Abstract
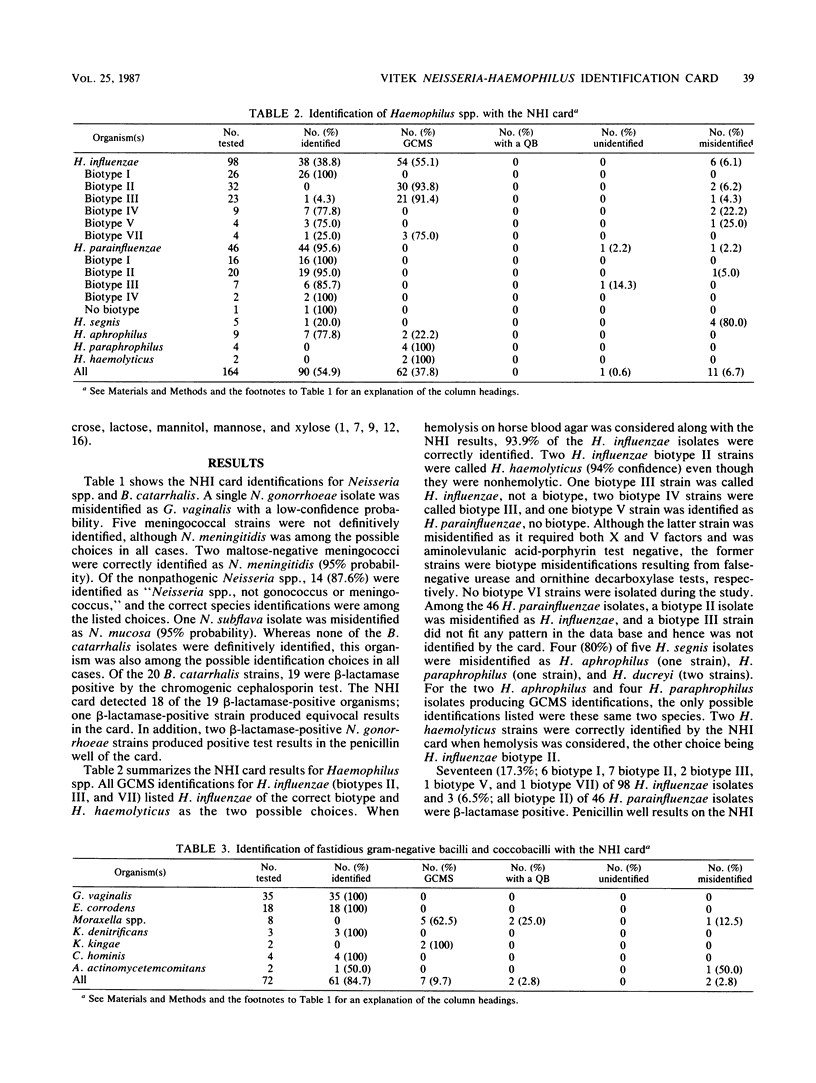
A clinical evaluation of the Vitek Neisseria-Haemophilus Identification (NHI) card (Vitek Systems, Inc., Hazelwood, Mo.) was performed with 480 clinical isolates and stock strains of Neisseria spp., Haemophilus spp., and other fastidious microorganisms included in the data base of the system. Identifications obtained with the NHI card were compared with those determined by conventional methods. The card identified 83.2% of 244 Neisseria spp. and Branhamella catarrhalis, 54.9% of 164 Haemophilus spp., and 84.7% of 72 fastidious gram-negative species with no further testing required. Some isolates produced good confidence-marginal separation identifications, in which the correct identification was listed with one or two other possible identifications and extra tests were required and suggested. When isolates producing good confidence-marginal separation identifications were included, correct identifications of these organism groups increased to 97.1, 92.7, and 94.4%, respectively. Among the commonly isolated microorganisms, the NHI card identified 99.1% of 110 N. gonorrhoeae, 98.5% of 68 N. meningitidis, 93.9% of 98 H. influenzae, and 95.6% of 46 H. parainfluenzae strains. All of these organisms produced excellent to very good confidence level identifications except for H. influenzae biotypes II, III, and VII, for which hemolytic reactions were required for differentiation from H. haemolyticus. The NHI card reliably identified other fastidious gram-negative species, including H. aphrophilus, Eikenella corrodens, Gardnerella vaginalis, and Kingella denitrificans.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Back A. E., Oberhofer T. R. Use of the Minitek system for biotyping Haemophilus species. J Clin Microbiol. 1978 Mar;7(3):312–313. doi: 10.1128/jcm.7.3.312-313.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlin B. W. Cellular elongation under the influence of antibacterial agents: way to differentiate coccobacilli from cocci. J Clin Microbiol. 1975 Jan;1(1):102–105. doi: 10.1128/jcm.1.1.102-105.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doern G. V., Morse S. A. Branhamella (Neisseria) catarrhalis: criteria for laboratory identification. J Clin Microbiol. 1980 Feb;11(2):193–195. doi: 10.1128/jcm.11.2.193-195.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edberg S. C., Melton E., Singer J. M. Rapid biochemical characterization of Haemophilus species by using the micro-ID. J Clin Microbiol. 1980 Jan;11(1):22–26. doi: 10.1128/jcm.11.1.22-26.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood J. R., Pickett M. J., Martin W. J., Mack E. G. Heamophilus vaginalis (Corynebacterium vaginal): method for isolation and rapid biochemical identification. Health Lab Sci. 1977 Apr;14(2):102–106. [PubMed] [Google Scholar]
- Hollis D. G., Sottnek F. O., Brown W. J., Weaver R. E. Use of the rapid fermentation test in determining carbohydrate reactions of fastidious bacteria in clinical laboratories. J Clin Microbiol. 1980 Oct;12(4):620–623. doi: 10.1128/jcm.12.4.620-623.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda W. M., Ulanday M. G., Bohnhoff M., LeBeau L. J. Evaluation of the RIM-N, Gonochek II, and Phadebact systems for the identification of pathogenic Neisseria spp. and Branhamella catarrhalis. J Clin Microbiol. 1985 May;21(5):734–737. doi: 10.1128/jcm.21.5.734-737.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp J. S., Totten P. A., Mulks M. H., Minshew B. H. Characterization of Neisseria cinerea, a nonpathogenic species isolated on Martin-Lewis medium selective for pathogenic Neisseria spp. J Clin Microbiol. 1984 Jan;19(1):63–67. doi: 10.1128/jcm.19.1.63-67.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton W. D., Battaglioli G. J. Gono Gen coagglutination test for confirmation of Neisseria gonorrhoeae. J Clin Microbiol. 1983 Nov;18(5):1264–1265. doi: 10.1128/jcm.18.5.1264-1265.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M. E., Blazevic D. J. Rapid speciation of Haemophilus with the porphyrin production test versus the satellite test for X. J Clin Microbiol. 1977 Feb;5(2):142–144. doi: 10.1128/jcm.5.2.142-144.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery K., Raymundo L., Jr, Drew W. L. Chromogenic cephalosporin spot test to detect beta-lactamase in clinically significant bacteria. J Clin Microbiol. 1979 Feb;9(2):205–207. doi: 10.1128/jcm.9.2.205-207.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse S. A., Bartenstein L. Adaptation of the Minitek system for the rapid identification of Neisseria gonorrhoeae. J Clin Microbiol. 1976 Jan;3(1):8–13. doi: 10.1128/jcm.3.1.8-13.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhofer T. R., Back A. E. Biotypes of Haemophilus encountered in clinical laboratories. J Clin Microbiol. 1979 Aug;10(2):168–174. doi: 10.1128/jcm.10.2.168-174.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. J., Oberhofer T. R. Identification of pathogenic Neisseria species with the RapID NH system. J Clin Microbiol. 1983 Mar;17(3):400–404. doi: 10.1128/jcm.17.3.400-404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totten P. A., Amsel R., Hale J., Piot P., Holmes K. K. Selective differential human blood bilayer media for isolation of Gardnerella (Haemophilus) vaginalis. J Clin Microbiol. 1982 Jan;15(1):141–147. doi: 10.1128/jcm.15.1.141-147.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]


