOI] [PubMed] [Google Scholar]
- 7.Croce, R., Remelli, R., Varotto, C., Breton, J., and Bassi, R. (1999) FEBS Lett. 456 1-6 [DOI] [PubMed] [Google Scholar]
- 8.Caffarri, S., Croce, R., Breton, J., and Bassi, R. (2001) J. Biol. Chem. 276 35924-35933 [DOI] [PubMed] [Google Scholar]
- 9.Ruban, A. V., Lee, P. J., Wentworth, M., Young, A. J., and Horton, P. (1999) J. Biol. Chem. 274 10458-10465 [DOI] [PubMed] [Google Scholar]
- 10.Dainese, P., and Bassi, R. (1991) J. Biol. Chem. 266 8136-8142 [PubMed] [Google Scholar]
- 11.Ben Shem, A., Frolow, F., and Nelson, N. (2003) Nature 426 630-635 [DOI] [PubMed] [Google Scholar]
- 12.Pascal, A., Gradinaru, C., Wacker, U., Peterman, E., Calkoen, F., Irrgang, K.-D., Horton, P., Renger, G., van Grondelle, R., Robert, B., and Van Amerongen, H. (1999) Eur. J. Biochem. 262 817-823 [DOI] [PubMed] [Google Scholar]
- 13.Giuffra, E., Cugini, D., Croce, R., and Bassi, R. (1996) Eur. J. Biochem. 238 112-120 [DOI] [PubMed] [Google Scholar]
- 14.Croce, R., Morosinotto, T., Castelletti, S., Breton, J., and Bassi, R. (2002) Biochim. Biophys. Acta 1556 29-40 [DOI] [PubMed] [Google Scholar]
- 15.Bassi, R., Croce, R., Cugini, D., and Sandona, D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10056-10061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gastaldelli, M., Canino, G., Croce, R., and Bassi, R. (2003) J. Biol. Chem. 278 19190-19198 [DOI] [PubMed] [Google Scholar]
- 17.Croce, R., Canino, G., Ros, F., and Bassi, R. (2002) Biochemistry 41 7334-7343 [DOI] [PubMed] [Google Scholar]
- 18.Pagano, A., Cinque, G., and Bassi, R. (1998) J. Biol. Chem. 273 17154-17165 [DOI] [PubMed] [Google Scholar]
- 19.Castelletti, S., Morosinotto, T., Robert, B., Caffarri, S., Bassi, R., and Croce, R. (2003) Biochemistry 42 4226-4234 [DOI] [PubMed] [Google Scholar]
- 20.Caffarri, S., Passarini, F., Bassi, R., and Croce, R. (2007) FEBS Lett. 581 4704-4710 [DOI] [PubMed] [Google Scholar]
- 21.Remelli, R., Varotto, C., Sandona, D., Croce, R., and Bassi, R. (1999) J. Biol. Chem. 274 33510-33521 [DOI] [PubMed] [Google Scholar]
- 22.Morosinotto, T., Castelletti, S., Breton, J., Bassi, R., and Croce, R. (2002) J. Biol. Chem. 277 36253-36261 [DOI] [PubMed] [Google Scholar]
- 23.Mozzo, M., Morosinotto, T., Bassi, R., and Croce, R. (2006) Biochim. Biophys. Acta 1757 1607-1613 [DOI] [PubMed] [Google Scholar]
- 24.Morosinotto, T., Mozzo, M., Bassi, R., and Croce, R. (2005) J. Biol. Chem. 280 20612-20619 [DOI] [PubMed] [Google Scholar]
- 25.Schmid, V. H. (2008) CMLS Cell. Mol. Life Sci. 65 3619-3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alboresi, A., Caffarri, S., Nogue, F., Morosinotto, T., and Bassi, R. (2008) PLoS ONE 3 e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganeteg, U., Kulheim, C., Andersson, J., and Jansson, S. (2004) Plant Physiol. 134 502-509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demmig-Adams, B., and Adams, W. W. (1992) Annu. Rev. Plant Physiol. Plant Mol. Biol. 43 599-626 [Google Scholar]
- 29.Kovacs, L., Damkjaer, J., Kereiche, S., Ilioaia, C., Ruban, A. V., Boekema, E. J., Jansson, S., and Horton, P. (2006) Plant Cell 18 3106-3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson, J., Wentworth, M., Walters, R. G., Howard, C. A., Ruban, A. V., Horton, P., and Jansson, S. (2003) Plant J. 35 350-361 [DOI] [PubMed] [Google Scholar]
- 31.Andersson, J., Walters, R. G., Horton, P., and Jansson, S. (2001) Plant Cell 13 1193-1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dall'Osto, L., Caffarri, S., and Bassi, R. (2005) Plant Cell 17 1217-1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn, T. K., Avenson, T. J., Ballottari, M., Cheng, Y. C., Niyogi, K. K., Bassi, R., and Fleming, G. R. (2008) Science 320 794-797 [DOI] [PubMed] [Google Scholar]
- 34.Mozzo, M., Dall'Osto, L., Hienerwadel, R., Bassi, R., and Croce, R. (2008) J. Biol. Chem. 283 6184-6192 [DOI] [PubMed] [Google Scholar]
- 35.Holt, N. E., Zigmantas, D., Valkunas, L., Li, X. P., Niyogi, K. K., and Fleming, G. R. (2005) Science 307 433-436 [DOI] [PubMed] [Google Scholar]
- 36.Nagai, K., and Thøgersen, H. C. (1987) Methods Enzymol. 153 461-481 [DOI] [PubMed] [Google Scholar]
- 37.Paulsen, H., Finkenzeller, B., and Kuhlein, N. (1993) Eur. J. Biochem. 215 809-816 [DOI] [PubMed] [Google Scholar]
- 38.Goodwin, T. W. (ed) (1981) Chemistry and Biochemistry of Plant Pigments, 2nd Ed., pp. 3-18, Academic Press, London
- 39.Gilmore, A. M., and Yamamoto, H. Y. (1991) Plant Physiol. 96 635-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amunts, A., Drory, O., and Nelson, N. (2007) Nature 447 58-63 [DOI] [PubMed] [Google Scholar]
- 41.Horn, R., and Paulsen, H. (2004) J. Biol. Chem. 279 44400-44406 [DOI] [PubMed] [Google Scholar]
- 42.Horn, R., Grundmann, G., and Paulsen, H. (2007) J. Mol. Biol. 366 1045-1054 [DOI] [PubMed] [Google Scholar]
- 43.Hoober, J. K., and Eggink, L. L. (1999) Photosynth. Res. 61 197-215 [Google Scholar]
- 44.Eggink, L. L., and Hoober, J. K. (2000) J. Biol. Chem. 275 9087-9090 [DOI] [PubMed] [Google Scholar]
- 45.Morosinotto, T., Baronio, R., and Bassi, R. (2002) J. Biol. Chem. 277 36913-36920 [DOI] [PubMed] [Google Scholar]
- 46.Jahns, P., and Schweig, S. (1995) Plant Physiol. Biochem. 33 683-687 [Google Scholar]
- 47.Demmig-Adams, B., Winter, K., Kruger, A., and Czygan, F.-C. (1989) in Photosynthesis: Plant Biology (Briggs, W. R., ed) Vol. 8, pp. 375-391, Alan R. Liss, New York [Google Scholar]
- 48.Matsubara, S., Morosinotto, T., Osmond, C. B., and Bassi, R. (2007) Plant Physiol. 144 926-941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frank, H. A., Das, S. K., Bautista, J. A., Bruce, D., Vasil'ev, S., Crimi, M., Croce, R., and Bassi, R. (2001) Biochemistry 40 1220-1225 [DOI] [PubMed] [Google Scholar]
- 50.Morosinotto, T., Caffarri, S., Dall'Osto, L., and Bassi, R. (2003) Physiol. Plant. 119 347-354 [Google Scholar]
- 51.Croce, R., Cinque, G., Holzwarth, A. R., and Bassi, R. (2000) Photosynth. Res. 221-231 [DOI] [PubMed]
- 52.Wentworth, M., Ruban, A. V., and Horton, P. (2000) FEBS Lett. 471 71-74 [DOI] [PubMed] [Google Scholar]
- 53.Noctor, G., Rees, D., Young, A., and Horton, P. (1991) Biochim. Biophys. Acta 1057 320-330 [Google Scholar]
- 54.Formaggio, E., Cinque, G., and Bassi, R. (2001) J. Mol. Biol. 314 1157-1166 [DOI] [PubMed] [Google Scholar]
- 55.Novoderezhkin, V. I., Palacios, M. A., van Amerongen, H., and van Grondelle, R. (2005) J. Phys. Chem. B 109 10493-10504 [DOI] [PubMed] [Google Scholar]
- 56.Young, A. J., and Frank, H. A. (1996) J. Photochem. Photobiol. B Biol. 36 3-15 [DOI] [PubMed] [Google Scholar]
- 57.Moya, I., Silvestri, M., Vallon, O., Cinque, G., and Bassi, R. (2001) Biochemistry 40 12552-12561 [DOI] [PubMed] [Google Scholar]
- 58.Crimi, M., Dorra, D., Bosinger, C. S., Giuffra, E., Holzwarth, A. R., and Bassi, R. (2001) Eur. J. Biochem. 268 260-267 [DOI] [PubMed] [Google Scholar]
- 59.Van Amerongen, H., and van Grondelle, R. (2001) J. Phys. Chem. B 105 604-617 [Google Scholar]
- 60.Ruban, A. V., Berera, R., Ilioaia, C., van Stokkum, I. H., Kennis, J. T. M., Pascal, A. A., Van Amerongen, H., Robert, B., Horton, P., and van Grondelle, R. (2007) Nature 450 575-578 [DOI] [PubMed] [Google Scholar]
- 61.de Bianchi, S., Dall'Osto, L., Tognon, G., Morosinotto, T., and Bassi, R. (2008) Plant Cell 20 1012-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Avenson, T. J., Ahn, T. K., Niyogi, K. K., Ballottari, M., Bassi, R., and Fleming, G. R. (2009) J. Biol. Chem. 284 2830-2835 [DOI] [PubMed] [Google Scholar]
- 63.Bonente, G., Howes, B. D., Caffarri, S., Smulevich, G., and Bassi, R. (2008) J. Biol. Chem. 283 8434-8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mozzo, M., Passarini, F., Bassi, R., van Amerongen, H., and Croce, R. (2008) Biochim. Biophys. Acta 1777 1263-1267 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.