Abstract
microRNAs (miRNAs) represent ∼4% of the genes in vertebrates, where they regulate deadenylation, translation, and decay of the target messenger RNAs (mRNAs). The integrated role of miRNAs to regulate gene expression and cell function remains largely unknown. Therefore, to identify the targets coordinately regulated by muscle miRNAs in vivo, we performed gene expression arrays on muscle cells sorted from wild type, dicer mutants, and single miRNA knockdown embryos. Our analysis reveals that two particular miRNAs, miR-1 and miR-133, influence gene expression patterns in the zebrafish embryo where they account for >54% of the miRNA-mediated regulation in the muscle. We also found that muscle miRNA targets (1) tend to be expressed at low levels in wild-type muscle but are more highly expressed in dicer mutant muscle, and (2) are enriched for actin-related and actin-binding proteins. Loss of dicer function or down-regulation of miR-1 and miR-133 alters muscle gene expression and disrupts actin organization during sarcomere assembly. These results suggest that miR-1 and miR-133 actively shape gene expression patterns in muscle tissue, where they regulate sarcomeric actin organization.
Keywords: microRNAs, muscle, target identification, zebrafish
microRNAs (miRNAs) are ∼22-nucleotide (nt) RNAs that control gene expression at the post-transcriptional level through various regulatory mechanisms, including messenger RNA (mRNA) deadenylation, translation, and decay of target mRNAs (Bushati and Cohen 2007; Filipowicz et al. 2008; Staton and Giraldez 2008). miRNAs play key roles in animal development where they regulate multiple cellular processes including cell fate specification, cell signaling, and tissue morphogenesis (Kloosterman and Plasterk 2006; Staton and Giraldez 2008). Despite the recent progress in defining the phenotypic functions of specific miRNAs, the mechanisms by which miRNAs regulate gene expression during different developmental processes are still poorly understood. A crucial step to elucidate the function of miRNAs is to identify their target mRNAs in vivo.
Recent studies have used two complementary strategies to systematically identify miRNA targets in animals; computational approaches involving nucleotide sequence and structural algorithms (Rajewsky 2006; Grimson et al. 2007; Long et al. 2007; Nielsen et al. 2007), and experimental approaches that examine changes in mRNA or protein expression patterns after manipulating the levels of individual miRNAs (Lim et al. 2005; Giraldez et al. 2006; Baek et al. 2008; Selbach et al. 2008). Computational approaches have identified between 100 and 200 putative targets per individual miRNA. These predictive methods assume considerable sequence conservation of miRNA complementary sites between orthologous genes and in particular between the structural features flanking the target site (Rajewsky 2006). However, estimates suggest that only ∼10%–20% of genes have targets that are conserved over a broad range of different animal species. Hence, the computational approach underestimates the ability of miRNAs to regulate gene expression through nonconserved targets.
Experimental approaches for identifying miRNA targets have included the production of miRNA gain and loss of function, both in cell culture (Lim et al. 2005) and in vivo (Giraldez et al. 2006), combined with either gene expression arrays or proteomics (Baek et al. 2008; Selbach et al. 2008) as read-outs to look for target mRNAs and proteins down-regulated by a given miRNA. These approaches can identify both evolutionarily conserved and nonconserved targets in an unbiased manner. Such studies have revealed ∼100 putative targets for several individual miRNAs, including miR-1, miR-124, miR-430, miR-155, and let-7 (Lim et al. 2005; Giraldez et al. 2006; Baek et al. 2008; Selbach et al. 2008).
While such experiments typically analyze the effect of manipulating a single miRNA, cells in any given tissue express a combination of several miRNAs. Furthermore, it is unclear how many of the putative targets identified in silico or in cell culture are actually regulated by miRNAs in vivo. Hence, developing an in vivo system to investigate the combined effect of miRNAs in cells within individual tissues could provide major insights into the role of miRNAs during tissue development and homeostasis.
Zebrafish embryos mutant for maternal and zygotic dicer function (MZdicer) are depleted of mature miRNAs (Giraldez et al. 2005). By comparing the gene expression profile of wild-type and MZdicer mutant embryos we were able to identify a large set of target mRNAs for the ubiquitously expressed miRNA miR-430 during early embryogenesis (Giraldez et al. 2006). Based on these findings, we hypothesized that analysis of the mRNA expression profile of a single tissue in wild type and MZdicer might provide a useful approach to identify a large set of tissue-specific targets in vivo.
Previous studies that combined target prediction methods and gene expression data have shown that target mRNAs tend to be present at low levels in domains expressing the cognate miRNAs (Farh et al. 2005; Lim et al. 2005; Stark et al. 2005; Sood et al. 2006). This results in quantitatively complementary expression patterns between the miRNAs and targets. Such results suggest a “mutual exclusion model” of miRNA regulation of gene expression (Stark et al. 2005; Bushati and Cohen 2007), wherein targets of the given miRNA are weakly transcribed or actively repressed in the tissues expressing that miRNA. In this model, the specific miRNA might support transcriptional repression, by ensuring repression post-transcriptionally. Alternatively, in an “instructive model,” miRNAs could shape gene expression patterns post-transcriptionally. In this scenario, the complementary expression pattern between the miRNA and its targets is mainly due to the accelerated degradation of the targets by the miRNA. However, few experiments have tested the ability of miRNAs to shape embryonic gene expression post-transcriptionally.
In the current study, we identify 245 target mRNAs that are post-transcriptionally regulated by muscle miRNAs. These targets tend to be expressed at lower levels in muscle compared with nonmuscle tissue. Two previously described muscle miRNAs, miR-1 and miR-133 (Sokol and Ambros 2005; Chen et al. 2006; van Rooij et al. 2008), appear to instruct embryonic muscle gene expression and to down-regulate these targets in muscle. We also identified a set of targets whose relative low muscle expression is miRNA-independent. These results suggest that two modes of target regulation coexist: one involving miRNAs to govern gene expression in muscle and the other that is primarily regulated at the transcriptional level and may be tuned by functional miRNA target sites. Furthermore, our gene ontology analysis of the muscle target mRNAs reveals that miR-1 and miR-133 regulate a number of actin-related and actin-binding proteins. Indeed, loss of Dicer or down-regulation of miR-1 and miR-133 altered muscle gene expression and disrupted actin organization during sarcomere assembly. Thus, miR-1 and miR-133 may actively shape gene expression patterns in muscle tissue, where they regulate sarcomeric actin organization.
Results
Identification of the muscle miRNA targets
To investigate the influence of miRNAs on muscle tissue, we first aimed to identify the muscle miRNA targets. Because miRNAs can accelerate target mRNA decay, we hypothesized that bona fide in vivo muscle miRNA targets would accumulate in the absence of muscle miRNAs. To identify the mRNAs that are up-regulated in the absence of miRNAs we integrated three experimental strategies. First, by making maternal-zygotic dicer mutants, we created embryos that were depleted of mature miRNAs (Giraldez et al. 2005). These mutants have gastrulation defects, which we rescued by injecting miR-430 at the one cell stage (MZdicer+miR-430) (Supplemental Fig. 1; Giraldez et al. 2005). Studying these rescued mutant zebrafish allowed us to focus on muscle-specific miRNAs. Next, to label muscle cells we crossed a muscle-specific transgene (α-actin GFP) into the wild-type and MZdicer mutant backgrounds (Tg: α-actin-GFP) (Fig. 1A; Higashijima et al. 1997). Third, we isolated muscle (GFP+) and nonmuscle (GFP−) cells from 24-h-old embryos using FACS. This enabled us to characterize the muscle gene expression profiles in wild type and MZdicer+miR-430 mutants (Supplemental Fig. 2).
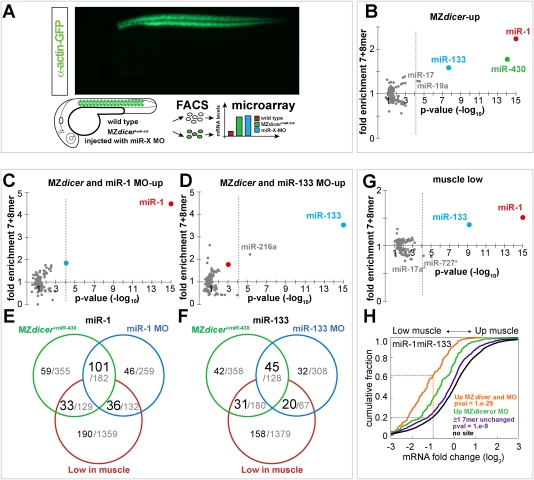
Figure 1.
Identification of the muscle miRNA targetome. (A) Identification of miRNA targets in zebrafish muscle. (Top right) Expression pattern of GFP-labeled muscle cells using the α-actin-GFP transgenic. GFP-labeled muscle cells (green, GFP+) and unlabeled nonmuscle cells (white, GFP−) were sorted by FACS for microarray analysis. Microarrays looked at wild-type (red), miRNA loss of function (green, MZdicer+miR-430), and embryos injected with single miRNA morpholino (blue, miR-X MO). (B–D,G) miRNA target site enrichment in different experimental conditions, as described in the header. The enrichment of 135 target sites compared with the control set (8mer + 7mer, Y-axis) and P-values (X-axis) is plotted. The P-value cutoff after Bonferroni multiple test correction (equivalent to 0.01) is shown as a dashed gray line. miR-1 and miR-133 target sites are shown with a red and blue dots, respectively (E,F). Venn diagram representing the different groups of genes present in the microarray. Each circle represents the number of genes up-regulated (up) in MZdicer+miR-430 (green), miRNA-MO (blue) or low in muscle (red). MZdicer and miRNA-MO analysis was performed using gene expression data from GFP+ cells. Low-in-muscle genes (G–F) were obtained by comparing their gene expression levels in GFP+ (muscle) to GFP− (nonmuscle), using a twofold cutoff. The number of genes with ≥7mer for miR-1 (E) or miR-133 (F) is shown in black. All genes present in each criterion are shown in gray. (H) Fraction of genes (Y-axis) and their relative expression in muscle versus nonmuscle (X-axis). (Orange) Genes with muscle miRNA target sites up-regulated (up) both in MZdicer+miR-430 and miRNA-MO experiment. (Green) Genes with muscle miRNA target sites up-regulated only in MZdicer or miRNA-MO experiment. (Purple) Genes with muscle miRNA target sites not up-regulated in the absence of muscle miRNAs. (Black) Genes that lack a 7mer seed for miR-1 or miR-133 in their 3′UTR. The dashed line represents the number of genes that are expressed at less than or equal to twofold (log2, −1) the level in muscle (GFP+) versus nonmuscle (GFP−). The P-value refers to the differences between the no site control and the orange or the blue line. Two-sided Mann-Whitney-Wilcoxon test was used to estimate the P-values.
Using Affymetrix gene expression arrays, we found that 907 mRNAs were up-regulated in MZdicer+miR-430 mutant muscle compared with wild-type muscle (≥1.3-fold, P-value ≤0.05, 5% false discovery rate [FDR]) (Materials and Methods; Supplemental Fig. 3). To determine whether these up-regulated mRNAs might be direct targets of muscle miRNAs, we analyzed whether specific miRNA target sites were overrepresented in this set. In zebrafish there are 337 known miRNAs corresponding to 135 distinct nucleotide seed sequences (Griffiths-Jones 2006). We searched the 3′ untranslated regions (3′UTRs) for all 7mer and 8mer sequences complementary to 135 zebrafish miRNA seeds (Fig. 1B; Materials and Methods; Lai 2002; Brennecke et al. 2005; Lewis et al. 2005). We initiated this analysis looking at the 3′UTRs because functional miRNA-binding sites in animals seem to be enriched in this portion of mature mRNAs (Giraldez et al. 2006; Grimson et al. 2007). We performed this analysis on all up-regulated mRNA that had experimentally identified 3′UTRs (666 out of 907). As a control, we used all mRNAs with an identified 3′UTR that were detected in the array (6825).
Analysis of the mRNAs that were up-regulated in MZdicer +miR-430 muscle compared with wild-type muscle, revealed a significant enrichment of the 7mers and 8mers complementary to just three particular miRNAs: miR-430, miR-1/206 and miR-133 (P-value <10−6; hereafter, all P-values in our analyses for target site enrichment were corrected for multiple testing using the Bonferroni correction) (Fig. 1B; Supplemental Figure 4D). Conversely, genes down-regulated in MZdicer muscle were not significantly enriched for any particular seed and were slighlty depleted of miR-1 target sites (Supplemental Fig. 5B). Previous studies have shown that miR-1/206 and miR-133 are expressed in muscle (Lagos-Quintana et al. 2003; Wienholds et al. 2005; Rao et al. 2006), whereas miR-430 is ubiquitously expressed (Giraldez et al. 2005). Although miR-430 duplex was injected at the one cell stage to rescue gastrulation defects in MZdicer, the enrichment of miR-430 seeds in the genes up-regulated in muscle suggests that this rescue did not persist during organogenesis. Next in our analysis, we aimed to distinguish between ubiquitous and muscle-specific miRNA targets. To do so, we focused just on the genes up-regulated in muscle (GFP+) but not outside the muscle (GFP−). The only motifs significantly enriched in genes up-regulated only in muscle were complementary to miR-1/206 and miR-133 (P-value <10−15 and <10−5, respectively) (Supplemental Fig. 5A). Indeed, 54% (176 out of 327) of the mRNAs up-regulated only in muscle had at least one 7mer site for miR-1 or miR-133, a frequency above that expected by chance, 19.1% (P-value <10−43). While muscle cells express other miRNAs, such as miR-214, and miR-216 (Wienholds et al. 2005; Flynt et al. 2007), our results suggest that miR-1/206 and miR-133 play a fundamental role regulating muscle gene expression during embryonic development.
To further investigate the contribution of miR-1/206 and miR-133 to muscle gene regulation, we interfered with the activity of each miRNA individually and repeated the above analysis. To inhibit each of the miRNAs, we injected a morpholino antisense oligonucleotide complementary to it (MO) (Supplemental Fig. 6; Leaman et al. 2005; Orom et al. 2006; Kloosterman et al. 2007). Since miR-1 and miR-206 have very similar sequence composition (18 of 22 nt) (Lagos-Quintana et al. 2003; Griffiths-Jones 2006) and expression patterns (Wienholds et al. 2005; Rao et al. 2006), hereafter we call this miRNA family miR-1 and the morpholino mix to inhibit it miR-1-MO (Supplemental Fig. 6G). Injecting antisense MO to miR-1 and miR-133 specifically inhibited expression and activity of the cognate miRNA (Supplemental Fig. 6 A–F). Motif analysis of the 3′UTRs of the mRNAs up-regulated in miRNA-MO-injected embryos revealed that a single seed sequence was enriched more than twofold in each experiment, corresponding to miR-1 and miR-133, respectively (P-value <10−44 and <10−14) (Supplemental Figs. 4D, 5C,D). To increase the specificity of our method, we pooled the results of the single miRNA morpholino and the MZdicer analysis. Genes up-regulated both in MZdicer +miR430 and single miRNA MO were enriched ∼4.4-fold and ∼3.5-fold for miR-1 and miR-133 target sites compared with the control set (P-value <10−40 and <10−12) (Fig. 1C,D). These criteria identified 101 miR-1 and 45 miR-133 putative targets (Fig. 1E,F).
miR-1 and miR-133 target mRNAs are expressed at lower level in muscle than in nonmuscle tissue
Previous studies using predicted miRNA targets suggested that miRNAs and their targets tend to be expressed in a mutually exclusive manner (Farh et al. 2005; Stark et al. 2005). To test this model, we compared the levels of miRNA target and nontarget mRNAs in wild-type muscle versus nonmuscle cells. Our analysis revealed the following: (1) The identified targets for miR-1 and miR-133 tend to be expressed at lower levels in muscle (GFP+) than nonmuscle (GFP−) (P -value <10−29, hereafter, all the cumulative plots were compared using two-sided Mann-Whitney-Wilcoxon test) (Fig. 1H). (2) Genes expressed at lower levels in muscle than in nonmuscle tissue (less than or equal to twofold, P-value ≤0.05) were enriched for miR-1 and miR-133 target sites (P-value <10−15 miR-1; P-value <10−6 miR-133) (Fig. 1G).
Based on these target site enrichments, we reasoned that including “low in muscle” as a criteria might allow us to identify additional targets that were below our initial cutoff in the MZdicer or single miRNA-MO experiments. Indeed, we found that genes whose expression is low in muscle and up-regulated in MZdicer +mir-430 or miRNA-MO, were enriched more than twofold for miR-1 and miR-133 target sites (Fig. 3A, below; Supplemental Fig. 5F–H). By these criteria, we defined 66 and 51 additional targets for miR-1 and miR-133, respectively (Fig. 1E,F). Moreover, this analysis revealed that (1) the relative expression levels in muscle versus nonmuscle reflect the activity of muscle miRNAs, and (2) genes expressed at low levels in muscle are enriched for muscle miRNA targets.
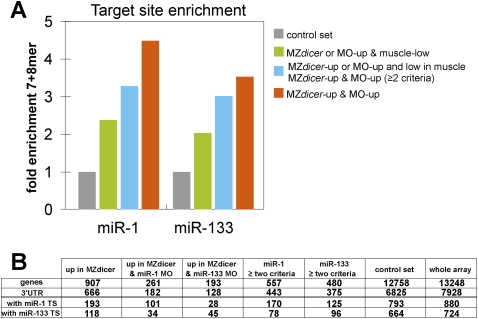
Figure 3.
Muscle miRNA targets are enriched for miR-1 and miR-133 target sites. (A) Fold enrichment for 7mer and 8mer sites for miR-1 or miR-133 in different groups shown on the right (colored bars) compared with the control set (gray bar). Target site frequency in the control group is set to 1. (B) Number of target genes with sites for miR-1 and miR-133 in each group. The columns summarize the number of genes, number of mRNAs with known 3′UTRs, and number of the 7mer and 8mer target sites for miR-1 or miR-133 found in the 3′UTRs for each criterion. The control set includes all the genes that were experimentally identified 3′UTRs and had a present call in the microarray.
Validation of miR-1 and miR-133 targets in vivo
To test whether the identified mRNAs are bona fide targets of miR-1 and miR-133 in vivo, we used three complementary approaches: (1) We tested whether miR-1 or miR-133 could regulate experimental targets engineered to contain putative miRNA target sequences. For this study, we cloned the 3′UTRs of 24 candidate targets downstream from firefly luciferase to create the reporter constructs. Each reporter mRNA was coinjected with a renilla luciferase control mRNA with or without the miRNA duplex (Fig. 2A). About 80% of the targets tested with at least one 7mer site were significantly repressed by the injected miRNA in vivo (P-value <0.05, n = 3) (Fig. 2A). For example, we validated 10 of 10 mRNAs with two or more seeds, six of six with one 8mer (100%) and four of six with one 7mer (67%). (2) Next, to test for direct miRNA-mediated regulation of the targets, we focused on miR-1 as an example, and mutated two nucleotides in the predicted target miR-1 site, going from CATTCC to CTATCC (miR-1 targets). In all of the cases tested (n = 4), mutation of the target site relieved miR-1-mediated repression (Fig. 2A). This suggests that most if not all validated 3′UTRs are likely to be directly regulated by the miRNA. (3) Finally, we tested whether the experimental targets are repressed by endogenously expressed miRNAs in muscle cells. We used a sensor construct with two promoters (De Pietri Tonelli et al. 2006); a reporter that expressed red fluorescent protein (RFP) mRNA with the miRNA target 3′UTR and a control reporter that expressed green fluorescent protein (GFP) mRNA lacking the miRNA target (Fig. 2B). In wild-type muscle cells, we observed repression of the RFP reporter compared with the GFP control. Nonmuscle cells expressed both GFP and RFP, consistent with the lack of expression of miR-1 and miR-133 in those cells (Fig. 2C–E). Conversely, injection of the cognate miRNA-MO disrupted miRNA-mediated repression of the RFP reporter by the endogenous miR-1 or miR-133 in muscle cells. Together, these experiments indicate that ∼80% of the mRNAs identified are likely to be bona-fide miR-1 and miR-133 targets in vivo.
Figure 2.
Muscle miRNA target validation. (A) Luciferase reporter assays to validate individual miR-1 and miR-133 targets. The reporter mRNA contains the Firefly luciferase (Fluc) ORF and the target 3′UTR sequence. Each reporter was coinjected with Renilla luciferase and buffer (gray bar) or a miRNA duplex (miR-1, red; miR-133, blue; both, violet). Fluc activity was normalized to Renilla luciferase and Fluc activity without miRNA duplex is set to 100%. Light-colored bar indicates repression by miRNA (P < 0.05, n = 3). Error bar shows SD. Gene name and number of target sites are shown below each bar; asterisks indicate targets expressed twofold lower in muscle versus nonmuscle. Dark gray bars (right), represent the level of repression when the seed sequences in the 3′UTR were mutated from CATTCC to CTATCC. Green bars show a positive control with three targets partially complementary to miR-1 or miR-133 (light green, IPTx3) and a negative control with no miRNA target sites (dark green, no site). (B) Dual miRNA reporter construct used to analyze the target regulation by endogenous miRNAs. A plasmid DNA containing GFP (control, green) and RFP-3′UTR (reporter, red) expressed from two SV40 promoters was injected into wild-type or miRNA-MO-injected embryos at the one-cell stage. Because injected DNA is inherited in a mosaic pattern, the dual reporter ensures that each cell that expressed GFP also expresses RFP. GFP and RFP expression in muscle was analyzed at 48 h post-fertilization (hpf) . (C–E) Control GFP expression (green) and reporter RFP expression (red) at 48 hpf. The panels show an enlarged view of somites. RFP expression is repressed in wild-type muscle cells (left) while the repression is abolished in the presence of miRNA-MO (right) (dashed boundaries). Arrowheads indicate muscle cells, which are elongated. Asterisks indicate nonmuscle cells. In all cases tested (n = 3), repressing the endogenous miRNA upon injection of the cognate miRNA-MO (+miR-1-MO or +miR-133-MO) abolished miRNA-mediated repression of the RFP reporter in muscle.
Defining the muscle miRNA targetome
The above analysis indicated that mRNAs that are up-regulated in the MZdicer/single miRNA knockdown animals (MO) or are expressed at low levels in muscle of wild-type zebrafish were enriched for miR-1 and miR-133 targets. Consistent with these mRNAs being enriched for bona fide targets, a fraction of the identified genes were validated in zebrafish embryos. Based on these results, we used two criteria to identify a set of muscle miRNA targets in vivo. The first criterion defined a nonredundant set of 143 mRNAs with 101 miR-1 and 45 miR-133 target mRNAs (Fig. 3B). These mRNAs included a target site for either miR-1 or miR-133 and were up-regulated in MZdicer +miR-430- and miRNA-MO-injected embryos. The second criterion selects “low in muscle” mRNAs that were up-regulated in MZdicer +miR-430- or miRNA-MO-injected embryos (Fig. 3B). This criterion defined additional 66 miR-1 and 51 miR-133 targets that combined resulted in 102 nonredundant set of target mRNAs. These groups were enriched between twofold and 4.5-fold for miR-1 and miR-133 target sites above the genomic background (Fig. 3A). Based on our target validation, these criteria defined 245 genes with miR-1 and miR-133 target sites that have ∼80% probability to be direct miR-1 and miR-133 targets termed the embryonic zebrafish muscle targetome (Supplemental Fig. 4C). This number is most likely an underestimate, in part because additional targets might be regulated only at the protein level. In addition, experimentally identified 3′UTRs were available only for approximately three out of four of the genes that fulfilled either of the two criteria, and the microarray covered only about half of the predicted genes in zebrafish. Taking these factors into consideration, we estimate that miR-1 and miR-133 likely regulate several hundred mRNAs during zebrafish muscle development (Supplemental Fig. 4C).
Muscle miRNAs regulate the spatial expression pattern of their target mRNAs
Having identified a set of mRNA targets for miR-1 and miR-133, we sought to define the role of these miRNAs in shaping the expression of target genes. Our results indicate that ∼63% of the identified targets in MZdicer- and MO-injected embryos are expressed at low levels in muscle cells (i.e., GFP+ cells) compared with nonmuscle cells (GFP−) (Fig. 4A; Supplemental Fig. 7A–I). Likewise, mRNAs with relatively low expression in muscle are enriched for miR-1 and miR-133 complementary sites. Two possible mechanisms might account for this inverse correlation between the expression of the miRNA and their target mRNAs. A prevailing view in the miRNA field suggests that differential expression of miRNAs and their targets occurs primarily by transcriptional repression of the targets (the mutual exclusion model; Stark et al. 2005). This model predicts that the expression bias of the targets would be maintained in the absence of miRNAs. In support of this model, we find that 779 mRNAs with putative miR-1 or miR-133 target sites are not up-regulated in MZdicer or morpholino backgrounds. Among these mRNAs, 24.5% tend to be expressed at lower levels in muscle versus nonmuscle when compared with the control set of genes that lack target sites for miR-1 or miR-133 (P-value <10−9) (Fig. 1H). Some of these mRNAs indeed have functional miRNA target sites in their 3′UTR, according to results of our luciferase analyses (Supplemental Fig. 8A). This suggests that genes in this set that are poorly expressed in muscle are predominantly regulated at the transcriptional level but have acquired functional miR-1 and miR-133 targets. Indeed, in situ analysis of some of these targets revealed that they are primarily expressed in nonmuscle tissue (Supplemental Fig. 8B–E), and their mRNA expression levels seem to be unaffected by the depletion of muscle miRNAs. In this context, the putative miRNA target sites may support transcriptional repression by repressing translation.
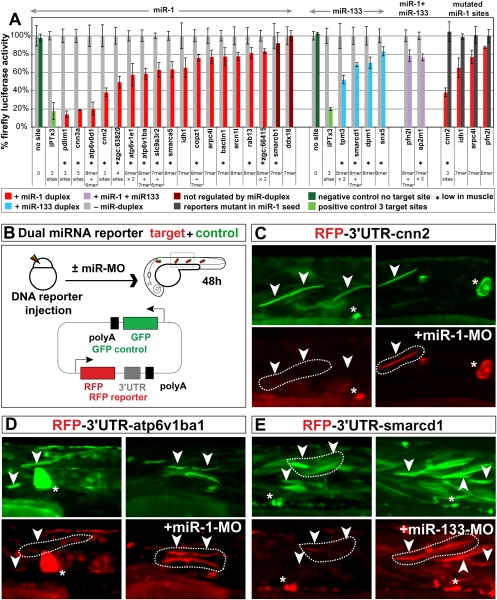
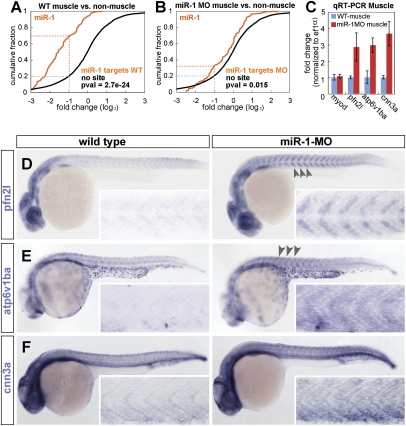
Figure 4.
Loss-of-muscle miRNAs affects the gene expression pattern of targets mRNAs. (A,B) Relative expression of miR-1 target and nontarget mRNAs in muscle versus nonmuscle tissue. Plotted is the fraction of genes (Y-axis) with a relative expression in muscle versus nonmuscle (X-axis) in wild-type (A), and miR-1 MO knockdown (B). Genes with muscle miRNA target sites up-regulated in both MZdicer+miR-430 and miR-1 MO experiments (orange). Genes that lack a 7mer target sites for miR-1 or miR-133 in their 3′UTR (black). Note that the expression bias of muscle miRNA targets (orange) toward low in muscle is strongly reduced when miR-1 is knocked down. (C) qRT–PCR of target mRNAs and a control mRNA (myoD) in muscle cells after FACS sorting of wild-type or miR-1 MO knockdown embryos. (D–F) In situ hybridization (purple) for miR-1 target genes in control MO-injected embryos (wild type) or miR-1 knockdown embryos (miR-1 MO). The insets show the enlarged view of the dissected muscle. Note the moderate increase in the expression levels of the targets when miR-1 is inhibited (arrowhead and insets).
A second model proposes that miRNA-mediated repression might constitute a primary mode of regulation to control gene expression patterns during development (the instructive model). In this model, the bias in target gene expression would be strongly reduced in the absence of miRNAs. To test this model, we first asked whether the expression bias of the muscle miRNA targets, toward low in muscle, was maintained in the absence of miRNAs. In wild-type embryos, a high percentage of the identified targets for miR-1 (∼63%) are less abundant in muscle than nonmuscle (less than twofold), while only 20% of the nontargets fulfill this criterion (P-value <10−24, two-sided Mann-Whitney-Wilcoxon test) (Fig. 4A). Next, we analyzed the relative expression of targets in muscle versus nonmuscle in embryos where miR-1 was knockdown. Inhibition of miR-1 revealed that the percentage of targets expressed at low levels in muscle was strongly reduced compared with the control set with no target sites for miR-1 or miR-133 (32% vs. 20% respectively, P-value 0.015) (Fig. 4B). The cumulative distribution for the miR-1 targets in wild type was significantly different (P-value <10−9) from that in miR-1 MO knockdown. Similar results were observed for the identified targets in MZdicer and miR-133 knockdown (Suplemental Fig. 7A–C). Interestingly, even after miRNA repression, the relative muscle expression was slightly weaker for targets compared with nontarget genes. This was shown by a slight displacement of the cumulative plot of the targets toward low in muscle compared with the control with no target sites (P-value <10−5). Second, we used miR-1 as a test case and analyzed the in situ expression pattern of 14 identified targets in wild-type and miR-1-MO-injected embryos. Blocking miR-1 function caused a significant increase in the muscle expression pattern of eight targets (Fig. 4D–F; Supplemental Fig. 10). For example, when miR-1 was inhibited, profilin 2-like, calponin 3, and atp6v1ba, were up-regulated in muscle as measured by quantitative RT–PCR (qRT–PCR) and in situ compared with wild-type control (Fig. 4C).
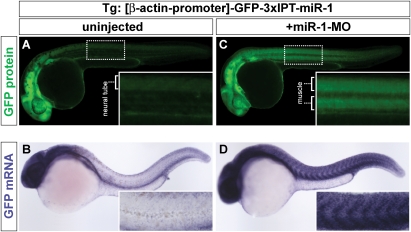
To test the instructive model directly, we examined the ability of miR-1 to shape the expression of GFP mRNA in a transgenic line expressing a GFP-miR-1 sensor. This sensor included a ubiquitous promoter (β-actin) (Higashijima et al. 1997), driving the expression of GFP mRNA with a 3′UTR containing three partially complementary target sites for miR-1 (3xIPT-miR-1) (Giraldez et al. 2005). Each target site is complementary to the seed and the 3′end of miR-1. It also includes three mismatches in the middle sequence that correspond to nucleotides 8–10 of the mature miR-1 and disrupt the slicer cleavage site (Giraldez et al. 2005). Despite GFP mRNA being expressed from a ubiquitous promoter, wild-type transgenic embryos showed low GFP mRNA and protein levels in the muscle tissue, and high expression levels in nonmuscle tissue (i.e., brain, neural tube, pronephros, etc.) (Fig. 5A,B). Interfering with miR-1 function by injecting the miR-1-MO at the one-cell stage caused a strong up-regulation of GFP mRNA and protein levels in muscle, comparable with the levels of the transgene in other tissues (Fig. 5C,D). These results demonstrate that blocking miR-1 function can alter the expression of its target, supporting a role for miRNAs in shaping the gene expression patterns of their targets post-transcriptionally, like transcription factors do at the transcriptional end.
Figure 5.
miR-1 shapes the embryonic expression pattern of a ubiquitously transcribed miR-1 sensor mRNA. (A–D) Whole mounts of control noninjected (A,B) or miR-1 MO knockdown (C,D) embryos at 30 hpf, expressing a GFP sensor transgene with three partially complementary targets for miR-1 (3xIPT-miR-1). GFP is expressed from the ubiquitously expressed promoter of the β-actin gene (β-actin-promoter). (A,C) Show fluorescent expression of the GFP protein (green). (B,D) Show expression levels of the GFP-mRNA (blue). The inset shows an enlarged view of the muscle. (C) The GFP sensor protein and mRNA observed in the trunk of noninjected embryos corresponds primarily to nonmuscle tissue (i.e., neural tube, the vasculature, the pronephros, and the skin) and is excluded form muscle. (D) Note that blocking miR-1 function leads to a strong up-regulation of the GFP mRNA levels in muscle.
Based on these results we propose that two modes of transcript regulation coexist. One set of targets is transcribed at lower levels in muscle than nonmuscle, independently of miRNA function. A second set of targets is strongly influenced by muscle miRNAs that regulate their degradation, shaping gene expression.
A subset of miR-1 and miR-133 targets regulate actin function
To determine whether the 245 genes that we identified in the muscle miRNA targetome coordinately modulate specific cellular processes in the muscle, we analyzed their Gene Ontology (GO) enrichment (DAVID bioinformatics resource) (Dennis et al. 2003). To this end, we compared the actual number of targets present in each GO category with the expected number, based on the frequency in the control set, using Fisher's exact test. The control set included all expressed transcripts with an identified 3′UTR that passed the quality control of the microarray (6825 genes).
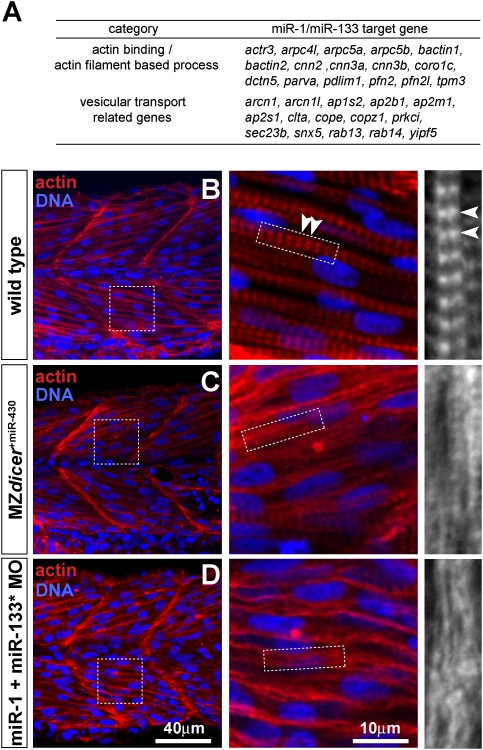
This analysis revealed that the “instructive targets” (245 genes) were significantly enriched for mRNAs encoding actin filament-based processes (GO:0030029, 6.9-fold, P-value 10−5) and vesicle transport (GO:0016192, fivefold, P-value 10−7) (Supplemental Figure 9A). In contrast, the “mutually exclusive transcriptional” targets were enriched in a variety of GO categories such as system development and organ development (data not shown). The differential GO enrichment in each set suggests that instructive and mutually exclusive targets may differ not only in their dependency on miRNAs for their regulation but also in their molecular function. Two types of evidence suggest that these GO enrichments were specific to the targets. First, those genes that fulfilled two of three criteria similar to the targets (up-regulated in the MZdicer, up-regulated in morpholino and low in muscle) but lacked miR-1 or miR-133 sites were mainly enriched for DNA replication and DNA metabolism compared with the control set. Second, the mRNAs that were expressed in muscle at low levels showed GO enrichment that was different from the target genes and involved primarily functions such as DNA replication. Among the actin-binding and actin-related genes that we identified in the instructive target set were profilin 2, profilin 2-like, calponin 2, calponin 3a, calponin 3b, tropomyosin 3, and coronin 1c, as well as actin-related proteins in the ARP2/3 complex such as arpc5a and arpc5b (Fig. 6A). Interestingly, there was a partial overlap between human and zebrafish targets, including calponin 3, profilin 2, coronin1c and tropomyosin (Supplemental Fig. 9D) The enrichment of actin-related and vesicular transport genes in the muscle miRNA targets supports the concept that these two processes may be governed by muscle miRNAs during embryonic development.
Figure 6.
Loss-of-muscle miRNAs disrupts sarcomeric actin organization. (A) Table showing the miR-1 and miR-133 target genes involved in actin-binding/actin-filament based processes and vesicular transport. (D) Muscle α-actin immunostaining (red) in control-MO, MZdicer+miR-430 and muscle miR-1 + miR-133 MO-injected embryos at 28 hpf. Left panels show actin staining of the fast muscle layer (10th somite). Dashed rectangles show insets magnified on the right. DNA is shown in blue. Note that actin staining that corresponds to the I-bands in the sarcomeres of control embryos (arrowheads) is disrupted in the fast muscle of MZdicer+miR-430 and miR-1/miR-133 knockdown embryos.
miR-1 and miR-133 regulate actin organization in the sarcomere
Actin assembly plays a crucial role in the formation of the sarcomere, the functional unit of the muscle; thus, we focused our analysis on the role of miR-1 and miR-133 in regulating sarcomeric actin. To test whether actin organization depends on miRNA function, we analyzed sarcomere assembly in embryos depleted of muscle-specific miRNAs. First, we compared the actin immunostaining in the skeletal muscle of MZdicer +miR-430 and wild-type embryos. In contrast to the regular array of actin that defines the I-bands in wild-type muscle (Fig. 6B, arrowhead), MZdicer +miR-430 mutants showed disorganized actin filaments and loss of the I-bands in fast skeletal muscle (Fig. 6C). Second, to test whether the actin disorganization was caused by depletion of mature miR-1 or miR-133, we injected miR-1-MO and miR-133-MO at the one-cell stage. Knockdown of either miR-1 or miR-133 caused a reduction in the fiber size and had a mild effect in the actin bands (Supplemental Fig. 11). Interestingly, knockdown of both miR-1 and miR-133 disrupted sarcomeric actin organization in fast muscle, as evidenced by the loss of I-bands of actin (Fig. 6D). Loss of miR-1/133 did not disrupt Z-lines, as visualized by actinin staining (Supplemental Fig. 12). This suggests that actin disorganization is likely a primary effect of muscle miRNA loss of function, rather than secondary consequence of sarcomere disorganization. Taken together, these results suggest that miR-1 and miR-133 regulate actin organization during sarcomere assembly.
Discussion
Our study of the muscle miRNA targetome in vivo provides four major insights into miRNA function. First, miR-1/206 and miR-133 are the two muscle miRNAs with the strongest influence on gene expression in embryonic muscle. Second, these muscle miRNAs shape the embryonic expression pattern of over a hundred target mRNAs whose levels are up-regulated in the absence of miR-1 and miR-133. Third, miR-1 and miR-133 target mRNAs are enriched for actin-related, actin-binding, and vesicle transport gene functions. Fourth, interfering with the function of miR-1 and miR-133 disrupts sarcomeric organization of actin in fast skeletal muscle. Taken together our results identify a set of embryonic muscle miRNA targets that are enriched for actin-binding and actin-related genes, providing a possible causal relationship between failure to regulate these transcripts and the actin organization defects in the sarcomeres of MZdicer mutants.
Our analysis indicates that ∼54% of the transcripts up-regulated in the absence of muscle miRNAs have a 7mer or 8mer for either miR-1 or miR-133. An additional 14% has at least one 6mer site for these miRNAs. The remaining mRNAs up-regulated in MZdicer muscle might lack canonical miR-1 or miR-133 seeds, be regulated by additional muscle miRNAs like miR-216, or be secondary targets of miR-1 and miR-133. These results suggest that miR-1 and miR-133 are likely to account for a large fraction of the miRNA-mediated regulation that takes place in embryonic muscle. The identified muscle miRNA targets tend to be expressed at relatively low levels in muscle compared with nonmuscle tissue. Inversely, genes expressed at low levels in muscle are enriched for muscle miRNA target sites. The mRNA expression levels in one tissue (i.e., muscle) reflect the impact of miRNA activity in that tissue (i.e., miR-1 and miR-133 in muscle) (Farh et al. 2005; Stark et al. 2005; Sood et al. 2006). Therefore, comparing the mRNA expression levels in the tissue of interest versus reference material—for example, tissue from a previous developmental stage (Giraldez et al. 2006) or a set of other tissues (i.e., nonmuscle cells)—in order to find those mRNAs that contain the relevant miRNA targets and that are expressed at low levels in the tissue of interest provides a strategy to discover the miRNAs that have substantial impact in that tissue (Farh et al. 2005; Sood et al. 2006). Such comparisons can also identify a set of mRNAs enriched for bona fide miRNA targets, even if miRNA loss-of-function experiments cannot be easily undertaken.
Previous studies based on target prediction and miRNA misexpression in vitro identified many targets that are absent or tend to be expressed at relative low levels in muscle (Farh et al. 2005; Lim et al. 2005; Stark et al. 2005). The predominant interpretation of such findings has been that many miRNAs and their mRNA targets show mutual exclusion in their expression pattern, mainly due to transcriptional repression (Bushati and Cohen 2007). In this “mutual exclusion” model, miRNAs have been proposed to serve a fail-safe mechanism to support regulation at the transcriptional level (Farh et al. 2005; Stark et al. 2005; Hornstein and Shomron 2006). While this mechanism seems to hold for a subset of the target mRNAs, we find that muscle miRNAs (1) strongly influence the expression bias of a large set of targets that are “low in muscle,” (2) shape the spatial expression pattern of half of the targets analyzed, and (3) can instruct the embryonic expression pattern of an ubiquitously transcribed target gene. These results indicate that miRNAs can also actively shape gene expression patterns during development.
Therefore, the mutual exclusion end state may result from two regulatory mechanisms operating simultaneously, one functioning predominantly at the transcriptional level and a second that is deeply influenced by miRNAs acting post-transcriptionally. As a set, the so-called instructive targets might be more directly connected to the function of the miRNA in physiological conditions, because they require muscle miRNAs to determine their expression levels in that tissue. Indeed, transcriptional and instructive targets may differ not only in their dependence on miRNAs for their regulation, but also in their functions. Consistent with this notion, Gene Ontology analysis suggested that instructive targets are enriched for distinct gene functions (actin- and vesicular transport-related) that differ from those regulated at transcriptional level (system development and organ development). Our results suggest that dissecting the different regulatory modes of the targets provides an important step to understand how miRNAs regulate cell function.
What advantages would favor a post-transcriptional regulation in the context of a tissue? Previous studies indicate that miRNAs play an important role in clearing the transcriptional legacy of an earlier developmental stage (Lee et al. 1993; Reinhart et al. 2000; Rhoades et al. 2002; Bartel and Chen 2004; Farh et al. 2005; Giraldez et al. 2006). In addition, it has been proposed that miRNAs might reduce transcriptional noise (Hornstein and Shomron 2006; Tsang et al. 2007). Our results suggest two additional contexts in which miRNAs might function. First, tissue-specific miRNAs can shape transcriptional outputs through differential regulation of actively transcribed genes. For instance, two different tissues exposed to the same signal or expressing the same transcription factor during development could transcribe the same set of genes. Tissue-specific miRNAs might shape gene expression post-transcriptionally by reducing the transcriptional output of a specific transcription factor or signaling pathway (Supplemental Fig. 16A). In a second scenario, miRNAs could regulate transcript homeostasis. Historically, transcription has been viewed as the main determinant of the mRNA levels in the cell while degradation is seen in many cases as a default steady-state process. However, miRNAs accelerate target deadenylation and degradation (Bagga et al. 2005; Lim et al. 2005; Giraldez et al. 2006; Wu et al. 2006). Gain or loss of miRNA target sites in the 3′UTR can have a dramatic influence on the degradation rate and is likely to provide a fundamental mechanism to control the steady state levels for hundreds of transcripts (Supplemental Fig. 16B). Our results support a model in which miRNAs dictate gene expression patterns and mRNA homeostasis post-transcriptionally.
In addition to presenting a global picture of miRNAs in muscle, our studies suggest a critical role for miR-1/miR-133 in regulating actin organization in the sarcomere of the fast skeletal muscle. While early studies have emphasized the role of miRNAs during differentiation (Chen et al. 2006), miRNAs as a class are expressed throughout the life of the organism, implying that members of this RNA class regulate specific cellular properties and tissue physiology (Poy et al. 2004; Krutzfeldt et al. 2005; Staton and Giraldez 2008; van Rooij et al. 2008). At present we cannot exclude the possibility that the phenotype observed might be due to the combined misregulation of targets affecting different biological processes. However, loss of Dicer function or knockdown of miR-1/206-miR-133 results in the misregulation of actin-binding and actin-related proteins along with the disorganization of sarcomeric actin. Interestingly, loss of dicer function in the heart leads to dilated cardiomyopathy and heart failure, and is also associated with defects in sarcomere structure (Chen et al. 2008). These results suggest that miRNA-mediated regulation of sarcomeric organization is likely conserved across vertebrates. Indeed, actin filaments must be tightly regulated in muscle cells. For example, sarcomere formation requires the assembly of linear actin, and muscle cells express specific actin nucleation factors such as Leiomodin (Chereau et al. 2008). Our finding that muscle miRNAs may regulate mRNAs for profilin 2, profilin 2-like, components of the arp2/3 complex such as arpc5a/b and coronin 1c is consistent with a model in which miR-1 and miR-133 down-regulate proteins that promote actin filament branching and cross-linking activity (Pollard 2007) to facilitate the assembly of linear actin to form thin filaments.
miRNA-mediated regulation of actin dynamics might be important not only during development but also during disease. Recent studies have shown that a member of the miR-1 family (miR-206) can inhibit tumor metastasis in vivo (Tavazoie et al. 2008). Given that a set of targets for miR-1/206 regulates the actin cytoskeleton, and given the important roles of actin dynamics and branching during cell migration, it is tempting to speculate that miRNA-mediated regulation of actin function might account at least in part for the inhibitory activity of miR-206 on metastatic tumors. Future experiments will be needed to uncover the molecular mechanisms underlying the regulatory potential of muscle miRNAs during metastatic migration and sarcomeric actin assembly.
Materials and methods
Zebrafish strains
MZdicer mutant embryos were generated by germline replacement as described previously (Giraldez et al. 2005). Early developmental phenotypes in MZdicer were rescued with miR-430 duplex injection (Giraldez et al. 2005). This allows us to reduce secondary defects that might appear in the muscle tissue due to early gastrulation phenotypes in MZdicer mutants. Tg: α-actin GFP embryos were obtained by crossing either Tg: α-actin GFP homozygous or heterozygous fish (Higashijima et al. 1997) with wild-type fish . To introduce α-actin GFP transgene into the MZdicer background, females transplanted with dicer homozygous germ cells (hu896/hu896, hu715/hu715, or hu715/hu896) were crossed with α-actin GFP/+; dicer hu715 or hu896/+ males. MZdicer +miR-430; Tg: α-actin GFP/+ embryos were identified by measuring GFP expression and developmental delay compared with Mdicer +miR-430 in the same batch at 24 h post-fertilization (hpf).
Tg: β-actin-GFP-3xIPT-miR-1 contain the ßactin promoter that drives expression ubiquitously; the ORF of GFP and three partially complementary sites for miR-1 (3xIPT; one target site: TACATACTTCTaatCATTCCAtagctaa) as described in Giraldez et al. (2005). Five independent transgenic lines were isolated, and all of them repressed GFP expression in muscle. As described previously (Higashijima et al. 1997), other transgenic lines using the same promoter that lacked miR-1 target sites were not repressed in muscle (A. Staton, unpubl.).
FACS analysis and RNA isolation
Tg: α-actin GFP embryos were collected at 24–25 hpf for wild type. Tg: α-actin GFP;MZdicer +miR-430 embryos were collected at 28–29 hpf to compensate developmental delay. Yolk was removed by pipetting in Ca2+-free Ringer's solution. Embryos were then incubated in 0.25% trypsin and 1 mM EDTA/PBS for 50 min at 28°C with frequent pipetting. Dissociated cells were collected by centrifuging at 3000 rpm for 2 min, washed with suspension medium (Leibovitz medium L-15 [Gibco] with 0.5 mM EDTA and 1% calf serum), and resuspended in the same medium. To distinguish live cells and dead cells, Propidium iodide (Sigma) was added at final concentration 0.01 mg/mL and incubated for ≥10 min. Cell sorting was performed at Yale University Cell Sorter Facility using Dako MoFlo. Purity of the sorted cells was tested by resorting GFP+ and GFP− cells after first sort. The purity was 92%–96%. For total RNA isolation, cells were directly sorted into Trizol. Total RNA was isolated from ∼100,000 cells and purified by RNeasy mini elute kit (Qiagen). RT–PCR was performed using primers for ef1a, myoD, pri-miR-206-1, fli1a, and huC.
Luciferase reporter constructs
The ORF of firefly luciferase (Fluc) was cloned between BamHI–StuI sites of pCS2+ (pCS2 + Fluc). 3xIPT miR-1 was excised from plasmid GFP miR-1 3xIPT (Giraldez et al. 2006) by XhoI–NotI enzyme digestions and inserted into XhoI–NotI site of pCS2 + Fluc. 3xIPT miR-133: Oligonucleotides 1a and 1b and 2a and 2b were annealed, phosphorylated, and cloned into the XhoI–XbaI site of pCS2 + F-luc by three-fragment ligation. 3′UTR sequences: The 3′UTRs of the gene of interest were amplified by RT–PCR from a cDNA library made from 0- to 36-h embryos. The sequences of 3′UTR and primers used are shown in the document called Mishima-targets-tested.doc. The PCR fragments were digested and cloned into pCS2 + Fluc between XhoI–XbaI site. Mutant luciferase reporter: The 3′UTR of the gene of interest was amplified in two fragments. These two fragments had a ∼35-nt overlap in the mutant region. The mutation in the miR-1 target site (CATTCC to CTATCC) was included in the bottom primer of the 5′ fragment and the top primer of the 3′ fragment. The full-length 3′UTR was obtained by PCR of the fragments one and two with the 5′ primer of fragment one and the 3′ primer of the second fragment.
Dual miRNA reporter constructs
Dual miRNA reporters containing DFRS-control and DFRS-miR-1 were obtained from Dr. Davide De Pietri Tonelli (De Pietri Tonelli et al. 2006) and modified as follows: First, oligonucleotides 3a and 3b, containing multiple cloning sites (XhoI–SalI–NheI), are annealed and inserted into EcoRI–NotI site of the DSFR plasmid. Second, the BglII–ScaI fragment of the DFRS plasmid was inserted into the XhoI–BglII site of pT2AL200R10G (Urasaki et al. 2006) to make the DFRS construct with tol2 sequence. miR-133 PTx2 sequence was cloned into XhoI–NheI site by annealing oligonucleotides 4a and 4b. 3′UTR sequences were excised from pCS2 + Fluc-3′UTR with XhoI–XbaI, and cloned into XhoI–NheI sites.
Microinjection
MOs directed against mature miRNA sequence (MO miR-X) or miRNA* sequence (MO miR-X*) were purchased from Gene Tool. MOs are dissolved in water and injected into one-cell-stage embryos. Concentrations were as follows: MO miR-1/206 mixture, 1 mM each; MO miR-1*/206* mixture, 1 mM each; MO miR-133, 0.25 mM; MO miR-133*, 1 mM; MO miR-1/206/133* mixture, 0.83 mM each. As a control experiment, a control MO from GeneTool was injected at same concentration to each MO. Approximately 1000 pL were injected.
For the dual miRNA reporter assay, a mixture of 50 ng/μL DNA and 50 ng/μL tol2 transposase mRNA was injected. MO was injected into half of those embryos at same one-cell stage. Injected volume was ∼1000 pL. miR-430 duplex injection was performed as described previously (Giraldez et al. 2005). For mRNA synthesis except for Rluc mRNA, template DNA was cut by NotI and mRNA was synthesized by mMessage mMachine SP6 kit (Ambion). For Rluc mRNA, template plasmid pRL-CMV was cut by BamHI and transcribed by T7 polymerase with the 2xNTP/CAP mixture provided with mMessage mMachine SP6 kit.
Target validation by luciferase assay
Zebrafish embryos were collected at the one-cell stage and injected with 100 pL of reporter/control mix that included firefly luciferase mRNA and renilla luciferase mRNA mixture (10 ng/μL each) using the same needle and settings. One-half of these embryos were injected with 1000 pL of miRNA duplex. Concentrations were as follows: 5 μM each for miR-1 and miR-206 duplex, 50 μM for miR-133 duplex. miR-1 and miR-206 duplexes were purchased as siRNA duplex, while miR-133 were purchased as ssRNA and annealed (IDT). In the case of miR-1/miR-133 coinjection, a mixture of 5 μM miR-1 duplex and 25 μM miR-133 duplex was used. Five to 10 embryos were collected at 9 hpf and homogenized in 50 μL of Passive Lysis Buffer (Promega). Luciferase assays were performed using Dual-Glo luciferase Assay System (Promega) and Modulus (Turner Biosystems) according to the manufacturer's instructions. After subtracting background, Fluc activity intensity (IFluc) was normalized by Rluc activity intensity (IRluc). The fold change expression of the reporter with and without the miRNA duplex was calculated as follows: Fold change = (IFluc + miRNA/IRluc + miRNA)/(IFluc − miRNA/IRluc − miRNA).
The luciferase ratio IFluc-miRNA/IRluc-miRNA was normalized to 100% in Figure 2. Experiments were repeated at least three times for each reporter, and P-value was calculated by Students’ t test.
In situ hybridization
In situ hybridization was performed as described previously (Mishima et al. 2006), with several modifications. To detect mature miRNAs, digoxigenin-labeled LNA probes (Exiqon) were used. For comparative in situ hybridization, wild-type or MO-injected embryos were marked, either by clipping the tip of tail, or by injecting GFP mRNA. After being fixed with 4% PFA and stored in methanol at −20°C, paired wild-type and MO-injected embryos were hybridized and developed in the same well/tube during the whole procedure, to avoid any possible technical differences. In the case of the embryos that were injected with GFP mRNA, GFP was detected by fluorescent immunostaining after developing the in situ hybridization. Embryos were cleared in benzyl benzoate/benzyl alcohol before they were photographed using a Zeiss Axioimager M1.
Immunostaining
Twenty-eight-hour-post-fertilzation embryos were fixed in 4% PFA for 2 h. Primary antibodies were used at following concentrations: MF20 1:50, A4.1025 1:50 (Developmental Studies Hybridoma Bank), anti-cardiac actin 1:10 (Progen), anti-actinin 1:250, anti-GFP rabbit 1:250 (Sigma). DNA was stained with Topro-3 (Invitrogen). Embryos were cleaned in 70% glycerol. Images were taken by Zeiss LSM 510 meta.
Microarray analysis
Total RNA was isolated from ∼10,000 sorted cells using Trizol (Invitrogen) and RNeasy mini elute (Qiagen). All microarrays were performed in triplicate except the GFP− from embryos injected with miR-1 or miR-133 that were done in duplicates. Quality of RNA was ensured before labeling by analyzing 5 pg of each sample using the RNA 6000 picoAssay and a Bioanalyzer 2100 (Agilent). Samples with a RNA Integrity Number (RIN) >7.0 were considered suitable for labeling. For samples meeting this standard, 20 ng of total RNA were labeled using the GeneChip two-cycle target labeling kit (Affymetrix). Ten micrograms of labeled and fragmented cRNA were then hybridized to the Zebrafish Genome Array (Affymetrix), for 16 h at 45°C. Automated washing and staining were performed using the Affymetrix Fluidics Station 400 according to the manufacturer's protocols. Finally, chips were scanned with a high-numerical aperture and flying objective (FOL) lens in the GS3000 scanner (Affymetrix). Raw expression data were analyzed using GCOS 1.4 (Affymetrix).
Microarray data processing
Raw expression CEL files were processed using R and BioConductor (Gentleman et al. 2004). Muscle (GFP+) and nonmuscle (GFP−) samples were separately normalized using RMA (Bolstad et al. 2003), followed by a robust quantile normalization of the log2 expression values. We used limma to fit linear models to every probe set, using an empirical Bayes approach to shrink the estimated variance (Smyth 2004). Each type of sample was treated as a factor, and since the arrays were run on two separate occasions, we added an extra factor to take this batch effect into account. MAS 5.0 absolute detection calls (Smyth 2004) were calculated for each probe set from the raw data, and the P-values for replicate samples were combined using Stouffer's inverse normal method (Stouffer et al. 1949). Probe sets that did not have at least one sample detected with P < 0.01 were considered absent from the study and were removed prior to adjusting for multiple testing. Differential expression P-values were adjusted for multiple testing (Benjamini and Hochberg 1995), and a cutoff corresponding to a 5% FDR was used in all cases.
Target analysis
Sylamer miRNA seed-enrichment analysis
The genes called present on the arrays that have a 3′UTR were ranked by fold change from up to down-regulated when comparing miR-1-MO, miR-133-MO, MZdicer +miR-430 and WT_GFP− against WT_GFP(+). We used van Dongen et al. (2008) to test each of these ranked gene lists for hypergeometric enrichment and depletion of all zebrafish miRNA-related 7mer words at 50 equally spaced fold change cutoffs. We confirmed that each experiment presented only the expected miRNA signals in the 3′UTR sequences. The enrichment curves were inspected to select the fold change cutoff that optimized the appropriate miRNA enrichment. We also tested coding sequences, using the same gene ranking showing that enrichment, if any, is barely significant. To avoid low-complexity and repetitive-sequence biases, all sequences were first dusted and purged as described in van Dongen et al. (2008). Since nucleotide composition can vary across the gene lists, we used a Markov model to correct for the observed frequency of words of length 4 at each cutoff, as recommended (van Dongen et al. 2008).
Target site enrichment
3′UTR sequences were obtained for all the genes in the array when available from Refseq or Ensemble transcripts with EST evidence. The control set used in this study contains all the genes with 3′UTR information and present call in at least one of the arrays. Up-regulated genes in MZdicer+ miR-430 muscle cells and MO-injected muscle cells fulfilled following criteria: (1) They have a present call in at least one experimental condition. (2) Their fold change compared with wild-type muscle cells is >1.3-fold (MZdicer and miR-133 MO) or 1.5-fold in miR-1 MO. (3) Their P-value is ≤0.05, leading to an estimate FDR of ∼5%. The fold change cutoff was selected using the sylamer plots to maximize the P-value for the top three seeds identified in the up-regulated genes by the sylamer algorithm. Up-regulated genes in MZdicer +miR-430 nonmuscle cells were identified similarly, but by comparing MZdicer +miR-430 nonmuscle cells to wild-type nonmuscle cells. Relative expression levels between muscle cells and nonmuscle cells were obtained by comparing wild-type muscle and nonmuscle cells. Genes expressed higher and lower in muscle cells compared with nonmuscle cells fulfilled the following criteria: (1) They have a present call in at least one experimental condition. (2) Their expression level in GFP+ is more than twofold the expression in nonmuscle cell. (3) Their P-value is ≤0.05, leading to an estimate FDR of ∼5%. Sequences complementary to each of the 135 unique miRNA seed sequences (8mer, 7mer-8m, 7mer-t1A, 6mer) were searched for in the 3′UTR of each group (NmiR-x) and in control set (CmiR- x). The total number of miRNA target site in a given set of 3′UTR was calculated by adding all 135 miRNA target sites (NmiR-total and CmiR-total). Fold change of miRNA target site frequency was calculated as (NmiR-x/NmiR-total)/(CmiR-x/CmiR-total). P-values for each miRNA target site was calculated by Fisher's exact test and corrected by Bonferroni's multiple test correction. miRNA target sites used in this study are provided in the file called Mishima-seed.xls.
Cumulative plots
The logFC values for control and target genes in all contrasts were first quantile normalized to make them comparable. A two-sided Mann-Whitney-Wilcoxon test was used to estimate the P-values.
Target gene identification
Genes with miR-1 and miR-133 target sites were identified by searching for 8mer target sites (miR-1; ACATTCCA, miR-133 GGACCAAA) or 7mer target sites (miR-1; ACATTCC and CATTCCA, miR-133; GGACCAA and GACCAAA) in the 3′UTRs. From these genes, miRNA target genes were up-regulated in MZdicer or miRNA-MO knocked down and were expressed lower in muscle compared with nonmuscle cells with the following criteria: Either (1) up-regulated (>1.3-fold) in MZdicer muscle cells compared with wild-type muscle cells, or (2) up-regulated in muscle cells of MO-injected embryos compared with muscle cells from wild-type embryos (>1.3-fold in miR-133-MO or >1.5-fold for miR-1 MO); (3) lower than twofold in muscle cells compared with wild-type nonmuscle cells, and (4) their P-value is ≤0.05, leading to an estimate FDR of ∼5%. Target gene lists are provided as Mishima-targets.xls.
Acknowledgments
We thank W. Huttner and D. De Pietri for the dual GFP-RFP reporter construct; K. Kawakami for the tol2 vector; and T. Pollard, S. Weatherbee, M. Hammarlund, M. Khokha, V. Greco, J. Noonan, R. Yi, H. Knaut, C. Takacs, D. Cifuentes, and B. Schachter for discussions and comments on the manuscript. We thank Robert Andrews for suggesting Stouffer's method for combining P-values. We thank the MSKCC for the microarray analysis and the FACS facility at Yale. We thank T.H. Kim for his advice during qRT–PCR analysis. Y.M. was supported by a JSPS post-doctoral fellowship for research abroad. A.J.G. is a Pew Fellow Lois and Franklin Top Yale Scholar. This project was funded by the Pew foundation, The Yale Scholar Program, and the Muscular Dystrophy Association. Individual contributions were as follows: Y.M. and A.J.G. designed the project; Y.M. performed the experiments, microarray analysis, and target analysis. Y.M. and A.J.G. wrote the paper. A.A.S. and C.S. generated the miR-1 reporter transgenic and analyzed their expression. A.J.E. and C.A.G. did the microarray analysis, sylamer plots, and initial target site enrichment, 3′UTR data set, and Gene ontology analysis. C.S., C.C., and M.G. performed PPIN analysis and GO analysis.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1760209.
Supplemental material is available at http://www.genesdev.org.
References
- Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel D.P., Chen C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. microRNA Functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.F., Murchison E.P., Tang R., Callis T.E., Tatsuguchi M., Deng Z., Rojas M., Hammond S.M., Schneider M.D., Selzman C.H., et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc. Natl. Acad. Sci. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D.B., Rebowski G., Lappalainen P., Pollard T.D., Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- De Pietri Tonelli D., Calegari F., Fei J.F., Nomura T., Osumi N., Heisenberg C.P., Huttner W.B. Single-cell detection of microRNAs in developing vertebrate embryos after acute administration of a dual-fluorescence reporter/sensor plasmid. Biotechniques. 2006;41:727–732. doi: 10.2144/000112296. [DOI] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Flynt A.S., Li N., Thatcher E.J., Solnica-Krezel L., Patton J.G. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: The microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S., Okamoto H., Ueno N., Hotta Y., Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- Hornstein E., Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Lagendijk A.K., Ketting R.F., Moulton J.D., Plasterk R.H. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs.’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Meyer J., Borkhardt A., Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Leaman D., Chen P.Y., Fak J., Yalcin A., Pearce M., Unnerstall U., Marks D.S., Sander C., Tuschl T., Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Long D., Lee R., Williams P., Chan C.Y., Ambros V., Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Mishima Y., Giraldez A.J., Takeda Y., Fujiwara T., Sakamoto H., Schier A.F., Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C.B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C.B. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom U.A., Kauppinen S., Lund A.H. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Pollard T.D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. microRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Sokol N.S., Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood P., Krek A., Zavolan M., Macino G., Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Staton A.A., Giraldez A.J. Encyclopedia of life sciences. Wiley: Chichester, UK: 2008. MicroRNAs in development and disease; pp. 1–10. [Google Scholar]
- Stouffer S.A., Suchman E.A., DeVinney L.C., Star S.A., Williams R.M., Jr . Studies in social psychology in World War II: The American soldier. Adjustment during army life. Princeton University Press; Princeton, NJ: 1949. [Google Scholar]
- Tavazoie S.F., Alarcon C., Oskarsson T., Padua D., Wang Q., Bos P.D., Gerald W.L., Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J., Zhu J., van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A., Morvan G., Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S., Abreu-Goodger C., Enright A.J. Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Liu N., Olson E.N. MicroRNAs flex their muscles. Trends Genet. 2008;24:159–166. doi: 10.1016/j.tig.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]