Abstract
Pneumocystis pneumonia (PcP) is the most common opportunistic disease in immunocompromised patients. Alveolar macrophages are responsible for the clearance of Pneumocystis organisms; however, they undergo a high rate of apoptosis during PcP due to increased intracellular polyamine levels. In this study, the sources of polyamines and mechanisms of polyamine increase and polyamine-induced apoptosis were investigated. The level of ornithine decarboxylase (ODC) was elevated in alveolar macrophages, and the number of alveolar macrophages that took up exogenous polyamines was increased 20-fold during PcP. Monocytes, B lymphocytes, and CD8+ T lymphocytes that were recruited into the lung during PcP expressed high levels of ornithine decarboxylase, suggesting that these cells are sources of polyamines. Both protein and mRNA levels of antizyme inhibitor (AZI) were increased in alveolar macrophages during PcP. This AZI overexpression correlated with increased polyamine uptake by alveolar macrophages, because AZI expression knockdown decreased the polyamine uptake ability of these cells. AZI expression knockdown also decreased the apoptosis rate of alveolar macrophages. Pneumocystis organisms and zymosan A were found to induce AZI overexpression in alveolar macrophages, suggesting that β-glucan, which is the major component of the Pneumocystis cell wall, induces AZI overexpression. The levels of mRNA, protein, and activity of polyamine oxidase were increased in alveolar macrophages during PcP, indicating that the H2O2 generated during polyamine catabolism caused alveolar macrophages to undergo apoptosis. Taken together, results of this study indicate that Pneumocystis organisms induce AZI overexpression in alveolar macrophages, leading to increased polyamine synthesis and uptake and apoptosis rate of these cells.
Pneumocystis is an opportunistic pathogen; it causes Pneumocystis pneumonia (PcP)3 in immunocompromised individuals, especially in patients with AIDS. Advanced PcP is characterized by infiltration of inflammatory cells in the lung, thickened alveolar septa, and foamy exudates that fill the alveoli. Although host inflammatory responses are important in the defense against Pneumocystis infection, they may also cause lung injury leading to impaired gas exchange and respiratory failure. CD4+ T lymphocytes are crucial in the defense against Pneumocystis infection, because a severe decrease in the number of CD4+ T lymphocytes predisposes patients to the infection (1). Therefore, prophylaxis with a combination of trimethoprim and sulfamethoxazole (TMP-SMX) is usually given to patients with fewer than 200 CD4+ T lymphocytes per microliter in the blood. TMP-SMX interrupts folate syntheses in Pneumocystis organisms and is also the most common regimen for treatment of PcP. Unfortunately, TMP-SMX-resistant Pneumocystis organisms have emerged due to mutations in the dihydropteroate synthase gene (2-4), and some PcP patients may fail TMP-SMX therapy (3).
The alveolar macrophage is the major cell type responsible for the clearance of Pneumocystis organisms (5). Several macrophage receptors are involved in recognition of the organism. Mannose receptor and Dectin-1 have been shown to mediate phagocytosis of Pneumocystis organisms (6, 7), whereas the interaction between TLR2 and Pneumocystis triggers NF-κB activation, leading to increased production of TNF-α and MIP-2 (8). However, the number of alveolar macrophages is decreased during PcP in humans (9) and animals (10, 11). This defect is at least partly due to increased rate of apoptosis of alveolar macrophages during PcP (11-13). We have found that the levels of polyamines, including spermidine, N1-acetylspermidine, and N1-acetylspermine are greatly increased in alveolar macrophages and in the alveoli during PcP (11). We have also found that bronchoalveolar lavage fluid (BALF) obtained from Pneumocystis-infected animals can induce normal macrophages to undergo apoptosis and that polyamine levels in BALF from animals with PcP are high (11). This BALF-induced alveolar macrophage apoptosis is diminished when the polyamines in the BALF are depleted and regained when polyamines are added back to the polyamine-depleted BALF (11). These data indicate that polyamines induce alveolar macrophage to undergo apoptosis during PcP.
Polyamines are polycationic amines present in all cells. The most common biological polyamines are putrescine, spermidine, and spermine. Ornithine decarboxylase (ODC) is the key enzyme in polyamine biosyntheses. ODC decarboxylates ornithine to putrescine, which is converted to spermidine by spermidine synthase (SRM). Spermidine is then converted to spermine by spermine synthase (SMS). Polyamine catabolism is activated in response to the increased intracellular polyamine levels to maintain the cellular polyamine homeostasis (14). This catabolism is mediated by spermidine/spermine N1-acetyltransferase (SSAT), polyamine oxidase (PAO), and spermine oxidase (SMO). SSAT acetylates spermidine to N1-acetylspermidine and spermine to N1-acetylspermine. N1-Acetylspermine and N1-acetylspermidine are excreted from cells or converted back to spermidine and putrescine, respectively, by PAO, whereas spermine can be back converted to spermidine by SMO. The catabolic reactions of PAO and SMO are accompanied by the generation of H2O2, a reactive oxygen species that can induce apoptosis (15).
Polyamines are involved in the regulation of many cellular functions, such as cell cycle progression (16), differentiation (17), oncogenesis (18), and apoptosis (19). Polyamines mediate G1 to S phase transition in the cell cycle by increasing the expression of cyclin A, which binds to cyclin-dependent kinase 2. The cyclin A-cyclin-dependent kinase 2 complex increases the rate of DNA synthesis during cell proliferation (16). A number of studies have demonstrated the essential role of polyamine in cell differentiation, but the mechanisms are not clear (17, 20, 21). The role of polyamines in cancer formation has been extensively studied, and a number of polyamine-mediated oncogenesis mechanisms have been reported (18, 22). For example, the oncoprotein c-Myc activates the expression of ODC, which produces polyamines to stimulate cell proliferation (22). Another oncoprotein, K-ras, inhibits polyamine catabolism by suppressing the transcription of SSAT, resulting in the accumulation of intracellular polyamines that cause increased cell proliferation and development of neoplasia (18).
In addition to transcriptional activation by c-Myc, ODC activity is also regulated at the level of protein degradation. ODC antizyme (OAZ) binds to and promotes transport of ODC to the 26 S proteasome for degradation by a ubiquitin-independent mechanism. In contrast, AZI prevents ODC from degradation by binding to OAZ. OAZ and AZI also regulate polyamine transport. Overexpression of OAZ decreases polyamine import (23), whereas AZI overexpression increases polyamine import (24). The mechanisms of OAZ- and AZI-mediated polyamine transport are still unknown.
In this study, we investigated the host sources of polyamines during PcP. The mechanisms by which polyamine levels are increased in alveolar macrophages were also examined. The results showed that inflammatory cells present in the lung during PcP are sources of polyamines and that the increase in intracellular polyamine levels in alveolar macrophages are due to both de novo synthesis of polyamines and increased uptake of exogenous polyamines.
EXPERIMENTAL PROCEDURES
Rat PcP Model—Pneumocystis carinii infection in rats was established as described previously (25). Briefly, female Sprague-Dawley rats (Harlan, Indianapolis, IN) of 120-140 g were divided into three groups designated Normal, Dex, and Dex-Pc. Normal rats were immunocompetent and uninfected. Dex rats were treated continuously with 1.8 mg/liter dexamethasone in drinking water to reduce the number of CD4+ T lymphocytes, mimicking the immunosuppression status in AIDS patients. Dex-Pc rats were immunosuppressed with dexamethasone and transtracheally inoculated with 7.5 × 106 Pneumocystis trophozoites 1 week after initiation of immunosuppression. To prevent other opportunistic infections, 10,000 units of penicillin (Butler, Dublin, OH) was given intramuscularly each week to each Dex and Dex-Pc rat. This antibiotic had no adverse effects on the growth of Pneumocystis organisms in infected rats. It took ∼8 weeks for Dex-Pc rats to develop severe Pneumocystis pneumonia after inoculation. When Dex-Pc rats were agonal, they were sacrificed for experiments. Age-matched Normal rats were used as controls, while age-matched Dex rats were used to control for the effects of the steroid treatment. Giemsa staining of lung impression smears was performed to determine the existence of Pneumocystis and other microorganisms. Dex-Pc lungs were excluded if they contained other microorganisms. All animal studies were approved by the Indiana University Animal Care and Use Committee and supervised by veterinarians.
Isolation of Alveolar Macrophages—To perform bronchoalveolar lavage, rats were first anesthetized by intramuscular injection of a 0.1-ml ketamine mixture (80 mg/ml ketamine hydrochloride, 0.38 mg/ml atropine, and 1.76 mg/ml acepromazine) and then sacrificed. The thoracic cavity and trachea were exposed by dissection. BALF was obtained by instilling 5 ml of sterile phosphate-buffered saline (PBS) into rat lungs via a 14-gauge angiocatheter (BD Biosciences, Bedford, MA) and then recovering the fluid. In experiments using BALF as supplement in cell cultures, Dulbecco's modified Eagle's medium (DMEM) instead of PBS was used for lavage. To isolate alveolar macrophages, multiple lavages were performed until a total of 50 ml of BALF was obtained. The cells in this 50-ml BALF were pelleted by centrifugation at 300 × g for 10 min and then resuspended in 5 ml of DMEM. Alveolar macrophages were isolated by adherence on plastic tissue culture dishes at 37 °C with 5% CO2 for 1 h followed by washing with warm PBS three times to remove unattached cells. The purity of alveolar macrophages was >97% as determined by anti-reactive macrophage antigen staining described previously (10, 11).
Real-time RT-PCR—Total RNA was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Reverse transcription was done using the iScript cDNA synthesis kit (Bio-Rad) with a thermal condition of 25 °C for 2 min, 42 °C for 30 min, and 85 °C for 5 min. RNA (0.2 μg) from each sample was reverse transcribed to cDNA, and 2 μl of each cDNA product was used as the template for real-time PCR. Real-time PCR analyses for expression of rat ODC, SSAT, SRM, OAZ, and AZI genes were performed using the Assays-on-Demand gene expression kits (Applied Biosystems, Foster City, CA); each of which contained two primers and a FAM-labeled probe. The mRNA levels of the ribosomal protein S8 gene was determined in an identical manner to serve as the internal control, because its expression is not affected by immunosuppression or infection (8). The primers and probe used for the rat ribosomal protein S8 real-time RT-PCR were described previously (8). Real-time PCR reactions were performed with a TaqMan Universal PCR Master Mix (Applied Biosystems) on a Smartcycler (Cepheid, Sunnyvale, CA) using the following temperature cycling conditions: 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 92 °C for 15 s and 60 °C for 1 min.
Western Blotting—Alveolar macrophages adhered in one well of a 6-well plate (∼2 × 106 cells) were scraped in 100 μl of cold (4 °C) cell lysis buffer (150 mm NaCl, 1.0% Triton X-100, 1% deoxycholate, 5 mm EDTA, 10 mm Tris, pH 7.2) containing 1% protease inhibitor mixture (Sigma #P8340). Protein samples for Western blot analysis were prepared by adding NuPAGE LDS Sample Buffer (Invitrogen) and NuPAGE Sample Reducing Agent (Invitrogen) to the cell lysates to a final concentration of 1 × of each reagent and then boiling for 5 min. Proteins in each sample were electrophoresed in a 10% polyacrylamide gel and then transferred onto a polyvinylidene difluoride membrane using the NuPAGE System (Invitrogen). Membranes were blocked by soaking in a buffer containing 3% bovine serum albumin, 0.9% NaCl, 100 mm Tris-HCl (pH 7.6) at 25 °C for 1 h and then reacted with the primary antibody anti-ODC (Lab Vision, Fremont, CA), anti-SSAT (Novus Biologicals, Littleton, CO), anti-OAZ (26, 27), anti-AZI (28), anti-cleaved or activated caspase 3 (Cell Signaling, Danvers, MA), or anti-glyceraldehyde-3-phosphate dehydrogenase (Research Diagnostics, Flanders, NJ) in blocking buffer for 2 h. After washing with TBST (100 mm Tris-HCl, 0.9% NaCl, 0.1% Tween 20, pH 7.6) six times, the blots were reacted with a secondary antibody anti-mouse IgG or anti-rabbit IgG conjugated with horseradish peroxidase (HRP) for 1 h at 25 °C followed again by six washes with TBST. The reaction signals on the blots were developed by chemiluminescence with the ECL Plus reagent kit (Amersham Biosciences) and revealed by exposing the blots to an x-ray film. Densitometry analysis of results on x-ray films was performed by using National Institutes of Health ImageJ (rsb.info.nih.gov/ij/).
Fluorescence Labeling of Spermidine—Labeling of spermidine with fluorescein was performed as previously described (29) with modifications. Due to poor stability in solution, the fluorophore 5-(and-6)-carboxyl fluorescein succinimidyl ester (FAM) (Invitrogen) was freshly prepared in DMSO at a final concentration of 100 mm immediately before use. Spermidine was dissolved in 200 mm NaHCO3 (pH 8.3) to a final concentration of 2 mm. The labeling reaction was performed by mixing 20 μl of FAM with 1 ml of spermidine in a 2-ml Eppendorf tube. The tube was fastened on a vortexer and shaken for 1 h at room temperature. The labeled products were separated by electrophoresis in a 1% agarose gel in 40 mm 2-(N-morpholino)ethanesulfonic acid monohydrate buffer (pH 6.0) at 100 V for 1 h and then visualized by UV transillumination. The FAM-labeled spermidine (FAM-SPD) moved toward the anode and exhibited an extra band in the gel as compared with the sample containing FAM only. The portion of the gel containing FAM-SPD or FAM was excised, chopped to small cubes, and placed at -80 °C until completely frozen. FAM-SPD was recovered by centrifugation at 10,000 × g for 2 h at 4 °C. Additional centrifugations were performed if the gel was not completely thawed. The concentration of recovered FAM-SPD was determined by spectrophotometry at 494 nm using known concentrations of FAM as standards. The yield of FAM-SPD was ≥30 μm. Purified FAM-SPD was stored in small aliquots at -80 °C until use.
Polyamine Uptake Assay—For in vivo polyamine uptake assay, each Normal, Dex, and Dex-Pc rat was anesthetized with 100 μl of ketamine mixture intramuscularly and then transtracheally instilled with 200 μl of 5 μm FAM-SPD or FAM. Alveolar macrophages were then isolated as described above 1 h after FAM-SPD or FAM instillation. The cells were mounted on slides in a mounting medium containing 4′,6-diamidino-2-phenylindole to stain the nuclei. The macrophages that took up FAM-SPD were detected and enumerated by fluorescence microscopy. To examine polyamine uptake in Pneumocystis-infected lungs, frozen lung sections of FAM-SPD-instilled rats were used. The lungs were first lavaged with 2 ml of PBS three times to remove free FAM-SPD or FAM and then instilled with 2 ml of OCT compound (Sakura, Tokyo, Japan) via a 14-gauge angiocatheter. Each lung was then placed in a base mold filled with OCT and frozen at -80 °C. The frozen lungs were sectioned with a cryostat microtome, and the sections were stored at -80 °C until use. Frozen lung sections were air-dried for 15 s and fixed in acetone for 2 min before staining. After an incubation in 5% bovine serum albumin for 1 h, the lung sections were reacted with phycoerythrin (PE)-conjugated anti-CD68 antibody in 5% bovine serum albumin in PBS for 1 h to stain macrophages followed by three washes with PBS. The nuclei were counterstained with 4′,6-diamidino-2-phenylindole, and the stained slides were examined by fluorescence microscopy. The polyamine uptake assay was also performed in isolated alveolar macrophages after AZI siRNA knockdown. This ex vivo polyamine uptake assay was performed by incubating alveolar macrophages with 2.5 μm FAM-SPD in serum-free DMEM for 1 h followed by fluorescence microscopy described above.
Immunohistochemical Staining—Rat lungs were fixed in 4% paraformaldehyde overnight and then embedded in paraffin. Tissue sections on slides were processed for immunohistochemical (IHC) staining by boiling the deparaffinized sections in High pH Target Retrieval Solution (DAKO, Carpinteria, CA) for 20 min to expose hidden antigens. After three PBS washes, sections were immersed in 3% hydrogen peroxide for 5 min to deplete the endogenous peroxidase activity. Immunostaining was performed using the UltraVision LP System with HRP Polymer (Lab Vision). Briefly, sections were incubated in 1% bovine serum albumin in PBS for 30 min followed by antibody staining at room temperature for 2 h. Monoclonal antibodies against rat ODC (MP16-2), CD68 (KP1), CD79a (HM47/A9), and CD3ζ (6B10) were purchased from Lab Vision, whereas polyclonal anti-spermidine antibody was acquired from Abcam (Cambridge, MA). The anti-spermidine antibody also cross-reacts with spermine, according to the manufacturer. Primary Antibody Enhancer and HRP polymer were used according to the manufacturer's instructions. Final chromogenic reaction was performed with the metal enhanced DAB substrate (Pierce).
Flow Cytometry—Isolation of single cells from Pneumocystis-infected rat lungs was performed as described previously (30). Each anesthetized rat was perfused with 30 ml of cold Dulbecco's PBS via the right ventricle. A total of 5 ml of Dispase II (neutral protease, 5 mg/ml, Sigma) was transtracheally injected into the lungs, and the trachea was ligated with suture silk to avoid leakage. Lungs containing Dispase II were incubated in Dulbecco's PBS for 45 min at 37 °C. The lungs were then cut into small pieces and incubated in 5 ml of Dulbecco's PBS containing 1 mg/ml collagenase/Dispase (Roche Applied Science) and 0.08 mg/ml DNase I Type II (Sigma) for 10 min at 37 °C with shaking. Digested lungs were forced through an 18-gauge needle 10 times and then mixed with 2 ml of fetal bovine serum. Single cells were isolated by filtering through a 70-μm nylon mesh and collected by centrifugation at 300 × g for 10 min. The pelleted cells were treated with 2 ml of red blood cell lysis buffer (Sigma) to remove red blood cells, followed by washing with 10 ml of PBS two times and then enumerated with a hemocytometer. Approximately one million cells were used for each flow cytometric analysis. Fluorescein isothiocyanate (FITC)-conjugated anti-ODC antibody was made by using the FITC Microscale Protein Labeling Kit (AnaSpec, San Jose, CA). PE-conjugated antibodies against CD4, CD8, and CD45RA were purchased from Cedarlane (Burlington, NC). PE-conjugated anti-CD68 antibody was acquired from AbD Serotec (Raleigh, NC). FITC- and PE-conjugated mouse IgG1 antibodies were used as isotype controls. The stained cells were analyzed with a FACScan flow cytometer (BD Biosciences), and the data were interpreted by using BD Biosciences CellQuest Software.
AZI Knockdown with siRNA—The rat AZI siRNA duplex and scrambled siRNA were synthesized by Integrated DNA Technologies (Coralville, IA). The sequence of the AZI siRNA was AAGAUCGUGAAGAAGCACAGU, which has been shown to be effective against AZI expression (31). To perform siRNA knockdown, ∼2 × 106 alveolar macrophages were placed in a well of a 6-well plate containing 2 ml of serum-free DMEM. The cells were transfected with the siRNAs by adding 2 μl of 1 μg/μl of siRNA molecules and 5 μl of TransMessenger (Qiagen) to the well. After 6 h of incubation at 37 °C with 5% CO2, the culture medium was supplemented with a final concentration of 10% fetal bovine serum, 1% antibiotic mixture (Sigma #P4333), and 2 ml of DMEM-based BALF to mimic the original lung environment. The cells were harvested after 3 days of incubation for real-time RT-PCR, Western blotting, and polyamine uptake assays described above.
TUNEL Assay—The TUNEL assay was performed using the DeadEnd Fluorimetric TUNEL System (Promega, Madison, WI) according to the manufacturer's instructions. Apoptotic cells were detected and enumerated by fluorescence microscopy with 4′,6-diamidino-2-phenylindole counterstaining.
Cytosolic Cytochrome c Assay—Determination of cytosolic cytochrome c levels was performed as described previously (11). Briefly, 2 × 106 alveolar macrophages were lysed, and the protein concentrations in the cell lysate were determined by the Bradford protein assay method (Bio-Rad). A total of 5 μg of protein in 100 μl of blocking buffer was used to determine the levels of cytochrome c by the Function ELISA cytochrome c assay kit (Active Motif, Carlsbad, CA). This kit uses the Sandwich ELISA technique with the first antibody to capture and the biotin-conjugated second antibody coupled with HRP-conjugated streptavidin to quantify cytochrome c.
PAO and SMO Activity Assay—PAO activity was assayed by the method of Suzuki et al. (32), which measures the levels of H2O2 formed. H2O2 converts homovanillic acid to a fluorescent compound in the presence of HRP. Alveolar macrophages (∼2 × 106 cells) from each rat were suspended in 1 ml of 83 mm sodium borate (pH 9.0) and broken up by passing through a 1-ml syringe with a 25-gauge needle 20 times. 500 μl of cell lysate was preincubated with 5 units of HRP for 20 min at 37 °C to remove endogenous substrates of hydrogen peroxide-producing enzymes. 100 μg of homovanillic acid and N1-acetylspermine (final concentration, 250 μm) were then added followed by an incubation at 37 °C for 30 min. The PAO reaction was stopped by adding 1 ml of 0.1 m NaOH solution. The intensity of fluorescence generated in the reaction was measured by fluorometry with an excitation wavelength of 323 nm and emission wavelength of 426 nm. Protein concentration was determined with a Bradford protein assay dye (Bio-Rad), and PAO activity was shown in the amount H2O2 produced per min per mg of protein. SMO activity was measured by the same method except that the substrate was replaced by spermine.
Statistical Analysis—Data are presented as means ± S.D. with the indicated number of experiments. The two-tailed Student's t test was used for statistical analysis, and the difference between the two groups analyzed was considered statistically significant if the p value was <0.05.
RESULTS
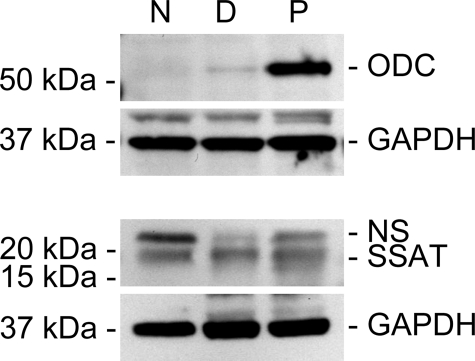
Polyamine Synthesis in Alveolar Macrophages during PcP—To examine whether the increased polyamine levels in alveolar macrophages during PcP are due to increased polyamine synthesis, the expression levels of ODC, SSAT, and SRM were determined by real-time RT-PCR and Western blotting. The results showed that the mRNA levels of ODC, SSAT, and SRM in alveolar macrophages from immunosuppressed (Dex) rats were 0.93-, 1.54-, and 1.21-fold, respectively, of those of Normal rats (data not shown). Because these differences were not statistically significant, the results indicated that long term treatment of rats with dexamethasone did not significantly affect polyamine synthesis mediated by ODC, SSAT, or SRM. In alveolar macrophages from Dex-Pc rats, ODC, SSAT, and SRM mRNA levels were 1.01-, 0.94-, and 1.01-fold that of Dex rats (data not shown), indicating that Pneumocystis infection also did not significantly affect the transcription of these genes. Western blotting analysis revealed that the level ODC protein in alveolar macrophages was not changed by immunosuppression but was increased ∼8-fold during Pneumocystis infection (Fig. 1). In contrast, the SSAT protein level was not altered during Pneumocystis infection (Fig. 1). The protein level of SRM was not determined, because anti-rat SRM antibody was not commercially available. The change in ODC protein but not mRNA levels indicated that ODC expression was regulated by a post-transcriptional mechanism in alveolar macrophages during PcP.
FIGURE 1.
Expression of polyamine synthesis enzymes in alveolar macrophages. Protein levels of ODC and SSAT in alveolar macrophages were analyzed by Western blotting. Results shown are representative of three independent experiments. N, normal rats; D, Dex rats; P, Dex-Pc rats; and NS, nonspecific band.
Polyamine Uptake by Alveolar Macrophages during PcP—Elevated intracellular polyamine levels could also be due to increased polyamine uptake; therefore, polyamine uptake assays were performed. Purified FAM-labeled spermidine (FAM-SPD) was transtracheally instilled into the lungs of Normal, Dex, and Dex-Pc rats. One hour after the transtracheal instillation, rats were sacrificed and lavaged to obtain alveolar macrophages. Fluorescence microscopy showed that <0.1% of alveolar macrophages from Normal and Dex rats but greater than 2% of those from Dex-Pc rats contained FAM-SPD (Fig. 2A), suggesting that the number of alveolar macrophages that can take up exogenous polyamines was increased by ∼20-fold during PcP (Fig. 2B). To confirm that these cells took up FAM-SPD and not FAM nonspecifically, a separate group of Dex-Pc rats were transtracheally instilled with FAM. This experiment revealed that <0.1% alveolar macrophages in this group of rats internalized FAM (Fig. 2B), confirming the specificity of polyamine uptake. To identify the cells that took up polyamines, frozen lung sections from FAM-SPD-instilled Dex-Pc rats were stained for the macrophage marker CD68, and all the cells that contained FAM-SPD were positive for CD68 (Fig. 2C), indicating that they were macrophages.
FIGURE 2.
Polyamine uptake assay. A, 200 μl of 5 μm FAM-SPD or FAM was transtracheally instilled into Normal, Dex, and Dex-Pc rats. Alveolar macrophages were isolated 1 h after the instillation and examined by fluorescence microscopy. Arrows indicate the cells with internalized FAM-SPD. B, a total of 1000 randomly selected cells of each sample shown in A were counted, and the percentages of cells with internalized FAM or FAM-SPD were determined and diagrammed. Results represent mean ± S.D. from four experiments. *, p < 0.05 as compared with Dex control. C, Dex-Pc rats were transtracheally instilled with FAM-SPD. 1 h after the instillation, rats were sacrificed, and frozen lung sections were made and stained for the macrophage marker CD68. Merged images show that the cells that took up FAM-SPD were exclusively alveolar macrophages, because every cell that took up polyamine was CD68-positive, and no CD68-negative cells took up polyamines.
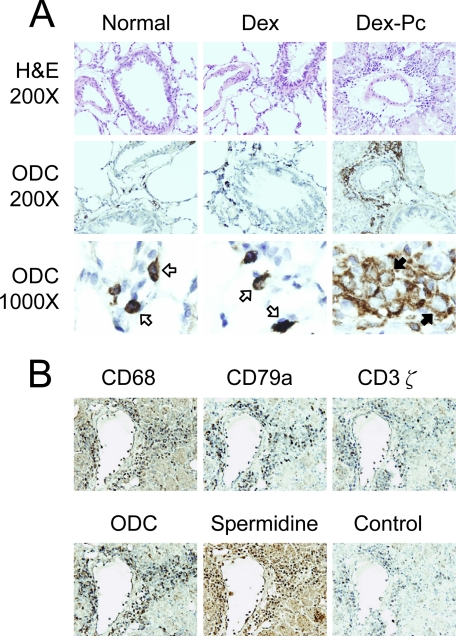
ODC Expression in Rat Lungs during PcP—To determine the sources of polyamines in the lung during PcP, IHC staining for ODC was performed on lung sections from Normal, Dex, and Dex-Pc rats. H&E and ODC staining revealed no differences in the microscopic features of the lungs from Normal and Dex rats, indicating that immunosuppression of rats with dexamethasone had no effect on ODC expression. However, ODC expression in the lung was greatly affected by Pneumocystis infection. The ODC protein was not detectable in most pulmonary cells, but was highly expressed in the mononuclear cells located around blood vessels and bronchioles (Fig. 3A), suggesting that these inflammatory cells were sources of polyamines during PcP. Interestingly, the intracellular location of ODC was changed during PcP. ODC was found distributed evenly in the cytoplasm of inflammatory cells from uninfected rats, but was located in pericytoplasmic areas in those from infected rats (Fig. 3A). To identify these cells, serial lung sections from Dex-Pc rats were stained with cell-specific markers CD68 (monocyte), CD79a (B lymphocyte), and CD3ζ (T lymphocyte). Results showed that these ODC-positive cells may be monocytes and B or T lymphocytes (Fig. 3B). Polyamine production determined by anti-spermidine staining also showed that the cells in this area contain high levels of polyamines (Fig. 3B).
FIGURE 3.
ODC expression in the lung. A, H&E staining (top) and IHC staining for ODC were performed on lung sections from Normal, Dex, and Dex-Pc rats. Cells reacted with anti-ODC antibody were examined at the magnifications of 200× (middle) and 1000× (bottom). Open and solid arrows indicate the cells with homogenous and pericytoplasmic ODC expression patterns, respectively. Results shown are representative of three sets of animals. B, IHC staining for CD68 (monocyte), CD79a (B cell), CD3ζ (T cell), ODC, and spermidine was performed on serial sections from Dex-Pc lungs. The control was processed in an identical manner except that no primary antibody was used. Magnification = 200×.
Flow Cytometry Analysis of ODC-positive Cells in the Lung with PcP—To quantify and further identify ODC-positive cells, total lung cells isolated from Dex-Pc rats were analyzed by flow cytometry. Anti-ODC antibody was labeled with FITC, and antibodies against CD4, CD8, CD45RA, and CD68 were labeled with PE. In this assay, 24.44% of total lung cells from Dex-Pc rats were ODC-positive (Table 1). Dual staining of the cells with ODC and CD4, CD8, CD45RA, or CD68 antibodies revealed that 1.83% of the cells were ODC+CD4+, 8.27% were ODC+CD8+, 9.79% were ODC+CD45RA+ (B lymphocytes), and 7.80% were ODC+CD68+ (monocytes/macrophages).
TABLE 1.
Flow cytometry analysis of lung cells from Pneumocystis-infected rats
| FITC(−)PE(−) | FITC(−)PE(+) | FITC(+)PE(−) | FITC(+)PE(+) | |
|---|---|---|---|---|
| Isotype control | 95.75 ± 0.81 | 0.04 ± 0.07 | 2.54 ± 0.10 | 1.66 ± 0.84 |
| ODC-FITC | 73.95 ± 5.43 | 0.00 ± 0.00 | 24.44 ± 6.78 | 1.61 ± 1.34 |
| ODC-FITC + CD4-PE | 77.04 ± 4.37 | 0.07 ± 0.06 | 21.06 ± 5.40 | 1.83 ± 1.09 |
| ODC-FITC + CD8-PE | 82.07 ± 2.59 | 0.02 ± 0.01 | 9.63 ± 1.60 | 8.27 ± 0.99a |
| ODC-FITC + CD45RA-PE | 82.73 ± 1.12 | 0.77 ± 0.58 | 6.71 ± 1.37 | 9.79 ± 1.91a |
| ODC-FITC + CD68-PE | 76.73 ± 3.19 | 0.00 ± 0.01 | 15.20 ± 8.01 | 7.80 ± 4.42a |
p < 0.05 compared to the FITC(+)PE(+) quadrant of the ODC-FITC group (n = 4).
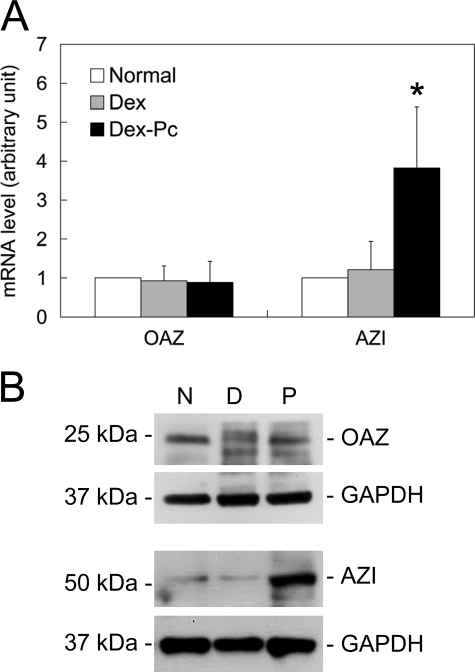
OAZ and AZI Expressions in Alveolar Macrophages during PcP—Because OAZ and AZI have been shown to regulate both polyamine production and transport (23, 24), their expressions in alveolar macrophages from Normal, Dex, and Dex-Pc rats were determined. Real-time RT-PCR results revealed that the mRNA expression levels of OAZ in alveolar macrophages from Dex and Dex-Pc rats were 0.93- and 0.96-fold, respectively, of those of Normal rats (Fig. 4A); these differences were not statistically significant. Western blotting results also showed that OAZ protein levels in alveolar macrophages were not affected by immunosuppression or Pneumocystis infection (Fig. 4B). In contrast, AZI mRNA levels in alveolar macrophages from Dex-Pc rats were 3.16-fold higher than those from Dex rats (Fig. 4A), and AZI protein levels in alveolar macrophages were increased by 6-fold during PcP as determined by Western blotting (Fig. 4B).
FIGURE 4.
OAZ and AZI expressions in alveolar macrophages. mRNA and protein levels of OAZ and AZI in alveolar macrophages from Normal (N), Dex (D), and Dex-Pc (P) rats were determined by real-time RT-PCR (n = 5) (A) and Western blotting (n = 3) (B). The average mRNA levels of OAZ and AZI genes in alveolar macrophages from Dex and Dex-Pc rats presented in panel A are shown as -fold increase relative to those of Normal rats, each of which was arbitrarily set to 1. *, p < 0.05 as compared with Dex control.
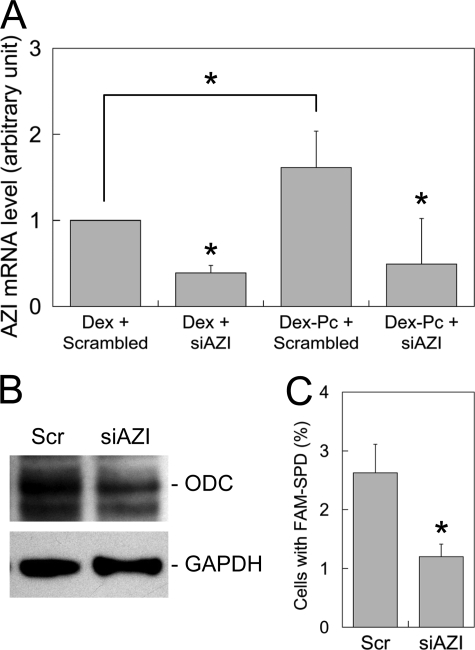
Polyamine Synthesis and Uptake after AZI Knockdown—To correlate AZI overexpression with alveolar macrophage apoptosis during PcP, alveolar macrophages isolated from Dex-Pc rats were analyzed for AZI expression, polyamine synthesis, polyamine uptake, and apoptosis after AZI siRNA knockdown. Results from real-time PCR showed that AZI siRNA successfully decreased AZI mRNA levels by ∼70% in alveolar macrophages from Dex-Pc rats (Fig. 5A). This AZI knockdown was also found to cause a 60% decrease in ODC protein level in these alveolar macrophages (Fig. 5B). In the ex vivo spermidine uptake assay, a 50% reduction in the number of cells taking up polyamines was observed in the group of cells transfected with AZI siRNA as compared with control transfectants (Fig. 5C).
FIGURE 5.
Polyamine synthesis and uptake after siRNA knockdown of AZI. A, real-time RT-PCR analysis of AZI mRNA levels after siRNA knockdown. Alveolar macrophages from Dex and Dex-Pc rats were transfected with AZI siRNA (siAZI) or scrambled siRNA (Scrambled) and then measured for AZI mRNA levels. B, the ODC protein level in alveolar macrophages from Dex-Pc rats was determined by Western blotting after transfection with AZI siRNA (siAZI) or scrambled siRNA (Scr). C, alveolar macrophages from Dex-Pc rats were assayed for FAM-SPD uptake after transfection with AZI siRNA (siAZI) or scrambled siRNA (Scr). A total of 1000 cells of each sample was counted, and the percentages of cells with internalized FAM-SPD were determined and diagrammed.
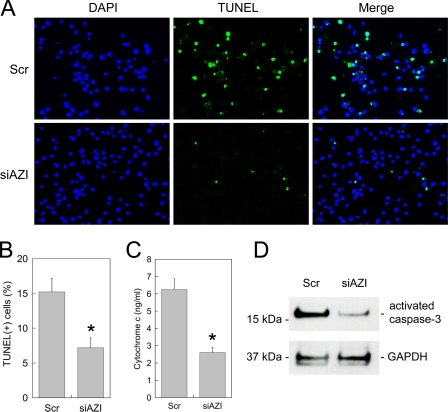
Macrophage Apoptosis after AZI Knockdown—BALFs from Pneumocytes-infected rats have been shown to induce macrophages to undergo apoptosis due to elevated levels of polyamines (11). To determine whether AZI plays a role in this BALF-induced apoptosis, 2 × 106 normal rat alveolar macrophages were transfected with AZI siRNA and then incubated with BALF from Dex-Pc rats 6 h after siRNA transfection. Three days after the transfection and BALF treatment, cells were analyzed for apoptosis by three different methods. Results from TUNEL assay showed that the number of apoptotic cells was decreased by 53% (Fig. 6, A and B). The levels of cytosolic cytochrome c showed a 58% reduction (Fig. 6C), and those of activated caspase-3 determined by Western blotting were decreased by 75% in macrophages transfected with AZI siRNA as compared with controls that were transfected with scrambled siRNA (Fig. 6D).
FIGURE 6.
Macrophage apoptosis after siRNA knockdown of AZI. A, Normal rat alveolar macrophages were transfected with AZI siRNA (siAZI) or scrambled siRNA (Scr) and then treated with BALF from Dex-Pc rats. Three days after transfection, a TUNEL assay was performed, and cells were examined by fluorescence microscopy. B, a total of 1000 randomly selected cells of each sample shown in A were counted, and the percentages of apoptotic cells were diagrammed. Results represent mean ± S.D. from three experiments. *, p < 0.05 as compared with scrambled control. C, cytosolic cytochrome c levels in cells treated with siRNA and BALF from Dex-Pc rats were measured. Results represent mean ± S.D. from three experiments. *, p < 0.05 as compared with scrambled control. D, the level of activated caspase-3 in alveolar macrophages from Dex-Pc rats was determined by Western blotting after transfection with AZI siRNA (siAZI) or scrambled siRNA (Scr).
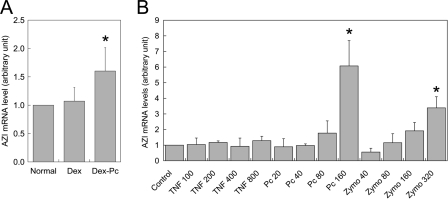
Substances That Induce AZI Overexpression—Because AZI was found overexpressed in alveolar macrophages from Dex-Pc rats, experiments were performed to determine whether certain substances capable of inducing the expression of AZI are present in BALF from Dex-Pc rats. Alveolar macrophages from normal rats were first treated with the BALF and then measured for AZI mRNA by RT-PCR. This treatment resulted in a 1.5-fold increase in AZI mRNA levels in alveolar macrophages compared with the same cells treated with BALF from Dex rats (Fig. 7A), suggesting that AZI expression in alveolar macrophages can be induced by a certain substance in Pneumocystis-infected lungs. To identify such substance, alveolar macrophages from normal rats were treated with Pneumocystis organisms and the cytokine TNF-α, which is present in high levels in Pneumocystis-infected lungs. Results showed that Pneumocystis organisms but not TNF-α increased AZI mRNA levels in a dose-dependent manner (Fig. 7B). The same result was observed when normal rat alveolar macrophages were treated with zymosan A (Fig. 7B). These results suggest that a component of the Pneumocystis cell wall is responsible for the increase in AZI expression during PcP.
FIGURE 7.
Induction of AZI overexpression. A, Normal rat alveolar macrophages were incubated with Normal, Dex, and Dex-Pc BALF for 24 h and then examined by real-time RT-PCR for AZI expression. Results represent mean ± S.D. from five experiments. *, p < 0.05 as compared with the Dex control. B, Normal rat alveolar macrophages were incubated with TNF-α (TNF, 100-800 pg/μl), P. carinii organisms (Pc, 20-160 nuclei/μl), or zymosan A (Zymo, 40-320 ng/μl) for 18 h and then examined by real-time RT-PCR for AZI expression. Results represent mean ± S.D. from three analyses. *, p < 0.05 as compared with mock control.
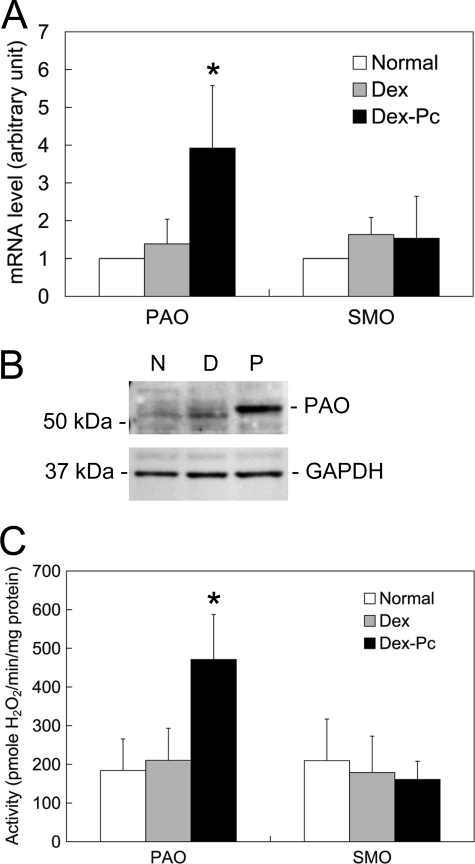
Polyamine Catabolism and Alveolar Macrophage Apoptosis—To investigate whether polyamine catabolism plays a role in alveolar macrophage apoptosis during PcP, the expression and activity levels of PAO and SMO in alveolar macrophages from Pneumocystis-infected rats were determined. Results of real-time RT-PCR showed that PAO mRNA levels in alveolar macrophages from Dex-Pc rats were 2.82-fold that of those from Dex rats, but SMO mRNA levels were not significantly changed (Fig. 8A). PAO protein levels were found increased ∼4-fold in alveolar macrophages during PcP (Fig. 8B), and PAO activity in alveolar macrophages from Pneumocystis-infected rats showed a 2.24-fold increase as compared with the Dex control (Fig. 8C). These results suggest that polyamine oxidation plays a role in alveolar macrophages apoptosis in Pneumocystis-infected lungs.
FIGURE 8.
Polyamine catabolism. A, the mRNA levels of PAO and SMO in alveolar macrophages form Normal (N), Dex (D), and Dex-Pc (P) rats were determined by real-time RT-PCR. The average mRNA levels of these genes in alveolar macrophages from Dex and Dex-Pc rats are shown as -fold increase relative to those of Normal rats, each of which was arbitrarily set to 1. Results represent mean ± S.D. from five experiments. *, p < 0.05 as compared with the Dex control. B, protein levels of PAO and SMO in alveolar macrophages were analyzed by Western blotting. C, the enzyme activities of PAO and SMO in alveolar macrophages from Normal, Dex, and Dex-Pc rats were determined. Results represent mean ± S.D. from five experiments. *, p < 0.05 as compared with Dex control.
DISCUSSION
We have previously found that the levels of spermidine, N1-acetylspermidine, and N1-acetylspermine in both the alveoli and alveolar macrophages are elevated leading to increased rate of alveolar macrophage apoptosis during PcP (11). In the present study, we investigated the possible mechanisms for such increase in polyamine levels. The transcription of the ODC gene, which encodes the key enzyme in polyamine synthesis, in alveolar macrophages was not affected by Pneumocystis infection, but the ODC protein level in alveolar macrophages was greatly (8-fold) increased during PcP (Fig. 1), indicating that the balance between ODC synthesis and degradation is altered during Pneumocystis infection. As described above, the ODC protein level is controlled by both OAZ and AZI. OAZ binds to ODC and transports it to the 26 S proteasome for degradation, whereas AZI competes with ODC for binding to OAZ. When AZI binds to OAZ, less ODC is degraded and thus a steady-state level of polyamines in the cells is maintained. Because OAZ mRNA and protein levels were not changed (Fig. 4) but both the mRNA (3-fold) and protein (6-fold) levels of AZI were greatly increased (Fig. 4), it is likely that the ODC turnover rate in alveolar macrophages during PcP is decreased due to AZI overexpression. The result that AZI knockdown by siRNA decreased the ODC protein level by 60% (Fig. 5B) confirmed this possibility. Higher polyamine synthesis during PcP was demonstrated by the IHC experiment in which ODC-positive cells, mainly inflammatory cells, were shown to react with the anti-spermidine antibody (Fig. 3 and Table 1), indicating polyamine synthesis in these cells.
An interesting finding in this study is that ODC distribution in inflammatory cells was changed from cytoplasmic to pericytoplasmic (Fig. 3A). ODC translocation has been shown to be associated with cell cycle progression (33) or cell activation and transformation (34). Because inflammatory cells are activated when they are recruited to the lung, it is very likely that the change in ODC distribution is due to cell activation during PcP and that this activation leads to elevated polyamine synthesis.
In addition to increased synthesis, increased uptake of exogenous polyamines is another possible cause of elevated polyamine levels in alveolar macrophages during PcP. However, polyamine transporters in mammalian cells remain largely unknown. Synthetic fluorescent polyamines have been used to explore mammalian polyamine transport systems (35-37), and a receptor-mediated endocytosis mechanism has been suggested (35, 36). Another study showed that polyamine uptake was unaffected in endocytosis-deficient cells and that the internalized polyamines were co-localized with acidic vesicles of the late endocytic compartment and the trans-Golgi (37). This finding suggests that polyamines are imported by a membrane carrier (37). The mammalian polyamine transport systems that have been described appear to have a broad substrate specificity and can transport a variety of polyamines (35). Therefore, it is possible that the spermidine uptake system (Fig. 2) is also responsible for the uptake of N1-acetylspermidine and N1-acetylspermine into alveolar macrophages during PcP. Polyamine uptake assays are commonly performed using radioactive polyamines (38-40). Aziz et al. (41) and Aouida et al. (29) used fluorescein-labeled polyamines to perform such assays in cultured cells and yeasts by fluorescence microscopy. It was demonstrated that the fluorescein moiety does not affect the transport of polyamines (41). In this study, we used fluorescein-labeled spermidine to investigate polyamine uptake in Pneumocystis-infected rats. We also modified Aouida's labeling process to increase the yield and showed for the first time that FAM-labeled spermidine can be used for polyamine uptake assay in vivo (Fig. 2).
Although the number of alveolar macrophages that took up fluorescent spermidine (FAM-SPD) in the polyamine uptake assay was found to be 20-fold higher in Pneumocystis-infected than in uninfected rats, only ∼2% of alveolar macrophages in Pneumocystis-infected rats internalized FAM-SPD (Fig. 2B). The reason for this low number of cells taking up FAM-SPD is unknown. It may be that most alveolar macrophages in Pneumocystis-infected lungs already contain high levels of polyamines (11) and are no longer able to take up exogenous polyamines. The alveolar macrophages that took up FAM-SPD in the polyamine uptake assay were probably those containing lower levels of intracellular polyamines. This possibility remains to be investigated. Because overexpression of AZI has been shown to increase polyamine uptake (24), it is conceivable that the increased level of AZI in alveolar macrophages contributes to this increased uptake of exogenous polyamines. The result that AZI expression knockdown by siRNA decreased the number of cells that can take up polyamines by 50% (Fig. 5C) strongly supports this possibility. A likely source of exogenous polyamines is Pneumocystis organisms, because they have been shown to produce and secrete polyamines, mainly N1-acetylspermine and N1-acetylspermidine (42). Because SSAT levels were not increased in alveolar macrophage during PcP (Fig. 1), the elevated levels of acetylated polyamines in alveolar macrophages are more likely due to uptake of such polyamines from exogenous sources. Like SSAT, the expression level of the spermidine synthase (SRM), which converts putrescine to spermidine, was also not increased in alveolar macrophage during PcP (data not shown), suggesting that SRM is not responsible for the increased spermidine level. It is possible that the increased spermidine level was due to conversion of N1-acetylspermine to spermidine.
In addition to Pneumocystis organisms, we believe that inflammatory cells are another source of polyamines, because they stained positively for ODC, and spermidine levels were high in these cells (Fig. 3B and Table 1). These cells were mainly located around blood vessels and bronchioles (Fig. 3A) and therefore are considered inflammatory cells that were recruited into the lung during PcP. Approximately 25% of the total lung cells from infected rats were ODC-positive. These inflammatory cells were shown by flow cytometry to be CD4+ T cells (1.83%), CD8+ T cells (9.79%), B cells (9.79%), and CD68+ cells (7.80%) that may be monocytes or macrophages. In this study, ODC was found to be present only in inflammatory cells by IHC staining (Fig. 3) and flow cytometry (Table 1), indicating that ODC levels were higher in inflammatory cells than in other types of pulmonary cells. The uninfected lungs were not analyzed for ODC-positive cells, because very few inflammatory cells were present.
Conversion of N1-acetylspermine to spermidine and N1-acetylspermidine to putrescine by PAO is accompanied by the production of H2O2 which may damage mitochondria leading to induction of apoptosis. This mechanism is thought to be responsible for the loss of alveolar macrophages during PcP, because reactive oxygen species levels are increased in alveolar macrophages during PcP (11). Because the mRNA, protein expression, and enzymatic activity levels of PAO were all found to be increased in alveolar macrophages during PcP (Fig. 8), it is conceivable that the reactive oxygen species generated by PAO plays an important role in alveolar macrophage apoptosis during Pneumocystis infection. It is possible that PAO overexpression in alveolar macrophages during PcP is induced by the increased intracellular levels of PAO substrates N1-acetylspermidine and N1-acetylspermine (11). An important finding in this study is that AZI expression knockdown by siRNA resulted in a 53% decrease in the number of apoptotic alveolar macrophages (Fig. 6B), a 58% decrease in cytosolic cytochrome c levels (Fig. 6C), and a 75% decrease in the level of activated caspase-3 (Fig. 6D). This result indicates that decreasing AZI protein levels also decreases the apoptosis rate of alveolar macrophages. It is possible that the reduction in AZI levels causes a decreased uptake of exogenous acetylated polyamines; therefore, less acetylated polyamines are available to be converted to regular polyamines, and thus less H2O2 is produced to trigger apoptosis. This is the first demonstration that AZI is involved in apoptosis. A previous study showed that transfection of the gene encoding OAZ, the negative regulator of AZI, can also induce apoptosis by activating mitochondrial depolarization and caspase cascade (43). It is not surprising that both OAZ and AZI have a pro-apoptotic activity due to the multiple roles of polyamines (18, 19).
Polyamines have also been found to be involved in the pathogenesis of Helicobacter pylori. H. pylori activates the transcriptional factor c-Myc, which induces ODC expression, resulting in increased intracellular polyamine pools in macrophages (44). This polyamine accumulation greatly reduces the uptake of l-arginine, which is the substrate of inducible nitric oxide synthase and is also the rate-limiting factor for inducible nitric oxide synthase translation (45). Therefore, increased polyamine levels consequently decrease the synthesis of inducible nitric oxide synthase in macrophages and result in loss of innate immune responses against H. pylori infection (46). H. pylori also activates SMO to convert spermidine to spermine accompanied by generation of H2O2, which causes macrophages to undergo apoptosis (15, 47). Therefore, induction of apoptosis by polyamines appears to be a survival mechanism for microbes to evade the clearance by immune systems.
Although both Pneumocystis and H. pylori cause macrophages to undergo apoptosis due to elevated levels of polyamines, the mechanisms in polyamine acquisition are different. Unlike H. pylori, Pneumocystis does not up-regulate ODC synthesis. Instead, it up-regulates AZI expression leading to increased uptake of exogenous polyamines and decreases the degradation of ODC thus increasing polyamine synthesis. Therefore, the increase in polyamine levels in alveolar macrophages during PcP is due to both increased uptake and polyamine synthesis. Up-regulation of AZI expression has also been shown in other diseases. A comparative clinical study showed that gastric cancer tissue removed during surgery expresses higher levels of AZI than the surrounding normal tissue (48). Oncogene Ras-transformed NIH3T3 cells also have increased AZI expression (24). Although AZI levels have been shown to be induced in a number of conditions, the mechanisms for AZI overexpression were largely unknown. In this study, we found that both Pneumocystis organisms and zymosan A induced AZI expression in a dose-dependent manner (Fig. 7B). Because the major component of zymosan A is β-glucan, which is also the major component of the Pneumocystis cell wall, it is very likely that β-glucan induces AZI overexpression in alveolar macrophages. The dectin-1 receptor has been shown to be the main receptor for β-glucan on alveolar macrophages (49), and binding of β-glucan to dectin-1 has been shown to activate the NF-κB signaling pathway (50). Therefore, it is possible that AZI expression is regulated by NF-κB. This possibility is being investigated. Because increased polyamine levels due to both increased synthesis and uptake are responsible for the apoptosis of alveolar macrophages during PcP, decreasing polyamine synthesis, uptake, or both may be potential therapeutic approach for PcP, because we have shown that prevention of alveolar macrophage apoptosis reduced the severity of PcP (13). Difluoromethylornithine, one of the most effective ODC inhibitors, has been used to treat rats with PcP, and the results showed that difluoromethylornithine decreases organism counts, although it was not as effective as TMP-SMX (51, 52). This partial success may be due to the compensatory effects of polyamine uptake. Recently, a synthetic compound, referred to as compound #7, has been shown to inhibit polyamine import with very low cytotoxicity (53). This type of polyamine transport inhibitor may be used alone or in combination with difluoromethylornithine for therapy of PcP by decreasing the intracellular polyamine levels and apoptosis rate of alveolar macrophages.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 HL65170 and RO1 AI062259. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PcP, Pneumocystis pneumonia; TMP, trimethoprim; SMX, sulfamethoxazole; TNF, tumor necrosis factor; BALF, bronchoalveolar lavage fluid; ODC, ornithine decarboxylase; SRM, spermidine synthase; SMS, spermine synthase; SSAT, spermidine/spermine N1-acetyltransferase; PAO, polyamine oxidase; SMO, spermine oxidase; OAZ, ODC antizyme; Dex, dexamethasone; PBS, phosphate-buffered saline; DMEM, Dulbecco's modified Eagle's medium; RT, reverse transcription; HRP, horseradish peroxidase; SPD, spermidine; PE, phycoerythrin; siRNA, small interference RNA; IHC, immunohistochemical; FITC, fluorescein isothiocyanate; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling; ELISA, enzyme-linked immunosorbent assay; FAM, fluorophore 5-(and-6)-carboxyl fluorescein succinimidyl ester; AZI, antizyme inhibitor.
References
- 1.Phair, J., Munoz, A., Detels, R., Kaslow, R., Rinaldo, C., and Saah, A. (1990) N. Engl. J. Med. 322 161-165 [DOI] [PubMed] [Google Scholar]
- 2.Huang, L., Crothers, K., Atzori, C., Benfield, T., Miller, R., Rabodonirina, M., and Helweg-Larsen, J. (2004) Emerg. Infect. Dis. 10 1721-1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nahimana, A., Rabodonirina, M., Bille, J., Francioli, P., and Hauser, P. M. (2004) Antimicrob. Agents Chemother. 48 4301-4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein, C. R., Poole, C., Kazanjian, P., and Meshnick, S. R. (2004) Emerg. Infect. Dis. 10 1760-1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limper, A. H., Hoyte, J. S., and Standing, J. E. (1997) J. Clin. Invest. 99 2110-2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezekowitz, R. A. B., Williams, D. J., Koziel, H., Armstrong, M. Y., Warner, A., Richards, F. F., and Rose, R. M. (1991) Nature 351 155-158 [DOI] [PubMed] [Google Scholar]
- 7.Steele, C., Marrero, L., Swain, S., Harmsen, A. G., Zheng, M., Brown, G. D., Gordon, S., Shellito, J. E., and Kolls, J. K. (2003) J. Exp. Med. 198 1677-1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang, C., Wang, S. H., Lasbury, M. E., Tschang, D., Liao, C. P., Durant, P. J., and Lee, C. H. (2006) Infect. Immun. 74 1857-1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleury, J., Escudier, E., Pocholle, M. J., Carre, C., and Bernaudin, J. F. (1985) Acta Cytol. 29 721-726 [PubMed] [Google Scholar]
- 10.Lasbury, M. E., Durant, P. J., Bartlett, M. S., Smith, J. W., and Lee, C. H. (2003) Clin. Diagn. Lab. Immunol. 10 293-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasbury, M. E., Merali, S., Durant, P. J., Tschang, D., Ray, C. A., and Lee, C. H. (2007) J. Biol. Chem. 282 11009-11020 [DOI] [PubMed] [Google Scholar]
- 12.Lasbury, M. E., Durant, P. J., and Lee, C. H. (2003) J. Eukaryot. Microbiol. 50 (suppl.) 630-631 [DOI] [PubMed] [Google Scholar]
- 13.Lasbury, M. E., Durant, P. J., Ray, C. A., Tschang, D., Schwendener, R., and Lee, C. H. (2006) J. Immunol. 176 6443-6453 [DOI] [PubMed] [Google Scholar]
- 14.Wang, Y., and Casero, R. A., Jr. (2006) J. Biochem. (Tokyo) 139 17-25 [DOI] [PubMed] [Google Scholar]
- 15.Chaturvedi, R., Cheng, Y., Asim, M., Bussiere, F. I., Xu, H., Gobert, A. P., Hacker, A., Casero, R. A., Jr., and Wilson, K. T. (2004) J. Biol. Chem. 279 40161-40173 [DOI] [PubMed] [Google Scholar]
- 16.Oredsson, S. M. (2003) Biochem. Soc. Trans. 31 366-370 [DOI] [PubMed] [Google Scholar]
- 17.Verma, D. S., and Sunkara, P. S. (1982) Cancer Res. 42 3046-3049 [PubMed] [Google Scholar]
- 18.Gerner, E. W., and Meyskens, F. L., Jr. (2004) Nat. Rev. Cancer 4 781-792 [DOI] [PubMed] [Google Scholar]
- 19.Schipper, R. G., Penning, L. C., and Verhofstad, A. A. (2000) Semin. Cancer Biol. 10 55-68 [DOI] [PubMed] [Google Scholar]
- 20.Erwin, B. G., Ewton, D. Z., Florini, J. R., and Pegg, A. E. (1983) Biochem. Biophys. Res. Commun. 114 944-949 [DOI] [PubMed] [Google Scholar]
- 21.Kufe, D. W., Griffin, J., Mitchell, T., and Shafman, T. (1984) Cancer Res. 44 4281-4284 [PubMed] [Google Scholar]
- 22.Nilsson, J. A., Keller, U. B., Baudino, T. A., Yang, C., Norton, S., Old, J. A., Nilsson, L. M., Neale, G., Kramer, D. L., Porter, C. W., and Cleveland, J. L. (2005) Cancer Cell 7 433-444 [DOI] [PubMed] [Google Scholar]
- 23.Mitchell, J. L., Judd, G. G., Bareyal-Leyser, A., and Ling, S. Y. (1994) Biochem. J. 299 19-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keren-Paz, A., Bercovich, Z., Porat, Z., Erez, O., Brener, O., and Kahana, C. (2006) Oncogene 25 5163-5172 [DOI] [PubMed] [Google Scholar]
- 25.Bartlett, M. S., Fishman, J. A., Queener, S. F., Durkin, M. M., Jay, M. A., and Smith, J. W. (1988) J. Clin. Microbiol. 26 1100-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsufuji, S., Miyazaki, Y., Kanamoto, R., Kameji, T., Murakami, Y., Baby, T. G., Fujita, K., Ohno, T., and Hayashi, S. (1990) J. Biochem. (Tokyo) 108 365-371 [DOI] [PubMed] [Google Scholar]
- 27.Matsufuji, S., Kanamoto, R., Murakami, Y., and Hayashi, S. (1990) J. Biochem. (Tokyo) 107 87-91 [DOI] [PubMed] [Google Scholar]
- 28.Murakami, Y., Ichiba, T., Matsufuji, S., and Hayashi, S. (1996) J. Biol. Chem. 271 3340-3342 [DOI] [PubMed] [Google Scholar]
- 29.Aouida, M., Leduc, A., Wang, H., and Ramotar, D. (2004) Biochem. J. 384 47-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, S. H., Zhang, C., Lasbury, M. E., Liao, C. P., Durant, P. J., Tschang, D., and Lee, C. H. (2008) Microbes Infect. 10 334-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S. W., Mangold, U., Waghorne, C., Mobascher, A., Shantz, L., Banyard, J., and Zetter, B. R. (2006) J. Cell Sci. 119 2583-2591 [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, O., Matsumoto, T., and Katsumata, Y. (1984) Experientia (Basel) 40 838-839 [DOI] [PubMed] [Google Scholar]
- 33.Schipper, R. G., Cuijpers, V. M., De Groot, L. H., Thio, M., and Verhofstad, A. A. (2004) J. Histochem. Cytochem. 52 1259-1266 [DOI] [PubMed] [Google Scholar]
- 34.Heiskala, M., Zhang, J., Hayashi, S., Holtta, E., and Andersson, L. C. (1999) EMBO J. 18 1214-1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cullis, P. M., Green, R. E., Merson-Davies, L., and Travis, N. (1999) Chem. Biol. 6 717-729 [DOI] [PubMed] [Google Scholar]
- 36.Soulet, D., Covassin, L., Kaouass, M., Charest-Gaudreault, R., Audette, M., and Poulin, R. (2002) Biochem. J. 367 347-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soulet, D., Gagnon, B., Rivest, S., Audette, M., and Poulin, R. (2004) J. Biol. Chem. 279 49355-49366 [DOI] [PubMed] [Google Scholar]
- 38.Hoet, P. H., and Nemery, B. (2000) Am. J. Physiol. 278 L417-L433 [DOI] [PubMed] [Google Scholar]
- 39.Uemura, T., Kashiwagi, K., and Igarashi, K. (2007) J. Biol. Chem. 282 7733-7741 [DOI] [PubMed] [Google Scholar]
- 40.Roy, U. K., Rial, N. S., Kachel, K. L., and Gerner, E. W. (2008) Mol. Carcinogr. 47 538-553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aziz, S. M., Yatin, M., Worthen, D. R., Lipke, D. W., and Crooks, P. A. (1998) J. Pharm. Biomed. Anal. 17 307-320 [DOI] [PubMed] [Google Scholar]
- 42.Merali, S. (1999) J. Biol. Chem. 274 21017-21022 [DOI] [PubMed] [Google Scholar]
- 43.Liu, G. Y., Liao, Y. F., Hsu, P. C., Chang, W. H., Hsieh, M. C., Lin, C. Y., Hour, T. C., Kao, M. C., Tsay, G. J., and Hung, H. C. (2006) Apoptosis 11 1773-1788 [DOI] [PubMed] [Google Scholar]
- 44.Cheng, Y., Chaturvedi, R., Asim, M., Bussiere, F. I., Scholz, A., Xu, H., Casero, R. A., Jr., and Wilson, K. T. (2005) J. Biol. Chem. 280 22492-22496 [DOI] [PubMed] [Google Scholar]
- 45.Chaturvedi, R., Asim, M., Lewis, N. D., Algood, H. M., Cover, T. L., Kim, P. Y., and Wilson, K. T. (2007) Infect. Immun. 75 4305-4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bussiere, F. I., Chaturvedi, R., Cheng, Y., Gobert, A. P., Asim, M., Blumberg, D. R., Xu, H., Kim, P. Y., Hacker, A., Casero, R. A., Jr., and Wilson, K. T. (2005) J. Biol. Chem. 280 2409-2412 [DOI] [PubMed] [Google Scholar]
- 47.Xu, H., Chaturvedi, R., Cheng, Y., Bussiere, F. I., Asim, M., Yao, M. D., Potosky, D., Meltzer, S. J., Rhee, J. G., Kim, S. S., Moss, S. F., Hacker, A., Wang, Y., Casero, R. A., Jr., and Wilson, K. T. (2004) Cancer Res. 64 8521-8525 [DOI] [PubMed] [Google Scholar]
- 48.Jung, M. H., Kim, S. C., Jeon, G. A., Kim, S. H., Kim, Y., Choi, K. S., Park, S. I., Joe, M. K., and Kimm, K. (2000) Genomics 69 281-286 [DOI] [PubMed] [Google Scholar]
- 49.Brown, G. D. (2006) Nat. Rev. Immunol. 6 33-43 [DOI] [PubMed] [Google Scholar]
- 50.Goodridge, H. S., Simmons, R. M., and Underhill, D. M. (2007) J. Immunol. 178 3107-3115 [DOI] [PubMed] [Google Scholar]
- 51.Chin, K., Merali, S., Saric, M., and Clarkson, A. B., Jr. (1996) Antimicrob. Agents Chemother. 40 2318-2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clarkson, A. B., Jr., Williams, D. E., and Rosenberg, C. (1988) Antimicrob. Agents Chemother. 32 1158-1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaur, N., Delcros, J. G., Imran, J., Khaled, A., Chehtane, M., Tschammer, N., Martin, B., and Phanstiel, O. (2008) J. Med. Chem. 51 1393-1401 [DOI] [PubMed] [Google Scholar]