Abstract
Nestin is a molecular marker for neural progenitor cells. Rat and human nestin genes possess a central nervous system-specific enhancer within their second introns. However, the transcription factors that bind to the nestin enhancer have not been fully elucidated. Here, we show that the second intron of the mouse nestin gene is sufficient to drive reporter gene expression in the developing nervous system. The core sequence of this central nervous system-specific enhancer localizes to the 3′ 320-bp region. The cis-elements for Sox and POU family transcription factors and the hormone-responsive element are essential for nestin expression during embryonic carcinoma P19 cell neural differentiation and in the developing chick neural tube. Interestingly, different transcription factors bind to the nestin enhancer at different stages of P19 cell neural differentiation and central nervous system development. Sox2 and SF1 may mediate basal nestin expression in undifferentiated P19EC cells, whereas Sox2, Brn1, and Brn2 bind to the enhancer in P19 neural progenitor cells. Similarly, in vivo, Oct1 binds to the nestin enhancer in embryonic day 8.5 (E8.5) mouse embryos, and Oct1, Brn1, and Brn2 bind to this enhancer in E10.5 and E12.5 mouse embryos. Our studies therefore suggest a temporal coordination of transcription factors in determining nestin gene expression.
The generation of the three germ layers, the ectoderm, the mesoderm, and the endoderm, is among the earliest and the most fundamental processes underlying animal development. The central nervous system is one of the main derivatives of ectoderm. During development, dorsal ectoderm gives rise to the neural plate, which is later transformed into the neural tube. The neural tube is further subdivided into the fore-, mid-, and hindbrain and the spinal cord along the anterior-posterior axis (1). Different types of neurons in the mature nervous system are generated from pools of mitotic progenitor cells located in the neural tube. Nestin, a type VI intermediate filament protein, is abundant in mammalian neurogenic progenitor cells and has been used as a marker for neural progenitor cells (2-5). To better understand the biology of neural progenitor cells, it is important to study the regulation of the nestin gene.
During the development of the central nervous system, early expression of nestin mRNA is observed in the neural plate at embryonic day 7.5 (E7.5).3 At E10.5, nestin mRNA is distributed predominantly in the neuroepithelial and radial glia cells of the neural tube of mouse embryos (2). The nestin protein is found in the neural tube prior to neurogenesis, but not post-neurogenesis. When neural progenitor cells (NPCs) in the neural tube differentiate into mature neurons and glial cells, nestin expression is down-regulated and replaced by the expression of neurofilament and glial fibrillary acidic protein (2, 6). nestin expression is barely detectable in adult central nervous system except for certain proliferative regions, such as the dentate gyrus of the hippocampus and subependymal zones of the brain and spinal cord (2, 7). Nestin expression is induced in response to central nervous system injury and in certain central nervous system tumors (8, 9). Interestingly, the onset of nestin expression occurs earlier than the neural plate stage (10). Embryonic stem cells initiate nestin expression immediately after differentiation (11), and pluripotent mouse P19 embryonic carcinoma cells (P19EC) cells maintain basal nestin expression (12), whereas both types of cells up-regulate nestin expression during their neural differentiation program. These observations suggest that there is a regulation switch for nestin expression between pluripotent stem cells and neural progenitor cells.
Compared with nestin expression, little is known about the function of this protein. Recently, nestin has been shown to play an important role in the phosphorylation-dependent disassembly of vimentin intermediate filaments during mitosis and to be a potential target for cdk5 and p35 kinase during development (13, 14). In addition, studies using chick transitin, the homolog of mammalian nestin, suggest that transitin may physically interact with Numb, determine the intracellular localization of Numb in mitotic avian neuroepithelial cells, and regulate neurogenesis (15).
Because nestin expression is closely related to central nervous system development, it is widely used as a specific marker for NPCs in the mammalian nervous system. Studies investigating the transcriptional regulation of the nestin gene could enhance our understanding of the regulatory network that links the nestin gene and the determination and maintenance of NPCs. The regulation of the rat and human nestin genes during development has been investigated in transgenic mice, and data from these mice suggested that the tissue-specific enhancer localized in the second intron of the nestin gene is sufficient to drive nestin expression in the developing central nervous system (5, 16). The second intron of the nestin gene was therefore widely used for deleting gene's expression from the neural progenitor state with the nestin-cre lines (17-20) and labeling the neural progenitor cells using the nestin-GFP transgenic mice (21-23). In this neural enhancer, a POU-binding site is required for general central nervous system expression, and an adjacent hormone response element is necessary for nestin expression in the dorsal midbrain and forebrain (24-26). Recently, Tanaka et al. (27) demonstrated that members of the Sox and POU family of transcription factors interact synergistically to transactivate the neural enhancer of nestin gene in vitro, and such an interaction might be involved in the determination of nestin expression in neural primordial cells. However, questions such as what transcription factors bind to the nestin enhancer to regulate its expression in vivo, and whether the same transcription factors regulate nestin expression at different stages of central nervous system development, need to be addressed.
Our previous studies described the cloning and expression pattern of the mouse nestin gene (28-30) and showed that the transcription factors Sp1 and Sp3 are important for the promoter activity of this gene (31). In this study, we found that a central nervous system-specific enhancer is located in the second intron of the mouse nestin gene. This enhancer and its core sequence were able to drive reporter gene expression in the developing nervous system of mouse and chick embryos, as well as during P19 cell neural differentiation. Moreover, different transcription factor complexes bind to the nestin enhancer at different stages of P19 cell neural differentiation and central nervous system development. Sox2 and SF1 may mediate basal nestin expression in undifferentiated P19EC cells, whereas Sox2, Brn1, and Brn2 regulate the enhancer to increase its expression in P19NPCs. Similarly, in vivo, Oct1 binds to the nestin enhancer in E8.5 mouse embryos, whereas Oct1, Brn1, and Brn2 bind to this enhancer in the brains of E10.5 and E12.5 mouse embryos.
EXPERIMENTAL PROCEDURES
Transgenic Mice—The second intron of the mouse nestin gene was inserted upstream of the basic herpes simplex virus thymidine kinase (tk) promoter in the pSaStk/lacZ vector (16). The plasmid was digested with the HindIII restriction enzyme to remove the vector backbone. A purified fragment containing the second intron of the mouse nestin gene, the tk promoter, and lacZ reporter gene was used for pronuclear injections of fertilized oocytes from ICR mice. Transgenic mice born after the injections were screened by PCR using the primers listed in the supplemental Table S1. One line for pSaStk2In43/1629 and three lines for pSaStk2In1310/1629 were identified as PCR-positive. These clones were propagated through breeding with ICR mice, and the progeny were used for subsequent analyses. One line for pSaStk2In43/1629 and two lines for pSaStk2In1310/1629 had identical patterns of lacZ expression throughout the developing central nervous system. Embryos were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside as previously described (25).
RNA Analysis—Total RNA was extracted from cultured cells or mouse embryos using TRIzol reagent (Invitrogen). Total RNA (10 μg) from P19 cells during neural differentiation was analyzed by Northern blot as described previously (32). Reverse transcription was performed with 5 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen). The PCR cycling conditions consisted of a denaturation step at 94 °C for 45 s, annealing for 45 s, and extension at 72 °C for 45 s. The primers for each gene are shown in supplemental Table S1. To quantify the relative expression of nestin mRNA during neural differentiation of P19 cells, quantitative reverse transcription PCR (qRT-PCR) was performed with the DNA Engine Opticon 2 Real-Time PCR Detection System (Bio-Rad) using Jumpstart Taq Readymix (Sigma) and EvaGreen dye (Biotium, Hayward, CA).
Plasmid Construction—To prepare luciferase reporter constructs for cell transfection, various fragments of the second intron of the mouse nestin gene were inserted into the pGL3-Px′ vector (Fig. 2C), which was modified within the multiple cloning sites from the pGL3-Promoter (Promega, Madison, WI). These inserts were either prepared by convenient digestions of plasmids or by PCR amplifications. The luciferase reporter construct pNesPE-Luc was used for stable transfection. It was constructed by first amplifying the 1587-bp (43/1629) second intron of the mouse nestin gene by PCR to generate the insert. The insert was then cloned into the pNESP-3809/+183 construct, which contains a 4-kb mouse nestin gene promoter and a luciferase reporter gene (31). The generation of site-mutated luciferase constructs was performed by PCR using two sets of primers, as described previously (31). All of these constructs were confirmed by enzymatic digestion and DNA sequencing. The primers used to generate the plasmid constructs are listed in supplemental Table S1. To prepare the GFP reporter constructs used for in ovo chick embryo electroporation, different fragments of the second intron of the mouse nestin gene were inserted into the pxtkEGFP vector. pxtkEGFP was obtained by inserting a KpnI-SacI fragment, which contains the multiple cloning sites in the vector pBluescript SKII into the KpnI and SacI sites in ptkEGFP (33). Site-mutated GFP constructs were generated by the recovery of insert fragments from corresponding luciferase constructs that carried the same site mutations, which were then ligated into the pxtkEGFP vector.
FIGURE 2.
The second intron of the mouse nestin gene directed nestin expression during RA-induced P19 neural differentiation. A, Northern blot analysis of nestin mRNA expression. Total RNA extracted from P19 cells during RA-induced neural differentiation was used for Northern blotting. 18 S rRNA was used as a control. Semi-quantitative analysis of the Northern blot results is shown in the right panel. B, qRT-PCR analysis of nestin mRNA expression. Total RNA extracted from P19 cells during neural differentiation was analyzed using qRT-PCR with primers specific for mouse nestin and β-actin. The amount of nestin mRNA was normalized to that of β-actin, and the value in undifferentiated P19EC cells (A0) was designated as 1. C, diagram of the pGL3-Px′ vector. Various fragments of the second intron of the mouse nestin gene were inserted upstream of the SV40 promoter. D, luciferase constructs containing the 5′ upstream promoter (pNESP-3809/+183), the first intron (pNes1In 1/1003), or the second intron (pNes2In32/1628) of the mouse nestin gene were transiently transfected into P19EC and P19NPC cells. The activity of each construct is shown relative to that of the pGL3-Px′ vector, which contains a SV40 promoter. Values were normalized for transfection efficiency by co-transfection with a Renilla luciferase expression plasmid, and they are shown as means ± S.D. for three independent experiments. E, luciferase activity during neural differentiation of NesPEL/P19 cells. P19 cells were transfected with pNesPE-Luc, pRL-TK, and pPGKneobpA and selected with G418. A stable cell line, NesPEL/P19-E, was induced for neural differentiation, and the luciferase activity was measured. Firefly luciferase activity was normalized with that of Renilla luciferase. Luciferase activity in undifferentiated NesPEL/P19-E cells (A0) was designated as 1, and the relative luciferase activity of NesPEL/P19-E cells at different stages was then calculated.
P19 Cell Culture and Retinoic Acid-induced Neural Differentiation—P19 cells were maintained in Dulbecco's modified Eagle's medium/F-12 1:1 medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) at 37 °C in 5% CO2. P19C6, a subclone of the mouse embryonic carcinoma P19 cell line, was used in this study. Neural differentiation of P19 cells treated with all-trans-retinoic acid (RA, Sigma) was performed as described previously (34, 35). Briefly, cells were allowed to aggregate in bacterial grade Petri dishes at a seeding density of 1 × 105 cells/ml in the presence of 1 × 10-6 m RA in 10% fetal bovine serum/α-minimal essential medium (Invitrogen). After 4 days of aggregation, cells were dissociated into single cells and replated on poly-l-lysine-coated tissue culture dishes at a density of 1 × 105 cells/cm2 in N2 serum-free medium (Dulbecco's modified Eagle's medium/F-12 supplemented with 5 μg/ml insulin, 50 μg/ml human transferrin, 20 nm progesterone, 60 μm putrescine, and 30 nm sodium selenite) supplemented with 1 μg/ml fibronectin. The cells were then allowed to adhere, and they were cultured with replacement of the medium every 48 h. The same protocol was used for neural differentiation of the P19 cell line stably transfected with pNesPE-Luc (named as the NesPEL/P19 cell line).
Cell Transfection and Luciferase Assays—For transient transfection in the P19EC cells, cells were plated onto 24-well plates in 10% fetal bovine serum/Dulbecco's modified Eagle medium/F-12 medium 1 day prior to transfection at a density of 2 × 104 cells/well. For transient transfection in P19NPCs, P19 cells were dissociated into single cells after 4 days of aggregation and replated into 12-well plates in N2 medium 5 h before transfection at a density of 5 × 105 cells/well. Cells were transfected with 0.4 μg of the various luciferase constructs using the Fugene6 reagent (Roche Applied Science), according to the manufacturer's instructions. The pRL-TK plasmid (Promega) containing a Renilla luciferase gene driven by the TK promoter was co-transfected with the constructs to normalize the luciferase activity. 40 h later, cell lysates were prepared, and the luciferase activity was measured using the dual reporter assay system (Promega) and a 20/20n luminometer (Turner Biosystems, Sunnyvale, CA) according to the manufacturer's instructions. All transfections were performed three times in triplicate. For the establishment of the stably transfected cell line NesPEL/P19, P19 cells were co-transfected with 6 μg of pNesPE-Luc, 1.5 μg of pRL-TK, and 2 μg of pPGKneobpA. After selection with G418 for 2 weeks, ∼30 G418-resistent clones were picked, expanded, and analyzed for luciferase activity.
In Ovo Chick Embryo Electroporation—Fertilized eggs were obtained from the Shanghai Academy of Agricultural Sciences (Shanghai, China). In ovo chick embryo electroporation was performed as previously described (36). Briefly, fertilized eggs were incubated at 38 °C, and the embryos were staged according to Hamburger and Hamilton (37). A DNA solution containing the GFP construct (1.0 μg/μl) as a reporter and pHcRed1-N1 (1.0 μg/μl) as a tracer was injected into the lumen of the closed neural tube of a chick embryo at HH stage 10 and electroporated into cells on one side of the neural tube by electrodes flanking the embryo. A square wave current (six 30-ms pulses of 25 V) was generated using a BTX ECM830 Electro Square Porator (Genetronics, San Diego, CA) connected to 4-mm platinum electrodes. Embryos were collected after a 24-h incubation (HH stage 17), fixed in 4% paraformaldehyde for 1 h at 4 °C, and they were examined with a fluorescence dissection microscope (Leica, Wetzlar, Germany). Neural tubes expressing HcRed1 were removed from the embryos, equilibrated in 20% sucrose overnight, embedded in Tissue-Tek OCT compound (Leica), and sectioned at a thickness of 10 μm. Fluorescence was visualized and photographed using the TCS SP2 Confocal Microscopy System (Leica).
Electrophoretic Mobility Shift Assays—Nuclear extracts (NE) from P19 cells and mouse embryos were prepared as described previously (31). NE (∼5 μg) was preincubated with 2 μg of poly(dG-dC) (Amersham Biosciences) and 10 μg of bovine serum albumin on ice for 30 min in 5 × binding buffer containing 50% glycerol, 60 mm HEPES, pH 7.9, 20 mm Tris-HCl, 300 mm KCl, 5 mm EDTA, and 5 mm dithiothreitol. A 32P-labeled DNA probe was added to the binding reaction mixture and incubated at 25 °C for 30 min. Competition experiments were performed by preincubation of a 100-fold molar excess of unlabeled probe together with NE. For supershift assays, specific antibodies were added to the reaction mixture before the addition of labeled probe. DNA-protein complexes were fractioned on a pre-run non-denaturing 6% polyacrylamide gel and subjected to electrophoresis in 0.5× TBE buffer (45 mM Tris-boric acid pH 8.0, and 1 mM EDTA). The gels were dried and exposed to x-ray film with an intensifying screen at -80 °C. The oligonucleotide sequences of the probes and competitors are reported in supplemental Table S1. The following antibodies were used for supershift assays: rabbit anti-human Oct1 (sc-232X, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-human Oct2 (sc-233X), goat anti-human Oct4 (sc-8628X), goat anti-human Oct6 (sc-11660X), goat anti-human Brn1 (sc-6028X), goat anti-human Brn2 (sc-6029X), goat anti-human Sox2 (sc-17320X), and rabbit anti-mouse SF1 (38).
Chromatin Immunoprecipitation—Chromatin immunoprecipitation (ChIP) was performed according to the manufacturer's protocol (Upstate Biotechnology, Charlottesville, VA) with minor modifications. Cellular protein-DNA complexes were cross-linked by incubating the cells in 1% formaldehyde for 10 min at room temperature. The reaction was stopped with the addition of 125 mm glycine. After washing with cold phosphate-buffered saline, the cells were lysed with cell lysis buffer (10 mm HEPES, pH 7.9, 0.5% Nonidet P-40, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Roche)) for 10 min at 4 °C. The pellets were resuspended in SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl pH 8.0, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture) and incubated for 20 min at 4 °C. Resuspended cells were sonicated with a Bioruptor (Diagenode, Liège, Belgium), and debris was removed by centrifugation. The supernatants were diluted 10-fold with dilution buffer (16.7 mm Tris-HCl, pH 8.0, 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 167 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture) and then precleared with 40 μl of protein A/G-Sepharose slurry (Upstate) for 1 h at 4 °C. Immunoprecipitation was performed with 4 μg of the specific antibody (the same antibodies were used as in the EMSA assays) incubated overnight at 4 °C. Normal rabbit or goat IgG was used as a negative control. Antibody complexes were pulled down for 2 h with 40 μl of protein A/G-Sepharose slurry. The Sepharose beads were then collected and washed sequentially for 5 min in low salt buffer (50 mm HEPES, pH 7.9, 0.1% SDS, 1% Triton X-100, 1 mm EDTA, 140 mm NaCl, 0.1% deoxycholate), high salt buffer (50 mm HEPES, pH 7.9, 0.1% SDS, 1% Triton X-100, 1 mm EDTA, 500 mm NaCl, 0.1% deoxycholate), and LiCl buffer (20 mm Tris-HCl, pH 8.0, 1 mm EDTA, 250 mm LiCl, 0.5% Nonidet P-40, 0.5% deoxycholate) three times. Beads were washed twice with 1× TE buffer and extracted with elution buffer (1% SDS, 100 mm NaHCO3). The eluates were heated at 65 °C for at least 5 h to reverse the cross-links. The DNA fragments were purified by phenol extraction, ethanol precipitation, and quantified by quantitative PCR using the DNA Engine Opticon 2 Real-Time PCR Detection System (Bio-Rad). The amounts of immunoprecipitated DNA were calculated by comparison to a standard curve generated by serial dilutions of input DNA, dividing the values obtained with normal rabbit or goat IgG. The data were plotted as the mean of at least two independent ChIP assays and three independent amplifications. The primer sets used to amplify the second intron of the mouse nestin gene were NesEn1 (second intron, 1311/1510, 200 bps) and NesEn2 (second intron, 1290/1510, 221 bps). The primer sets used as control to amplify the promoter of the mouse nestin gene were NesPro1 (5′ flanking, -40/+187, 227 bps). The primer sequences are listed in supplemental Table S1.
Immunofluorescent Staining—Immunofluorescent staining was performed as described previously (34). The primary antibodies and their final concentrations were as follows: rabbit anti-human Oct1 (1:1000, Santa Cruz Biotechnology), mouse anti-human Oct4 (1:200, Santa Cruz Biotechnology), goat anti-human Brn1 (1:1000, Santa Cruz Biotechnology), goat anti-human Brn2 (1:1000, Santa Cruz Biotechnology), rabbit anti-mouse Sox2 (1:200) (27), rabbit anti-mouse SF1 (1:1000) (38), rabbit anti-mouse nestin (1:200) (39), mouse anti-rat nestin (1:200, BD Biosciences, San Jose, CA). Fluorescein isothiocyanate- and Cy3-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). The fluorescence was visualized and photographed on a BX51 fluorescence microscope (Olympus, Tokyo, Japan) and a TCS SP2 Confocal Microscope (Leica).
RESULTS
The Second Intron of the Mouse nestin Gene Contains a Central Nervous System-specific Enhancer—Previous studies have shown that the second introns of the rat and human nestin genes contain a central nervous system-specific enhancer (5, 16). To examine if the second intron of the mouse nestin gene also contains this central nervous system-specific enhancer, we generated transgenic mice with the pSaStk2In43/1629 construct, which contains the second intron of the mouse nestin gene, a tk promoter, and lacZ reporter. At embryonic day (E) 12.5, the pSaStk2In43/1629 transgenic embryos exhibited lacZ expression in most parts of the developing central nervous system, including the spinal cord, myelencephalon, metencephalon, mesencephalon, diencephalon, and telencephalon (Fig. 1, A-C). Interestingly, about one-third of the lacZ-positive embryos exhibited lacZ expression in the whisker follicle sheath progenitor cells (Fig. 1C). No other area of expression was reproducibly observed. These results demonstrate that, like the rat and human nestin genes (2, 5, 16), the second intron of the mouse nestin gene confers a spatial and temporal pattern of expression in the developing central nervous system that is similar to that of the endogenous nestin gene.
FIGURE 1.
LacZ expression in transgenic embryos carrying mouse nestin second intron-lacZ constructs. Whole mount lacZ staining was performed using transgenic embryos expressing the pSAStk2In43/1629 construct at E12.5 (A-C) and the pSAStk2In1310/1629 construct at E10.5 (D), E12.5 (E), and E14.5 (F). LacZ was expressed in the spinal cord (sc), myelencephalon (my), metencephalon (mt), mesencephalon (ms), diencephalon (di), and telencephalon (te) (B-F). A is the dorsal view, and B and C are the lateral views of the same embryo. LacZ expression in whisker follicle sheath progenitor cells is indicated by the arrow (C). G and H, the transverse sections of the lumbar spinal cord from pSAStk2In1310/1629 transgenic mice, indicated by lines in the area of D and E, respectively. LacZ expression was restricted to both sides of neural tube (G and H). I, endogenous nestin expression in the neural tube was examined by immunostaining on a transverse section from wild-type mouse embryos at E12.5.
Nestin Expression Is Regulated by the Second Intron during P19 Cell Neural Differentiation—RA-induced P19 cell neural differentiation is a well established in vitro model to study the molecular mechanisms involved in the development of mammalian central nervous system (40-43). This process may be divided into two stages: the induction stage, where P19 cells were aggregated and induced by RA for 4 days, and the differentiation stage, in which the induced P19 cells were replated as single cell suspensions and differentiated for up to 2 weeks (40). In this study, we named the days after aggregation and RA treatment as A1, A2, A3, and A4 and the days after replating as R1, R2, R3, etc. Northern blotting was used to detect nestin expression during neural differentiation of the P19 cells (Fig. 2A, left panel). We found that nestin mRNA was present at low levels in undifferentiated P19 cells (A0), and the expression was up-regulated upon RA induction, with an expression peak at replating day 1 (R1) (Fig. 2A, right panel). As neural differentiation proceeded, nestin expression was down-regulated to the basal level at R9. Consistently, the qRT-PCR analysis also showed a transient increase in nestin mRNA, with an expression peak at R1 (Fig. 2B). These results suggest that the nestin gene has a very similar expression pattern in P19 cell neural differentiation and central nervous system development in vivo (2), suggesting that this process could be used as an in vitro system to study transcriptional regulation of the nestin gene during central nervous system development. Based on this observation, we proposed that the non-induced P19EC cells (A0) represented the pluripotent stem cells and that the replating day 1 or 2 (R1 or R2) RA-induced P19 cells (P19NPCs) represented the neural progenitor cells of the developing central nervous system (Fig. 2B).
To identify the regulatory sequence of the nestin gene responsible for nestin expression during P19 cell neural differentiation, we scanned the genomic sequence of the mouse nestin gene using a luciferase reporter assay in P19EC cells and P19NPCs. The second intron (pNes2In32/1628) increased the luciferase activity by ∼3-fold in P19EC cells, and up to 8-fold in P19NPCs compared with the control pGL3-Px′ vector, whereas the 5′ upstream promoter (pNESP -3809/+183) and the first intron (pNes1In1/1003) failed to show any enhancer activity (Fig. 2, C and D). To confirm the function of the second intron, we generated a pNesPE-Luc construct containing a 4-kb upstream region and the second intron of nestin gene, and established stable P19 cell lines with this construct (NesPEL/P19 cells). Representative clones, such as NesPEL/P19-E, were induced for neural differentiation, and the luciferase activity was measured. The expression of the luciferase reporter also transiently increased along with neural differentiation and had an expression peak at R2, 1 day later than the nestin mRNA peak at R1 (Fig. 2E). Similar results were obtained from other clones of the NesPEL/P19 cells (data not shown). In conclusion, the second intron of mouse nestin gene is responsible for regulation of nestin expression during P19 cell neural differentiation.
Identification of the Minimal Enhancer and cis-Elements—To identify the core sequence(s) in the second intron of the mouse nestin gene, a series of deletion constructs was transiently transfected into P19EC cells and P19NPCs (Fig. 3A). We found that a 320-bp region at the 3′ end of the second intron (pNes2In1310/1629) displayed enhancer activity comparable to that of the full-length second intron (pNes2In32/1629) in both P19EC cells and P19NPCs. The constructs lacking this 320-bp fragment showed a significant loss of enhancer activity in P19EC cells and P19NPCs, confirming the importance of this region to the augmented enhancer activity during neuronal differentiation. We further divided this 320-bp fragment into a 5′-proximal 151-bp fragment (pNes2In1310/1460) and a 3′-proximal 171-bp fragment (pNes2In1459/1629) and found that the upstream 151-bp fragment possessed full enhancer activity in P19EC cells and partial activity in P19NPCs. However, the downstream 171-bp region had no enhancer activity. These results suggest that the 151-bp fragment (1310/1460) contains the majority, but probably not all, of the regulatory elements (Fig. 3A). Some other elements in the 320-bp region may confer either repression or activation activity in P19EC cells or P19NPCs, although these experimental states will not be represented as the physiological state by the reporter vectors (Fig. 3A).
FIGURE 3.
Identification of the core sequence and functional cis-elements in the second intron of the mouse nestin gene. A, the minimal enhancer region was localized to the 3′ 320 bp (1310/1629) region of the second intron. P19EC cells or P19NPCs were transfected with deletion constructs as indicated in the left panel, and the luciferase activity was determined. The minimal enhancer containing 3′ 320 bp is indicated by an asterisk. B, potential cis-elements in the 3′ 320 bp of the second intron. The position of the first nucleotide of the second intron was designated as 1. The consensus sequences of putative cis-elements are shown in boxes. C, alignment of the nucleotide sequence, which contains the binding sites for Sox, POU, HRE, and SF1 factors in the 320-bp region with its counterparts in the rat and human nestin genes. D, identification of functional cis-elements. P19EC or P19NPC cells were transfected with site-mutated constructs as indicated in the left panel, and the luciferase activity was determined. The activity of each construct was shown relative to that of the enhancerless pGL3-Px′ vector. Values were normalized for transfection efficiency by co-transfection with a Renilla luciferase expression plasmid, and they are shown as means ± S.D. for three independent experiments.
To validate the enhancer activity of this 320-bp region in vivo, we generated the pSaStk2In1310/1629 construct and examined the pattern of lacZ expression in the transgenic mice. At E10.5 and E12.5, this 320-bp minimal enhancer could also initiate lacZ expression that was restricted to the proliferating central nervous system progenitor cells (Fig. 1, D and E). Although the enhanced lacZ staining was observed on the 320-bp enhancer transgenic mice versus the full-length second intron enhancer transgenic mice, the central nervous system-specific pattern was the same. As these two lines were produced at different time, the intensity difference may result from difference in the integration of the 320-bp region and the full-length second intron enhancer. For the human or rat nestin enhancers, the conserved regions of the second intron of the nestin genes have been shown to function indistinguishably as the full-length second intron (5, 16). In sections of transgenic embryos at E10.5 and E12.5, intense lacZ expression was observed proximal to the ventricular zone of the neural tube (Fig. 1, G and H), similar to the pattern of endogenous nestin expression (Fig. 1I). At E14.5, weak expression could be detected in relatively confined regions (Fig. 1F), consistent with the observation that most of the NPCs have differentiated into neurons and there is a decrease in nestin expression (2). Therefore, the central nervous system-specific enhancer of mouse nestin gene resides in the 3′ 320-bp region of the second intron.
The 320-bp minimal enhancer contains several putative cis-elements, such as HRE and binding sites for Sox and the POU family transcription factors (24, 27, 44, 45). By carefully examining this sequence, we also found two overlapping binding sites for the steroidogenic factor-1 (SF1, also known as Ad4BP and NR5A1), which are in perfect agreement with the consensus sequence for SF1 (Fig. 3B) (46). It is noteworthy that the binding sites for Sox, POU, HRE, and SF1 factors in the enhancer are conserved among the mouse, rat, and human nestin genes (Fig. 3C). The POU site and HRE from the three species are perfectly conserved (no mismatch). The SOX and SF1 sites are also perfectly conserved between the mouse and the rat nestin genes, either of which has two bases mismatch with the human counterpart. Nevertheless, the sequence of SOX site of the human nestin gene is also a good binding site for SOX (47), and the SF1 site of the human nestin gene has only one base mismatch with SF1 consensus binding sites (46). To further evaluate the importance of these regulatory elements, reporter constructs carrying mutations in the minimal enhancer element were generated and transfected into P19EC and P19NPC cells. As shown in Fig. 3D, mutations in the Sox-binding site (pNes2InSoxm) or POU-binding site (pNes2InPOUm), as well as double mutations in both sites (pNes2InSoxmPOUm), could attenuate luciferase activity in both P19EC cells and P19NPCs. Similar results were obtained when the HRE (pNes2InHREm) was mutated. Interestingly, mutation of SF1-binding sites caused a severe decrease in luciferase activity in P19EC cells, but not in P19NPCs. Taken together, these results demonstrate that Sox- and POU-binding sites, and HRE are important cis-elements for nestin enhancer activity in P19EC and P19NPC cells and are responsible for the activation of the enhancer during neural differentiation, while SF1-binding sites are important in P19EC cells.
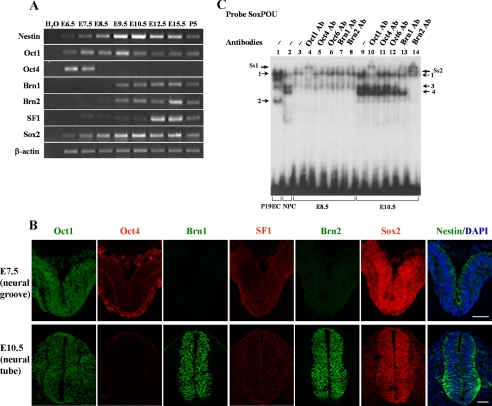
The Expression Pattern of Putative Transcription Factors during P19 Cell Neural Differentiation—To identify the putative transcription factors involved in nestin gene regulation, RT-PCR analysis was used to examine the expression patterns of the SF1, Sox, and POU family members during P19 cell neural differentiation (Fig. 4A). The results showed that SF1 and Oct4 were expressed in the undifferentiated P19EC cells (A0), and the levels of their expression began to decrease immediately after RA induction. The transcripts of Brn1, Brn2, Sox1, and Sox11 could not be detected in P19EC cells; however, the expressions of these genes were induced at A1 or A2, and they continued to be expressed in P19NPCs from R1 to R3. Sox2 was weakly expressed in P19EC cells, and it was transiently up-regulated during neural differentiation, which was similar to the pattern of nestin expression observed. In contrast to the other factors, Oct1 and Sox3 were continuously expressed throughout this process. We could not detect the expression of thyroid transcription factor-1 (TTF1, also known as Nkx2.1) and Brn4 in either undifferentiated or differentiating P19 cells (data not shown).
FIGURE 4.
Expression of SF1, Sox, and POU family transcription factors during neural differentiation of P19 cells. A, RT-PCR analysis of expression of transcription factors during neural differentiation of P19 cells. Total RNA extracted from P19 cells during RA-induced neural differentiation was used for RT-PCR. β-actin was used as a control. B, immunostaining of transcription factors in P19EC and P19NPC cells. For P19NPCs, P19 cells after 4 days of aggregation were dissociated into single cells and replated in N2 medium. At day 2 after replating (R2), the cells were analyzed by immunostaining. Bar, 10 μm.
Immunostaining was used to confirm the expression of putative transcription factors in P19EC cells and P19NPCs (Fig. 4B). Consistent with the RT-PCR results, the patterns of Oct4 and SF1 protein expression in P19EC and P19NPC were observed in the nuclei of P19EC cells, but not in P19NPCs. However, the Brn1 (data not shown) and Brn2 proteins were found in the nuclei of P19NPCs, but not in P19EC cells. The Oct1 and Sox2 proteins were expressed in both P19NPCs and P19EC cells. Taken together, these results suggest that the putative transcription factors expressed in P19EC cells include Oct1, Oct4, Sox2, Sox3, and SF1, and putative transcription factors expressed in P19NPCs are Oct1, Brn1, Brn2, Sox1-3, and Sox11.
The nestin Enhancer Is Occupied by Different Transcription Factors during P19 Cell Neural Differentiation—EMSA was utilized to characterize the transcription factors that potentially bind to the minimal enhancer of the mouse nestin gene. The 1367/1437 region within the 320-bp minimal enhancer, which contains all four cis-elements (Fig. 3B), was divided into two oligonucleotides for EMSA: oligonucleotides 1360-1395 as the Sox-POU probe and 1409-1441 as the HRESF1 probe (Fig. 3B, underlined). A time-course analysis of the proteins that bound to the Sox and POU cis-elements was first performed using the radiolabeled Sox-POU probe with the same quantity of P19 cell NE prepared at different time points of neural differentiation. Poly(dG-dC) was used instead of poly(dI-dC) as a nonspecific DNA competitor to allow binding by high-mobility group box proteins (48). Two prominent complexes (bands 1 and 2) were found in extracts from P19EC cells at A0 (Fig. 5A, lane 1); complex 2 disappeared after A2, while complex 1 became weaker but was still detectable until R5 (Fig. 5A, lanes 3-7). Another two complexes (bands 3 and 4) were obvious at A4, and they peaked by R1 and R3, decreasing and eventually disappearing after R7 (Fig. 5A, lanes 5-9). The complexes 1-2 in P19EC cells and complexes 1, 3, and 4 in P19NPCs were specific to particular DNA sequences since they were successfully competed away with 100-fold molar excess of non-radiolabeled probe (Fig. 5B, lanes 3 and 10) but not with an irrelevant competitor such as CIE (cis-inducible element) (Fig. 5B, lanes 2 and 9). As a control, we also used the POU-binding site in the upstream enhancer of the mouse Sox2 gene, POU1(Sox2) (49), which also competed away these bands in P19EC and P19NPC cells (Fig. 5B, lanes 4 and 11). To identify the important binding sites in this SoxPOU probe, DNA probes with mutations in the Sox (SoxmPOU) or POU sites (SoxPOUm) or mutations at both sites (SoxmPOUm) were used as competitors. The SoxmPOU probe effectively abolished the formation of bands 1-2 in P19EC cells (Fig. 5B, lane 5), as well as bands 1, 3, and 4 in P19NPCs (Fig. 5B, lane 12), whereas the other two probes failed to compete away any band (Fig. 5B, lanes 6 and 7, and 13 and 14). Consistently, the radiolabeled SoxmPOU probe could form complexes with the same mobility as the wild-type probe in P19 cell NE, although no complexes were formed with the radiolabeled SoxPOUm or SoxmPOUm probes (data not shown). These observations suggest that different transcription factors are specifically recruited to POU factor-binding sites in the nestin gene minimal enhancer to generate complexes 1 and 2 in P19EC cells and complexes 1, 3, and 4 in P19NPCs, respectively.
FIGURE 5.
Different transcription factors are recruited to the nestin enhancer during neural differentiation of P19 cells. A, a 32P-labeled SoxPOU probe (1360/1395) was incubated with NE from P19 cells. EMSA was performed with the same amount of NE from P19 cells (5 μg). Specific gel-shifted bands (1-4) formed in P19EC cells and P19NPCs are indicated by arrows. B, 32P-labeled SoxPOU probe was incubated with NE from P19EC cells (lanes 1-7) or P19NPCs (lanes 8-14). The competition experiment was performed with 100-fold excess of unlabeled non-relevant probe CIE (lanes 2 and 9), SoxPOU probe (lanes 3 and 10), POU1(Sox2) probe (lanes 4 and 11), Sox-mutated probe SoxmPOU (lanes 5 and 12), POU-mutated probe SoxPOUm (lanes 6 and 13), or a probe with both the Sox and POU sites mutated probe SoxmPOUm (lane 7 and 14). C, a 32P-labeled SoxPOU probe was incubated with NE from P19EC cells (lanes 1-4) or P19NPCs (lanes 5-7). The supershift assay was performed with an antibody against Oct1 (lane 2), Oct2 (lane 3), Oct4 (lane 4), Brn1 (lane 6), or Brn2 (lane 7). Note that there were supershifted bands in lanes 2 and 7. D, 32P-labeled HRESF1 (1409/1441) probe was incubated with NE from P19 cells. EMSA was performed with the same amount of NE from P19 cells (5 μg). Specific gel-shifted bands (5-7) formed in P19EC cells and P19NPCs are indicated by arrows. E, a 32P-labeled HRESF1 probe was incubated with NE from P19EC cells. The competition experiment was performed with 100-fold excess of unlabeled probe HRESF1 (lane 2) or the non-relevant probe CIE (lane 3). The supershift assay was performed with an antibody against SF1 (lane 4). Note that there were supershifted bands in lanes 4. F, in vivo occupancy of the nestin enhancer by different transcription factors. In ChIP assays, antibodies against Brn1, Brn2, Sox2, or SF1 were used to immunoprecipitate cross-linked chromatin fragments prepared from P19EC cells or P19NPCs. Immunoprecipitates were analyzed for the abundance of the nestin enhancer by quantitative PCR. The data were normalized relative to nestin enhancer occupancy by pre-immune IgG. Ss, supershift.
To identify the transcription factors present in the complexes 1-4, supershift assays were performed (Fig. 5C). In P19EC cells, complex 1 could be supershifted by an Oct1 antibody, and complex 2 was diminished by an Oct4 antibody, while the Oct2 antibody had no effect (Fig. 5C, lanes 2-4). In P19NPCs, formation of complex 3 was abolished by a Brn1 antibody, and complexes 3 and 4 were supershifted by a Brn2 antibody (Fig. 5C, lanes 6 and 7). Complex 4 could be gradually supershifted with an increasing amount of Brn2 antibody when a series dilution of Brn2 antibody was performed (supplemental Fig. S1A, lanes 6-8). To determine whether Sox2 protein was present in these complexes, we compared the complexes formed with the nonspecific DNA competitor poly(dI-dC) (supplemental Fig. S1A, lane 1) versus poly(dG-dC) (supplemental Fig. S1A, lanes 2-9) with a longer exposure time. We found an additional band in the presence of poly(dG-dC) but not poly(dI-dC) (supplemental Fig. S1A, lane 1 versus 2). This complex could be supershifted with an antibody against Sox2 (supplemental Fig. S1A, lane 9). Together, these results indicate that the cis-element for POU factors was bound by Oct1 and Oct4 in P19EC cells, and Oct1, Brn1, and Brn2 in P19NPCs.
A time-course analysis of proteins binding to the HRESF1 probe was also performed (Fig. 5D). There were two bands (complexes 5 and 6) present in P19EC cells (A0), and the intensity of these bands gradually decreased upon neural differentiation. Meanwhile, the third band (complex 7) could be observed from R1 to R3 in P19NPCs. The specificity of the two bands in P19EC cells was confirmed by a competition assay (Fig. 5E, lanes 1-3), and the binding of SF1 protein to this probe was validated by the observation of a supershift with treatment with an SF1 antibody (Fig. 5E, lane 4). These data suggest that two overlapping SF1-binding sites recruit the SF1 protein in P19EC cells.
To further validate the binding of these transcription factors to the endogenous nestin gene enhancer, we immunoprecipitated chromatin from P19EC cells and P19NPCs with antibodies against Brn1, Brn2, Sox2, and SF1 (Fig. 5F). The precipitated chromatin was quantified by quantitative PCR with primers for the nestin enhancer, flanking the HRE-, POU-, Sox-, and SF1-binding sites. We found that the binding of Brn1 and Brn2 to the nestin enhancer was markedly enhanced in P19NPCs when compared with P19EC cells. In contrast, the levels of SF1 binding were high in P19EC cells and low in P19NPCs. The Sox2 protein bound to this enhancer in both P19EC and P19NPC cells. Together, these results suggest that different transcription factors are recruited to the endogenous nestin gene enhancer during the process of P19 cell neural differentiation: Sox2 and SF1 in undifferentiated P19EC cells, and Sox2, Brn1, and Brn2 in P19NPCs.
Different Transcription Factors Bind to the nestin Enhancer at Different Stages of Central Nervous System Development in Vivo—The above results led us to hypothesize that different transcription factors might be recruited to the nestin enhancer to regulate its expression during central nervous system development in vivo. To investigate this, we first examined the expression pattern of transcription factors in developing mouse embryos (Fig. 6). RT-PCR analysis showed that low levels of nestin mRNA were detected in E6.5 embryos, and nestin expression started to increase in E7.5 mouse embryos, with an expression peak in E9.5 and E10.5 mouse brains (Fig. 6A). Oct1 was continuously expressed at all stages examined. Oct4 was expressed at a high level in E6.5 embryos, but down-regulated at E7.5, and it ceased to be expressed from E8.5. In contrast, Brn1 and Brn2 were not expressed in E6.5 and E7.5 embryos, and their expression was only initiated at E9.5. SF1 was not expressed in E6.5 embryos, started to be expressed at a very low level from E7.5 to E10.5, and was up-regulated after E12.5. Sox2 showed a very similar expression pattern to that of nestin, suggesting that Sox2 might regulate nestin transcription at both the neural plate and neural tube stages. Immunostaining was used to detect the pattern of protein expression of these transcription factors at the thoracic spinal level (Fig. 6B). Consistent with the RT-PCR results, the Oct1 protein was expressed in both the E7.5 neural fold and E10.5 neural tube. However, the levels of Oct4 protein expression were very low in the neural fold, and Oct4 expression was completely lost in the neural tube of E10.5 embryos. Brn1 and Brn2 were not expressed in E7.5 embryos, but these proteins were strongly and specifically expressed in E10.5 neural tube. Sox2 was expressed strongly in both the E7.5 neural fold and E10.5 neural tube. In contrast, weak expression of SF1 was detected in the E7.5 neural fold, and its expression was undetectable in the E10.5 neural tube.
FIGURE 6.
Expression of SF1, Sox and POU family transcription factors in developing mouse embryos. A, RT-PCR analysis for the expression of transcription factors. Total RNA extracted from mouse embryos at E6.5 and E7.5, mouse brain at E8.5, E9.5, E10.5, 12.5, and E15.5 or mouse cerebellum at P5 was used as the template for reverse transcription. Specific primers used for PCR amplifications are listed in supplemental Table S1. B, immunostaining of transcription factors was performed on transverse sections from E7.5 neural groove and E10.5 neural tube at the thoracic spinal level. Bar, 50 μm. C, a 32P-labeled SoxPOU probe was incubated with NE from mouse embryos at E8.5 (lanes 3-8) or mouse brain at E10.5 (lanes 9-14). NE from P19EC cells or P19NPCs was used as a control (lanes 1 and 2). The supershift assay was performed with an antibody against Oct1 (lanes 4 and 10), Oct4 (lanes 5 and 11), Oct6 (lanes 6 and 12), Brn1 (lanes 7 and 13), or Brn2 (lanes 8 and 14). Gel-shifted bands are indicated by arrows.
To determine whether these transcription factors could bind to the nestin gene enhancer, we prepared nuclear extracts from E8.5 mouse embryos, E10.5 and E12.5 mouse brain, and performed EMSA with a radiolabeled SoxPOU probe. As shown in Fig. 6C, we could detect one band from E8.5 embryo nuclear extract, and this band could be supershifted by an Oct1 antibody, but not by any other antibodies (Fig. 6C, lanes 3-8). The nuclear extract of E10.5 mouse brain contained three major bands, one slowly migrating band, and two rapidly migrating duplicate bands (Fig. 6C, lane 9). Oct1 antibody could supershift the slowly migrating band, but antibodies against Oct4 and Oct6 had no effect (Fig. 6C, lanes 10-12). A Brn1 antibody supershifted the upper band of the duplex, and treatment with a Brn2 antibody eliminated both bands (Fig. 6C, lanes 13 and 14). Brn1 and Brn2 proteins in E12.5 brain and spinal cord could also bind to the SoxPOU probe (supplemental Fig. S1B). Due to a difficulty in collecting sufficient quantities of nuclear extracts, we were unable to perform the EMSA analysis with E6.5 and E7.5 embryos. Taken together, these results show that different transcription factors were expressed differently in the neural stem cells of the neural fold of E7.5 and the neural tube of E10.5 mouse embryos. These transcription factors were recruited to the nestin enhancer at different stages, Oct1 in the neural epithelium of E8.5 mouse embryos, and Oct1, Brn1, and Brn2 in the neural tube of E10.5 embryos.
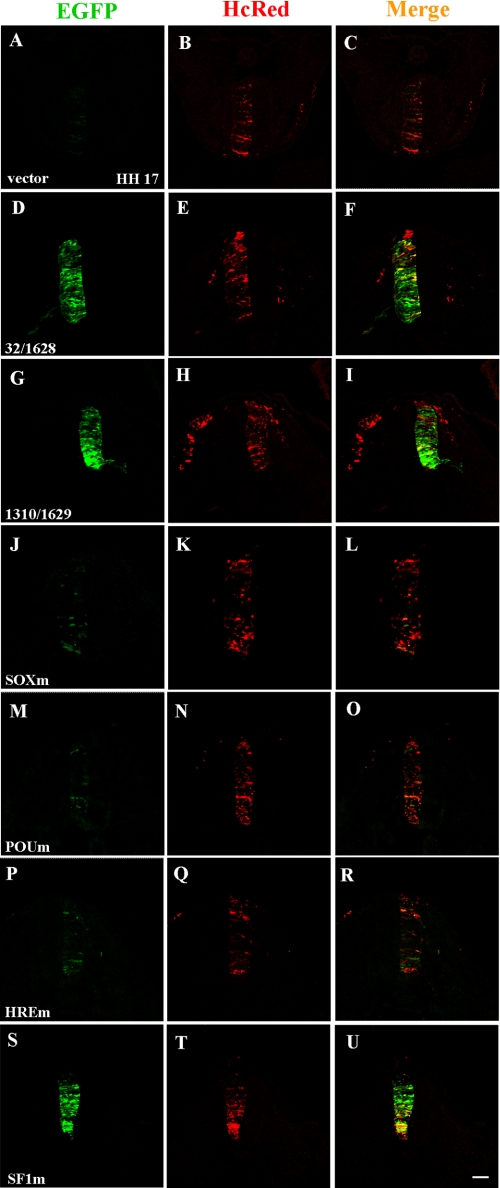
Different cis-Elements within the nestin Enhancer Are Used in Chick Embryo Neural Tube Development—To further confirm the function of different cis-elements in the mouse nestin gene enhancer during central nervous system development in vivo, we examined the enhancer activity in the neural tube of chick embryos. The wild-type and mutated enhancer-driven GFP reporter constructs were electroporated into one side of neural tube of chick embryos at HH stage 10, and GFP expression in the thoracic spinal cord was detected at HH stage 17. The HcRed1 expression vector was used as a tracer. We found that the expression vector itself could not induce GFP expression (Fig. 7, A-C), whereas the wild-type full-length enhancer (Fig. 7, D-F) or 320-bp minimal enhancer (Fig. 7, G-I) could activate strong GFP expression in the neural tube. However, this enhancer could not drive GFP expression in the surrounding somite, in which co-electroporated HcRed1 was expressed (Fig. 7, G versus H), indicating the neural tissue specificity of the nestin enhancer in chick embryos. Mutations of the cis-elements for Sox (Fig. 7, J-L), POU (Fig. 7, M-O), and HRE (Fig. 7, P-R) in the minimal enhancer greatly decreased GFP expression. In contrast, mutation of the SF1-binding sites did not disrupt reporter gene expression (Fig. 7, S-U). These results indicate that the Sox, POU, and HRE cis-elements are essential for nestin enhancer activity in the developing central nervous system, but SF1 sites were not functional in chick embryo neural tubes.
FIGURE 7.
In vivo characterization of the functional cis-elements in the second intron of the mouse nestin gene. The pxtkEGFP vector containing a TK promoter and GFP coding sequence was used as a negative control. The second intron of the mouse nestin gene with wild-type or mutated cis-elements was placed upstream of the TK promoter to generate a series of GFP reporter gene constructs. The GFP constructs were electroporated into chick neural tubes together with the tracer plasmid, pHcRed1-N1, at HH stage 10. For G and H, a GFP construct and the tracer plasmid, pHcRed1-N1, were also injected and electroporated into the surrounding somite. The expressions of GFP and HcRed were detected 24 h later (at HH stage 17). Neural tubes expressing HcRed were selected and cut from chick embryos for cryosection, and the expression of GFP in the thoracic spinal cord was examined under a confocal microscope. The numbers of independent electroporated embryos that expressed GFP and HcRed for different constructs were as follows: 0/10 for the vector pxtkEGFP (A-C), 9/9 for the full-length second intron construct pxtkEGFP2In32/1628 (D-F), 30/31 for the 320-bp minimal enhancer construct pxtkEGFP2In1310/1629 (G-I), 5/21 for the Sox-mutated construct pxtkEGFP2InSOXm (J-L), 0/7 for the POU-mutated construct pxtkEGFP2InPOUm (M-O), 5/19 for the HRE-mutated construct pxtkEGFP2InHREm (P-R), and 13/13 for the SF1-mutated construct pxtkEGFP2InSF1m (S-U). Bar, 50 μm.
DISCUSSION
Nestin is a neural progenitor cell-specific marker, and its expression is strictly regulated during central nervous system development. Studies in transgenic mice showed that the central nervous system-specific enhancer of the human and rat nestin genes resides in the second intron (5, 16). In this study, we confirm that the second intron of the mouse nestin gene also contains a central nervous system-specific enhancer, because it can drive reporter gene expression in the developing central nervous system of transgenic mice (Fig. 1) and in the neural tube of chick embryos (Fig. 7). To reveal the molecular mechanisms of transcriptional regulation of the mouse nestin gene in neural progenitor cells, we took advantage of the neural differentiation program of mouse embryonic carcinoma P19 cells. Analogous to neural induction and neurogenesis in vivo (5, 16), nestin expression showed a transient up-regulation during P19 cell neural differentiation (Fig. 2). nestin expression was at a basal level in undifferentiated P19EC cells; the expression increased along with P19EC cells differentiated into P19NPCs and decreased when NPCs terminally differentiated into neurons and glial cells (Fig. 2). Luciferase assays showed that the second intron of the mouse nestin gene could induce a transient up-regulation of the reporter gene, which was reminiscent of the endogenous nestin expression during P19 cell neural differentiation (Fig. 2E). These results suggested that the neural differentiation of P19 cells could be used to study the function of nestin enhancer and to dissect the detailed mechanisms of nestin gene regulation in vitro and in vivo.
Series deletion analysis led to the identification of a minimal enhancer located in the 3′ 320-bp region of the second intron (Fig. 3A), and this minimal enhancer could also drive reporter gene expression in developing NPCs of transgenic mice (Fig. 1). Transgenic analysis showed that the 3′ 320-bp region in the second intron of the mouse nestin gene harbors a general central nervous system-specific enhancer for the nestin expression, similar to what have been reported in the human and rat nestin genes. By analogy, some regional expression of the mouse nestin gene might require other cis-elements outside of the 320-bp region (24-26). Like the 257-bp region of the rat nestin enhancer and the 120-bp region of the human nestin gene enhancer, this 320-bp region contains HRE-, Sox-, POU-, and SF1-binding sites (24-26). Mutations in all of these sites, except SF1, strongly reduced the enhancer activity of the mouse nestin gene in P19EC cells and P19NPCs (Fig. 3D), indicating that these sites are important to both the basal and central nervous system-specific enhancer activities of mouse nestin gene. Interestingly, mutations in two overlapping SF1-binding sites abolished enhancer activity in P19EC cells, but they had no effect on reporter gene expression in P19NPCs, suggesting that the SF1 sites are involved in basal enhancer activity in undifferentiated EC cells, but not in central nervous system-specific enhancer activity in NPCs. Consistently, chick embryo in ovo electroporation also showed that mutations in HRE, Sox, and POU sites caused a significant decrease in reporter gene expression, whereas mutations in the SF1-binding sites had no effect (Fig. 7). The central nervous system-specific enhancer with mutations in the SF1-binding sites could still drive reporter gene expression in the NPCs of E12.5 transgenic mice embryos.4 These results support the notion that the functions of cis-elements in the central nervous system-specific enhancer are the same in P19NPCs as in the developing central nervous system in vivo.
The EMSA time-course analysis showed that different protein-DNA-binding complexes were formed for the Sox-POU and HRE-SF1 probes during P19 cell neural differentiation (Fig. 5). A reasonable hypothesis is that different transcription factors expressed at different stages of neural differentiation bind to these cis-elements to regulate nestin gene expression. To address this possibility, we performed supershift assays and found that oligonucleotides containing Sox- and POU-binding sites were bound by Oct1 and Oct4 proteins in P19EC cells but by the Oct1, Brn1, and Brn2 factors in P19NPCs (Fig. 5). The observation that the same POU-binding site is occupied by different POU factors at different differentiation stages is very interesting. The Sox2 gene also showed a similar regulation mechanism, where its upstream promoter was bound by the Oct4 protein in embryonic stem cells and by Brn1 and Brn2 in NPCs (49). The probe that contains the HRE- and SF1-binding sites was bound by SF1 and unidentified protein(s) in P19EC cells and by different protein(s) in P19NPCs (Fig. 5). It has been reported that a variety of nuclear hormone receptors are able to bind to the nestin enhancer, including the thyroid hormone receptor, retinoic acid receptor, retinoid X receptor, and chicken ovalbumin upstream promoter-transcription factor, and these receptors may have positive or negative effects upon nestin expression (25). Consistent with the EMSA results, ChIP assays confirmed binding of the Sox2 and SF1 proteins to the nestin enhancer in P19EC cells and also confirmed binding of Brn1, Brn2, and Sox2 in P19NPCs (Fig. 5). However, we were unable to detect any Sox2 binding to the SoxPOU probe in the EMSA assay, and the ChIP analysis did not indicate that the Oct1 and Oct4 proteins were recruited to the nestin enhancer. One possible explanation for the absence of Sox2 binding may be that our EMSA conditions do not favor the formation of the Sox2-DNA complex, even though poly(dG-dC) was used instead of poly(dI-dC) (48).
Our observations suggest that regulation of the expression of the mouse nestin gene during P19 cell neural differentiation may recapitulate the events in the developing central nervous system in vivo. To test this possibility, we examined the expression pattern of putative transcription factors during the neural development of mouse embryos and found that the expression patterns of transcription factors were very similar to those observed during the neural differentiation of P19 cells (Figs. 4 and 6). EMSA assays also showed that the nestin enhancer could be bound by different transcription factors at different developmental stages, e.g. Oct1 in E8.5 and Oct1, Brn1, and Brn2 in E10.5 and E12.5 mouse embryos (Fig. 6 and supplemental Fig. S1B). Consistently, Miyagi et al. showed the binding of Brn1, Brn2, and Sox2 to the nestin enhancer in E12.5 brain and spinal cord using ChIP assays (50). Difficulties obtaining sufficient quantities of tissue prevented us from performing EMSA and ChIP assays from early mouse embryo stages, because our efforts to detect transcription factor occupancy at these stages of development were not successful.
Cultured cell lines often resemble particular cell types found in part of the embryonic anatomy at a certain developmental stage. P19EC cells are derived from a teratocarcinoma formed after transplantation of a 7.5 days post coitum embryo into testis (51), and they have a number of features in common with cells of the epiblast (52). Interestingly, an Oct4/lacZ transgenic study revealed two regulatory elements in the Oct4 gene: one drives Oct4 expression in preimplantation embryos and is specifically active in embryonic stem and embryonic germ cells; the other directs the epiblast-specific expression pattern, and its activity is restricted to P19EC cells (53). Consistently, we found that P19EC cells express epiblast marker FGF5, and a high population of P19EC cells could be converted into neural stem cells in N2B27 serum-free medium. We further showed that N2B27-induced neural stem cells bear anterior neuroectoderm characters and could be efficiently caudalized by RA (54). Therefore, the P19NPCs generated from RA-treated P19EC cells likely are the neural progenitors of the caudal nervous system, i.e. the hindbrain and spinal cord.
It has been proposed previously that a change in POU proteins expression is required for the acquisition of neural potential (24). For the nestin gene, which is a molecular marker for neural progenitors, we provide evidence here that the cis-element for POU factors was bound by Oct1 and Oct4 in P19EC cells, and Oct1, Brn1, and Brn2 in P19NPCs. The observation that a same DNA regulatory element could be bound by different members of POU family strongly supports the idea that a switch for POU proteins works in controlling gene regulation. A recent study showed that nestin is regulated in a cell-cycle-dependent manner: the cell-cycle-dependent phosphorylation of an upstream regulator, Brn2, reduces its binding activity to the nestin core enhancer element and is responsible for the decreased nestin transcription in G2-M phase (55). This line of evidence suggests a scenario that the absence of nestin expression in postmitotic neurons may simply be due to the lack of Brn2 activity.
In conclusion, we identified a central nervous system-specific enhancer in the 3′ region of the second intron of the mouse nestin gene. This region could drive reporter gene expression in the NPCs of the developing nervous system of transgenic mice and regulate nestin expression during P19 cell neural differentiation. The HRE, Sox, and POU cis-elements in this enhancer are important for nestin gene expression in NPCs in vitro and in vivo. Different transcription factors regulate nestin gene expression during P19 cell neural differentiation and central nervous system development. Sox2 and SF1 may mediate basal nestin expression in undifferentiated P19EC cells, whereas Sox2, Brn1, and Brn2 are recruited to maintain nestin expression in P19NPCs. Different factors were recruited to the nestin enhancer at different stages of mouse embryo development, Oct1 in E8.5 neural fold and Oct1, Brn1 and Brn2 in the neural tube at E10.5. Our studies therefore provide evidence that a temporal combination of transcription factors bind to the central nervous system-specific enhancer of nestin gene and control its expression.
Supplementary Material
Acknowledgments
We thank Dr. U. Lendahl for pSaStk/lacZ vector, Dr. H. Kondoh for ptkEGFP vector and Sox2 antibody, and Dr. K. Morohashi for the SF1 antibody.
This work was supported by the National Natural Science Foundation of China (Grants 30623003, 30721065, and 30830034 to N. J.), the National Key Basic Research and Development Program of China (Grants 2005CB522704, 2006CB943902, 2007CB947101, 2008KR0695, and 2009CB941100 to N. J.; Grants 2006CB943900 and 2007CB947102 to L. C.), the National High-Tech Research and Development Program of China (Grant 2006AA02Z186 to N. J.), the Shanghai Key Project of Basic Science Research (Grants 06DJ14001, 06DZ22032, and 08DJ1400501 to N. J.), and the Council of Shanghai Municipal Government for Science and Technology (Grants 05814578 to N. J. and 07pj14098 to L. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
Footnotes
The abbreviations used are: E, embryonic day; HRE, hormone-responsive element; EC, embryonic carcinoma; NPC, neural progenitor cell; POU, pit, oct, and unc; TK, thymidine kinase; RA, retinoic acid; EMSA, electrophoretic mobility shift assay; NE, nuclear extract; ChIP, chromatin immunoprecipitation; GFP, green fluorescent protein; RT, reverse transcription; qRT, quantitative RT; SF1, steroidogenic factor-1.
Z. Jin and N. Jing, unpublished observation.
References
- 1.Itoh, K., and Sokol, S. Y. (2007) in Principles of Developmental Genetics (Moody, S. A., ed) pp. 241-257, Elsevier Inc., Academic Press, Burlington, MA
- 2.Dahlstrand, J., Lardelli, M., and Lendahl, U. (1995) Brain Res. Brain Res. Rev. 84 109-129 [DOI] [PubMed] [Google Scholar]
- 3.Lendahl, U., Zimmerman, L. B., and McKay, R. D. (1990) Cell 60 585-595 [DOI] [PubMed] [Google Scholar]
- 4.Wiese, C., Rolletschek, A., Kania, G., Blyszczuk, P., Tarasov, K. V., Tarasova, Y., Wersto, R. P., Boheler, K. R., and Wobus, A. M. (2004) Cell. Mol. Life Sci. 61 2510-2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmerman, L., Parr, B., Lendahl, U., Cunningham, M., McKay, R., Gavin, B., Mann, J., Vassileva, G., and McMahon, A. (1994) Neuron 12 11-24 [DOI] [PubMed] [Google Scholar]
- 6.Frederiksen, K., and McKay, R. D. (1988) J. Neurosci. 8 1144-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu, H., Wang, S., Messam, C. A., and Yao, Z. (2002) Brain Res. 943 174-180 [DOI] [PubMed] [Google Scholar]
- 8.Dahlstrand, J., Collins, V. P., and Lendahl, U. (1992) Cancer Res. 52 5334-5341 [PubMed] [Google Scholar]
- 9.Frisen, J., Johansson, C. B., Torok, C., Risling, M., and Lendahl, U. (1995) J. Cell Biol. 131 453-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenka, N., Lu, Z. J., Sasse, P., Hescheler, J., and Fleischmann, B. K. (2002) J. Cell Sci. 115, 1471-1485 [DOI] [PubMed] [Google Scholar]
- 11.Smukler, S. R., Runciman, S. B., Xu, S., and van der Kooy, D. (2006) J. Cell Biol. 172 79-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin, Z. G., Liu, L., Zhong, H., Zhang, K. J., Chen, Y. F., Bian, W., Cheng, L. P., and Jing, N. H. (2006) Acta Biochim. Biophys. Sin. 38 207-212 [DOI] [PubMed] [Google Scholar]
- 13.Chou, Y. H., Khuon, S., Herrmann, H., and Goldman, R. D. (2003) Mol. Biol. Cell 14 1468-1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahlgren, C. M., Mikhailov, A., Vaittinen, S., Pallari, H. M., Kalimo, H., Pant, H. C., and Eriksson, J. E. (2003) Mol. Cell. Biol. 23 5090-5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakamatsu, Y., Nakamura, N., Lee, J. A., Cole, G. J., and Osumi, N. (2007) Development (Camb.) 134 2425-2433 [DOI] [PubMed] [Google Scholar]
- 16.Lothian, C., and Lendahl, U. (1997) Eur. J. Neurosci. 9 452-462 [DOI] [PubMed] [Google Scholar]
- 17.Dubois, N. C., Hofmann, D., Kaloulis, K., Bishop, J. M., and Trumpp, A. (2006) Genesis 44 355-360 [DOI] [PubMed] [Google Scholar]
- 18.Isaka, F., Ishibashi, M., Taki, W., Hashimoto, N., Nakanishi, S., and Kageyama, R. (1999) Eur. J. Neurosci. 11 2582-2588 [DOI] [PubMed] [Google Scholar]
- 19.Petersen, P. H., Zou, K., Hwang, J. K., Jan, Y. N., and Zhong, W. (2002) Nature 419 929-934 [DOI] [PubMed] [Google Scholar]
- 20.Tronche, F., Kellendonk, C., Kretz, O., Gass, P., Anlag, K., Orban, P. C., Bock, R., Klein, R., and Schutz, G. (1999) Nat. Genet. 23 99-103 [DOI] [PubMed] [Google Scholar]
- 21.Kawaguchi, A., Miyata, T., Sawamoto, K., Takashita, N., Murayama, A., Akamatsu, W., Ogawa, M., Okabe, M., Tano, Y., Goldman, S. A., and Okano, H. (2001) Mol. Cell. Neurosci. 17 259-273 [DOI] [PubMed] [Google Scholar]
- 22.Mignone, J. L., Kukekov, V., Chiang, A. S., Steindler, D., and Enikolopov, G. (2004) J. Comp. Neurol. 469 311-324 [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi, M., Saito, H., Suzuki, M., and Mori, K. (2000) Neuroreport 11 1991-1996 [DOI] [PubMed] [Google Scholar]
- 24.Josephson, R., Muller, T., Pickel, J., Okabe, S., Reynolds, K., Turner, P. A., Zimmer, A., and McKay, R. D. (1998) Development (Camb.) 125 3087-3100 [DOI] [PubMed] [Google Scholar]
- 25.Lothian, C., Prakash, N., Lendahl, U., and Wahlstrom, G. M. (1999) Exp. Cell Res. 248 509-519 [DOI] [PubMed] [Google Scholar]
- 26.Yaworsky, P. J., and Kappen, C. (1999) Dev. Biol. 205 309-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka, S., Kamachi, Y., Tanouchi, A., Hamada, H., Jing, N., and Kondoh, H. (2004) Mol. Cell. Biol. 24 8834-8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, J., Cheng, L., Yan, Y., Bian, W., Tomooka, Y., Shiurba, R., and Jing, N. (2001) Biochim. Biophys. Acta 1520 251-254 [DOI] [PubMed] [Google Scholar]
- 29.Yang, J., Bian, W., Gao, X., Chen, L., and Jing, N. (2000) Mech. Dev. 94 287-291 [DOI] [PubMed] [Google Scholar]
- 30.Cheng, L. P., Yang, J., and Jing, N. H. (2000) Acta Biochim. Biophys. Sin. 32 649-65212058181 [Google Scholar]
- 31.Cheng, L., Jin, Z., Liu, L., Yan, Y., Li, T., Zhu, X., and Jing, N. (2004) FEBS Lett. 565 195-202 [DOI] [PubMed] [Google Scholar]
- 32.Hou, Q., Gao, X., Zhang, X., Kong, L., Wang, X., Bian, W., Tu, Y., Jin, M., Zhao, G., Li, B., Jing, N., and Yu, L. (2004) Eur. J. Neurosci. 20 1593-1603 [DOI] [PubMed] [Google Scholar]
- 33.Uchikawa, M., Ishida, Y., Takemoto, T., Kamachi, Y., and Kondoh, H. (2003) Dev. Cell 4 509-519 [DOI] [PubMed] [Google Scholar]
- 34.Gao, X., Bian, W., Yang, J., Tang, K., Kitani, H., Atsumi, T., and Jing, N. (2001) Biochem. Biophys. Res. Commun. 284 1098-1103 [DOI] [PubMed] [Google Scholar]
- 35.Tang, K., Yang, J., Gao, X., Wang, C., Liu, L., Kitani, H., Atsumi, T., and Jing, N. (2002) Biochem. Biophys. Res. Commun. 293 167-173 [DOI] [PubMed] [Google Scholar]
- 36.Yan, Y., Bian, W., Xie, Z., Cao, X., Le Roux, I., Guillemot, F., and Jing, N. (2004) Dev. Dyn. 231 248-257 [DOI] [PubMed] [Google Scholar]
- 37.Hamburger, V., and Hamilton, H. L. (1951) J. Morphol. 88 49-92 [PubMed] [Google Scholar]
- 38.Leers-Sucheta, S., Morohashi, K., Mason, J. I., and Melner, M. H. (1997) J. Biol. Chem. 272 7960-7967 [DOI] [PubMed] [Google Scholar]
- 39.Tomooka, Y., Kitani, H., Jing, N., Matsushima, M., and Sakakura, T. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 9683-9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bain, G., Ray, W. J., Yao, M., and Gottlieb, D. I. (1994) BioEssays 16 343-348 [DOI] [PubMed] [Google Scholar]
- 41.Wang, C., Xia, C., Bian, W., Liu, L., Lin, W., Chen, Y. G., Ang, S. L., and Jing, N. (2006) Mol. Biol. Cell 17 3075-3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai, G., Sheng, N., Xie, Z., Bian, W., Yokota, Y., Benezra, R., Kageyama, R., Guillemot, F., and Jing, N. (2007) Dev. Cell 13 283-297 [DOI] [PubMed] [Google Scholar]
- 43.Ding, S., Wu, T. Y., Brinker, A., Peters, E. C., Hur, W., Gray, N. S., and Schultz, P. G. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7632-7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lonigro, R., Donnini, D., Zappia, E., Damante, G., Bianchi, M. E., and Guazzi, S. (2001) J. Biol. Chem. 276 47807-47813 [DOI] [PubMed] [Google Scholar]
- 45.Pelizzoli, R., Tacchetti, C., Luzzi, P., Strangio, A., Bellese, G., Zappia, E., and Guazzi, S. (2008) Int. J. Dev. Biol. 52 55-62 [DOI] [PubMed] [Google Scholar]
- 46.Barnea, E., and Bergman, Y. (2000) J. Biol. Chem. 275 6608-6619 [DOI] [PubMed] [Google Scholar]
- 47.Kamachi, Y., Uchikawa, M., and Kondoh, H. (2000) Trends Genet. 16 182-187 [DOI] [PubMed] [Google Scholar]
- 48.Dailey, L., Yuan, H., and Basilico, C. (1994) Mol. Cell. Biol. 14 7758-7769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Catena, R., Tiveron, C., Ronchi, A., Porta, S., Ferri, A., Tatangelo, L., Cavallaro, M., Favaro, R., Ottolenghi, S., Reinbold, R., Scholer, H., and Nicolis, S. K. (2004) J. Biol. Chem. 279 41846-41857 [DOI] [PubMed] [Google Scholar]
- 50.Miyagi, S., Nishimoto, M., Saito, T., Ninomiya, M., Sawamoto, K., Okano, H., Muramatsu, M., Oguro, H., Iwama, A., and Okuda, A. (2006) J. Biol. Chem. 281 13374-13381 [DOI] [PubMed] [Google Scholar]
- 51.McBurney, M. W., and Rogers, B. J. (1982) Dev. Biol. 89 503-508 [DOI] [PubMed] [Google Scholar]
- 52.Robertson, E. J. (1987) Teratocarcinomas and Embryonic Stem Cells: A Practical Approach, IRL Press, Oxford/Washington DC
- 53.Yeom, Y. I., Fuhrmann, G., Ovitt, C. E., Brehm, A., Ohbo, K., Gross, M., Hubner, K., and Scholer, H. R. (1996) Development (Camb.) 122 881-894 [DOI] [PubMed] [Google Scholar]
- 54.Xia, C., Wang, C., Zhang, K., Qian, C., and Jing, N. (2007) Differentiation 75 912-927 [DOI] [PubMed] [Google Scholar]
- 55.Sunabori, T., Tokunaga, A., Nagai, T., Sawamoto, K., Okabe, M., Miyawaki, A., Matsuzaki, Y., Miyata, T., and Okano, H. (2008) J. Cell Sci. 121, 1204-1212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.