Abstract
MicroRNAs and trans-acting siRNAs (ta-siRNAs) have important regulatory roles in development. Unlike other developmentally important regulatory molecules, small RNAs are not known to act as mobile signals during development. Here, we show that low-abundant, conserved ta-siRNAs, termed tasiR-ARFs, move intercellularly from their defined source of biogenesis on the upper (adaxial) side of leaves to the lower (abaxial) side to create a gradient of small RNAs that patterns the abaxial determinant AUXIN RESPONSE FACTOR3. Our observations have important ramifications for the function of small RNAs and suggest they can serve as mobile, instructive signals during development.
Keywords: Small RNA, miRNA, Arabidopsis, leaf, polarity, pattern fomation
Small RNAs, such as microRNAs (miRNAs), trans-acting siRNAs (ta-siRNAs), and endogenous siRNAs, regulate diverse developmental processes in multicellular organisms. The comprehensive role of these small RNAs in mediating development is best visualized by their complex and varied expression patterns (Juarez et al. 2004; Wienholds et al. 2005; Nogueira et al. 2007). However, the mechanisms underlying the origins of these discrete accumulation patterns remain largely obscure. Direct localization of miRNA primary transcripts suggests that this complexity arises in part from transcriptional control (Aboobaker et al. 2005; Nogueira et al. 2009). Additionally, the post-transcriptional processing of small RNA precursors is contingent upon the presence of necessary biogenesis factors, the restricted activity of which can contribute to the final spatiotemporal localization of mature small RNAs (Viswanathan et al. 2008). The intercellular movement of small RNAs could also feasibly contribute to their final localization patterns. However, even though the movement of siRNAs during systemic silencing is a well-documented phenomenon (Dunoyer et al. 2005; Voinnet 2005), no instances of intercellular movement of endogenous small RNAs have as of yet been reported (Parizotto et al. 2004; Alvarez et al. 2006).
ta-siRNAs, a plant-specific small RNA class with roles in development, depend for their biogenesis on both miRNA activity and siRNA pathway components (Peragine et al. 2004; Vazquez et al. 2004; Allen et al. 2005). miRNA-guided cleavage triggers entry of ta-siRNA precursor transcripts into RNA-DEPENDENT RNA POLYMERASE6 (RDR6)-dependent and SUPPRESSOR OF GENE SILENCING3 (SGS3)-dependent pathways and sets the register for phased, 21 nucleotide ta-siRNA production by DICER-LIKE4 (DCL4). The biogenesis of TAS3-derived ta-siRNAs requires a subspecialized RNAi pathway (Allen et al. 2005; Adenot et al. 2006; Fahlgren et al. 2006). TAS3 transcripts are cleaved at a miR390 target site near the 3′ end, but also interact with miR390 in a noncleavage mode near the 5′ end (Axtell et al. 2006; Montgomery et al. 2008). At both sites, miR390 functions in a complex with ARGONAUTE7 (AGO7), which is relatively specialized for interaction with miR390 (Montgomery et al. 2008).
Mutants defective in TAS3 ta-siRNA biogenesis exhibit aberrant floral morphologies and accelerated juvenile-to-adult phase transition phenotypes (Peragine et al. 2004; Adenot et al. 2006). A subset of TAS3-derived ta-siRNAs, termed tasiR-ARFs, targets the AUXIN RESPONSE FACTOR (ARF) family members ARF3 and ARF4, which together are necessary for specifying abaxial fate in Arabidopsis (Allen et al. 2005; Pekker et al. 2005). Phenotypes resulting from mutations in TAS3 ta-siRNA biogenesis factors are mediated through the misregulation of tasiR-ARF targets, especially ARF3. Expression of ARF3 trangenes insensitive to tasiR-ARF regulation produce vegetative phase change defects similar to tasiR-ARF pathway mutations (Fahlgren et al. 2006; Hunter et al. 2006). The range of pleiotropic defects observed in such plants demonstrates the importance of this pathway to multiple aspects of leaf development.
Considering that ta-siRNA biogenesis factors are uniquely required for the production of mobile siRNAs in systemic silencing (Dunoyer et al. 2005; Voinnet 2005), we sought to determine whether the tasiR-ARF pathway in Arabidopsis contributes to leaf development via mobility of small RNA components. We therefore compared the localization patterns of tasiR-ARFs with those of their biogenesis factors. Although miR390 accumulates broadly throughout the shoot apex (the shoot apical meristem [SAM] plus developing leaf primordia), tasiR-ARF biogenesis is restricted to the few most adaxial cell layers of leaves by the localized expression of AGO7 and TAS3A. tasiR-ARFs accumulate outside this defined domain of biogenesis and form a gradient across leaves that maintain the polarized accumulation of ARF3. Incongruencies in the accumulation patterns of miR390, tasiR-ARFs, and their biogenesis factors are also observed within the specific developmental context of the SAM. These observations have important implications for the mechanisms through which ta-siRNAs and miRNAs pattern their targets and for their capacity to act as mobile, instructive signals during development.
Results and Discussion
tasiR-ARF activity is patterned across the leaf
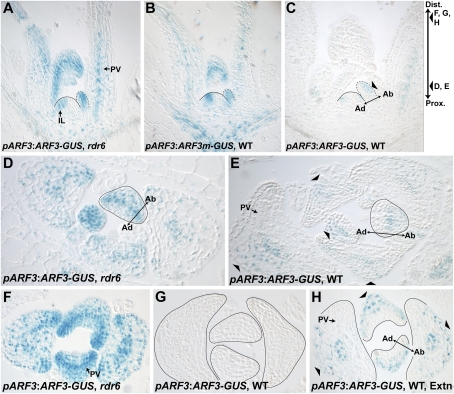
As a first approach to analyze the accumulation pattern of tasiR-ARFs in Arabidopsis, we used a sensor of their activity, similar to those routinely used to monitor miRNA expression (Parizotto et al. 2004). Patterns of tasiR-ARF activity were visualized by comparing expressions of an ARF3-GUS translational fusion reporter driven by the ubiquitously expressed ARF3 promoter (Supplemental Fig. S1) in the presence or absence of tasiR-ARF regulation. In the absence of tasiR-ARF activity, in an rdr6-15 mutant background, the pARF3:ARF3-GUS reporter is expressed in the vasculature and pith region below the SAM, as well as throughout the incipient and developing leaf primordia (Fig. 1A,D,F). Within leaf primordia, the reporter expresses most strongly in the provasculature, a broad band of cells that spans the middle of the leaf and later develops into vasculature and parenchyma (I. Sussex, pers. comm.). The near-ubiquitous expression of the pARF3:ARF3-GUS reporter in rdr6-15 unlikely results from developmental changes in the rdr6-15 mutant, as a pARF3:GUS transcriptional fusion shows similar activity in wild type (Supplemental Fig. S1). Expression of a nontargeted ARF3m-GUS transgene, in which the tasiR-ARF target sites are mutated (Fahlgren et al. 2006), also results in a similar ubiquitous pattern of expression (Fig. 1B). This shows that the expression of the targeted pARF3:ARF3-GUS reporter in rdr6-15 results from a lack of tasiR-ARF regulation specifically, rather than from indirect effects of other ta-siRNAs. Both the ARF3-GUS and ARF3m-GUS fusion proteins yield punctate expression patterns, consistent with ARF3 being a transcription factor that localizes to the nucleus.
Figure 1.
Differential tasiR-ARF activity across the adaxial-abaxial axis of leaf primordia. (A) In rdr6-15, pARF3:ARF3-GUS is expressed throughout leaf primordia and below the SAM. (IL) Incipient leaf; (PV) provasculature. (B) The tasiR-ARF insensitive pARF3:ARF3m-GUS reporter in wild type is expressed similarly. (C) pARF3:ARF3-GUS expression in wild type is reduced and abaxially restricted (arrowhead). (D,E) Transverse sections as indicated in C, showing pARF3:ARF3-GUS expression throughout primordia in rdr6-15 (D), but abaxially restricted in wild type (E). (F,G) Distal sections as indicated in C showing strong pARF3:ARF3-GUS activity throughout rdr6-15 leaves (F), but no activity in wild type (G). (H) Section identical to G but with extended GUS staining time shows abaxial pARF3:ARF3-GUS activity (arrowheads).
To determine the effects of tasiR-ARF activity on expression of the pARF3:ARF3-GUS reporter, we out-crossed this reporter from an rdr6-15 into a wild-type background. The overall activity of the pARF3:ARF3-GUS reporter is greatly reduced in wild-type vegetative apices (Fig. 1C). Expression is lost in the vasculature and pith region below the SAM, indicating strong tasiR-ARF activity in these regions. Expression of the pARF3:ARF3-GUS reporter is reduced in the incipient leaf, indicating the presence of tasiR-ARFs also within the SAM. In young leaf primordia, expression of the reporter is notably absent from the adaxial sides, margins, and the provasculature, but reporter activity persists on the abaxial sides of leaves (Fig. 1E). The specific abaxial expression of the reporter and the lack of an arf3 mutant phenotype in plants harboring the tasiR-ARF sensor indicates that the reporter is responding to tasiR-ARF activity rather than systemic transgene-induced gene silencing (Parizotto et al. 2004). Although reporter activity persists on the abaxial side of young primordia, this activity is reduced when compared with expression in the rdr6-15 background, indicating that tasiR-ARFs act throughout the developing leaf, but stronger on the adaxial side than the abaxial side (Fig. 1C,E).
That tasiR-ARFs act throughout the leaf is best visualized in distal regions of older leaf primordia. The pARF3:ARF3-GUS reporter in rdr6-15 (Fig. 1F) shows strong expression throughout distal leaf regions; whereas, under normal staining conditions, pARF3:ARF3-GUS reporter activity in a wild-type background is not detected in equivalent distal tissue sections (Fig. 1G). To detect potential residual expression of the transgene in distal regions, we allowed the GUS reaction to proceed for an extended period of time. Under these conditions, pARF3:ARF3-GUS reporter activity is detectable on the abaxial side of leaves, but not on the adaxial side (Fig. 1H). The disparity in tasiR-ARF activity along the adaxial–abaxial axis of leaves thus persists throughout primordium development. The absence of observable reporter activity on the adaxial side of both young and older leaf primordia suggests tasiR-ARFs accumulate strongly adaxially. This observation is also consistent with the role of ARF3 in promoting abaxial identity (Pekker et al. 2005) and the redundant role of the tasiR-ARF and ASYMMETRIC LEAVES pathways in organ polarity (Garcia et al. 2006). The weaker but measurable activity of tasiR-ARFs on the abaxial side of leaves is also significant, as it suggests tasiR-ARFs accumulate differentially throughout leaves (Fig. 1G,H).
tasiR-ARFs accumulate as a gradient in leaves
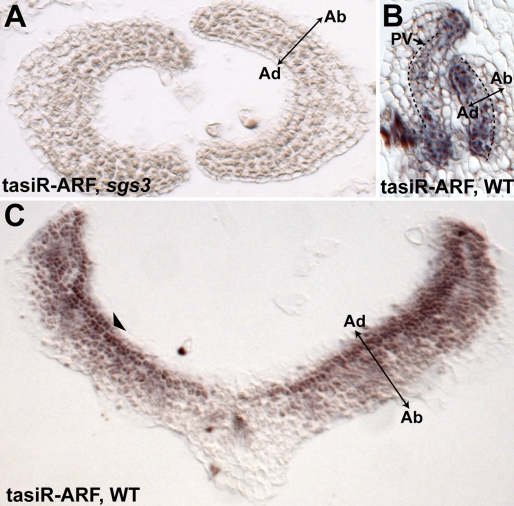
To independently verify that the pattern of tasiR-ARF activity in the vegetative shoot apex recapitulates the pattern of tasiR-ARF accumulation, we used in situ hybridization with a locked nucleic acid (LNA) probe that recognizes both tasiR-ARF variants (Nogueira et al. 2007). In an sgs3 background, in which tasiR-ARF biogenesis is blocked, this LNA probe yields no hybridization signal above background, indicating specificity of the probe for tasiR-ARFs (Fig. 2A). In wild-type plants, the tasiR-ARF in situ hybridization pattern is consistent with the pattern of tasiR-ARF activity detected using the sensor. In young primordia near the SAM, tasiR-ARFs accumulate predominantly on the adaxial side of leaves, especially within the margins and the provasculature, regions in which their activity is high as well (Figs. 1E, 2B). In more distal regions of the leaf, tasiR-ARFs remain most abundant on the adaxial side; however, they accumulate also on the abaxial side, forming a gradient that abaxially dissipates (Fig. 2C). The accumulation of tasiR-ARFs on the abaxial side of distal leaf regions likely accounts for our inability to detect the pARF3:ARF3-GUS reporter in these regions under normal staining conditions (Fig. 1G).
Figure 2.
tasiR-ARFs differentially accumulate along the adaxial-abaxial axis. (A) In situ hybridization in an sgs3 background using the tasiR-ARF LNA probe yields no hybridization signal above background, indicating the probe's specificity for tasiR-ARFs. (B) tasiR-ARFs accumulate on the adaxial side of young primordia near the SAM. Note their predominance in the margins and provasculature (PV, arrow). (C) In distal regions of older leaves, tasiR-ARFs accumulate strongest adaxially (arrowhead) and form a gradient that dissipates toward the abaxial side of the leaf.
Mature miR390 accumulates throughout the shoot apex
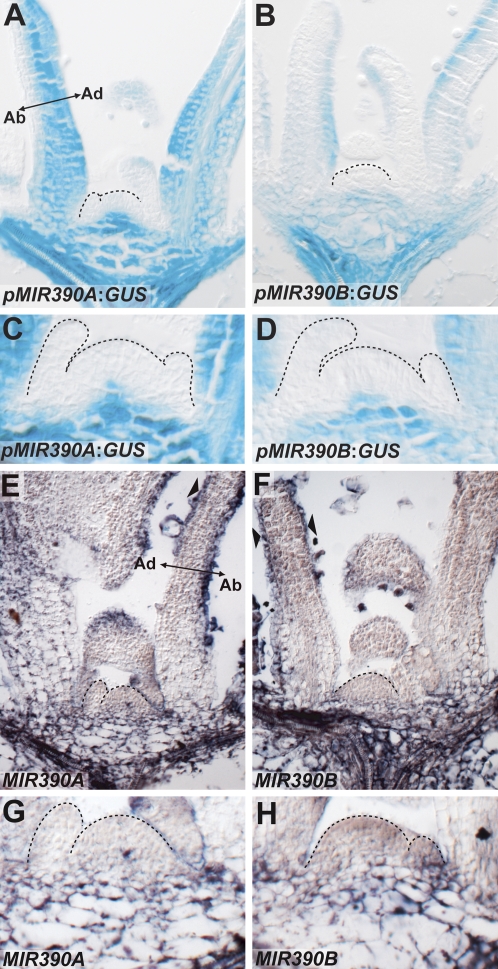
To better understand how the pattern of tasiR-ARF accumulation and activity along the adaxial–abaxial axis of leaves arises, we determined the expression patterns of upstream tasiR-ARF biogenesis components. tasiR-ARF biogenesis depends uniquely on the processing of TAS3 transcripts by a miR390–AGO7 complex (Montgomery et al. 2008). To determine the transcriptional activity of the MIR390 precursor loci, we histologically characterized previously described MIR390A and MIR390B reporters (Fig. 3; Montgomery et al. 2008). Both MIR390:GUS reporters are active in the vasculature and pith region below the SAM, but not within the meristem itself or within the youngest leaf primordia (P1–P3) (Fig. 3A–D). In older primordia, MIR390B is expressed in a more restrictive pattern compared with MIR390A, but both reporters act stronger on the adaxial than abaxial side (Fig. 3A,B; Supplemental Fig. S2A,B). In situ hybridization experiments, which reveal the standing accumulation of precursors, support the activity of the reporter lines. Both MIR390A and MIR390B precursors accumulate in older leaf primordia, where the prominence of precursor transcripts in the epidermis may be reflective of differential processing between the epidermis and underlying cell layers (Fig. 3E,F; Supplemental Fig. S2C,D). Moreover, both MIR390 precursors accumulate in the vasculature and pith region below the SAM and no in situ hybridization signal is detectable in the meristem or youngest leaf primordia (Fig. 3G,H).
Figure 3.
Absence of MIR390 precursor transcripts in the meristem and young leaf primordia. (A,B) Reporters for MIR390A (A) and MIR390B (B) show transcriptional activity for these loci in the vasculature and pith region beneath the SAM, but not within the meristem or youngest leaf primordia. In older leaf primordia, MIR390A:GUS is expressed more broadly than MIR390B:GUS. (C,D) Close-ups illustrating the lack of MIR390A:GUS (C) and MIR390B:GUS (D) reporter activity in the SAM and young leaf primordia. (E,F) In situ hybridizations show accumulation of MIR390A (E) and MIR390B (F) precursor transcripts in patterns similar to the expression patterns of their respective reporters. (G,H) Close-ups of meristem regions showing MIR390A (G) and MIR390B (H) accumulation beneath the SAM but not within the meristem or young leaf primordia. Note, sense probes for MIR390A and MIR390B yield no hybridization signal (Suppelemental Fig. S3).
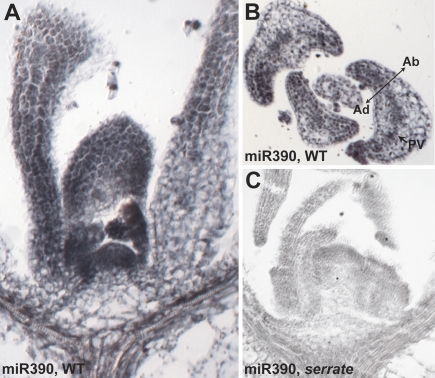
The absence of MIR390 precursors in the SAM and P1-P3 leaf primordia (Fig. 3), where tasiR-ARF activity is detected (Fig. 1C), suggests the presence of a mobile component in the tasiR-ARF pathway. To assess whether small RNAs might constitute such mobile components, we first localized mature miR390 by in situ hybridization (Fig. 4). For this we used a riboprobe that contains the antisense miR390 sequence connected to sense MIR390B sequence, which itself yields no detectable signal (Supplemental Figs. S3B, S4). Unlike its precursors, mature miR390 accumulates in the SAM and youngest leaf primordia, as well as in the pith region below the meristem (Fig. 4A). In older leaf primordia, miR390 localizes to both the adaxial and abaxial sides and accumulates predominantly in the provasculature and margins (Fig. 4B). Consistent with the riboprobe specifically detecting mature miR390, only a weak hybridization signal is observed in serrate-2 (se-2), a miRNA biogenesis mutant in which miR390 levels are reduced fourfold to fivefold (Fig. 4C; Supplemental Fig. S5). To further verify that miR390 accumulates ubiquitously throughout the vegetative apex, we performed in situ hybridization using LNA probes. A probe complementary to miR390 yields a similar localization pattern to that obtained with the riboprobe (Supplemental Fig. S6A), whereas an LNA probe detecting a murine miRNA yields no specific hybridization signal (Supplemental Fig. S6B).
Figure 4.
Mature miR390 accumulates throughout the shoot apex. (A) In situ hybridization shows miR390 accumulates robustly in the SAM and young primordia as well as in the vasculature and pith region below the SAM. (B) In leaves, miR390 accumulates throughout the leaf, but especially within the margins and provasculature (PV, arrow). (C) In serrate-2, the miR390 hybridization signal is barely detectable.
AGO7 and TAS3A expression restricts miR390 activity
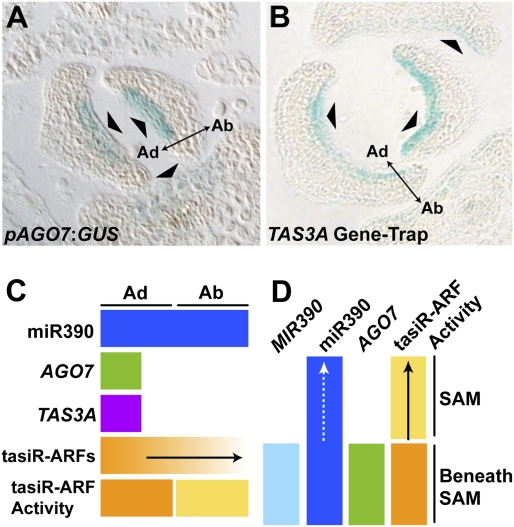
The uniform accumulation of miR390 throughout the SAM and young leaf primordia raises questions as to how miRNA activity might be regulated to control developmental patterning within the apex. Likewise, the nonpolarized accumulation of miR390 throughout leaves (Fig. 4B; Supplemental Fig. S6C) cannot account for the polarized activity of tasiR-ARFs (Fig. 1), or for the gradient of tasiR-ARFs observed by in situ hybridization (Fig. 2C). One possibility is that the activity of miR390 is spatially restricted by the localization of AGO7 or TAS3A. To determine the extent of miR390 activity, we localized AGO7 expression using an AGO7 promoter-GUS reporter. The cis-regulatory elements of this construct, when fused to AGO7, complement the ago7 mutant phenotype (Montgomery et al. 2008). AGO7 is expressed in the pith region just below the SAM, but not within the meristem proper (Supplemental Fig. S7A). Additionally, AGO7 is expressed within the adaxial-most cells of developing leaf primordia (Fig. 5A). Likewise, histological characterization of a previously described gene-trap line (Garcia et al. 2006) shows that TAS3A, the functional and molecular contributor of tasiR-ARFs during leaf development (Adenot et al. 2006; Addo-Quaye et al. 2008), is expressed only in the adaxial-most cells of leaves (Fig. 5B). In situ hybridization analyses verify the gene-trap expression pattern (Supplemental Fig. S7B), demonstrating that TAS3A accumulates cell-autonomously and is a restrictive factor of tasiR-ARF biogenesis. This notion is further supported by the observation that misexpression of AGO7 from the MIR390A promoter (Supplemental Fig. S2A) does not condition a loss-of-function arf3 arf4 phenotype (M. Howell and J. Carrington, unpubl.).
Figure 5.
Localized expression of AGO7 and TAS3A restricts the domain of tasiR-ARF biogenesis. (A) AGO7 expression is restricted to a central region of the adaxial-most cell layers of leaves (arrowheads). (B) Expression of TAS3A is similarly adaxially restricted in leaves (arrowheads). (C,D) Summary of the different localization patterns of tasiR-ARF pathway components and a model of small RNA mobility (arrows) in leaves (C) and the SAM (D).
Small RNAs as mobile, inductive signals
The localization of all known tasiR-ARF-specific biogenesis factors suggests a model whereby the activity of the tasiR-ARF pathway, which is required for normal leaf development, is patterned along the adaxial–abaxial axis through the movement of small RNAs (Fig. 5C). The seemingly ubiquitous accumulation of miR390 in leaves is tempered by the highly restricted localization of AGO7 and TAS3A, which curtail the extent of miR390 activity and delimit tasiR-ARF biogenesis to the adaxial-most cell layers (Fig. 5). tasiR-ARFs accumulate and act strongly adaxially within the margins and the provasculature of leaf primordia (Figs. 1E, 2B), a region slightly broader than that to which AGO7 and TAS3A localize (Fig. 5A,B), while weaker tasiR-ARF activity extends throughout the leaf into the abaxial side (Fig. 1F–H). This polarized activity reflects the pattern of tasiR-ARF accumulation. tasiR-ARFs accumulate most strongly near their source of biogenesis, on the adaxial side of the leaf and dissipate away to the abaxial side of the leaf as a gradient (Fig. 2C). The presence of tasiR-ARFs outside their domain of biogenesis is indicative of mobility (Fig. 5C), and provides an explanation for the recently suggested non-cell-autonomous silencing of transcripts that are targeted by multiple miRNAs (Tretter et al. 2008).
tasiR-ARFs possibly move within the context of the SAM as well. Expression of both MIR390 precursors is limited to below the SAM, whereas mature miR390 accumulation extends into the meristem proper and young leaf primordia (Figs. 3, 4, 5D). However, only miR390 that colocalizes with AGO7 functions to produce tasiR-ARFs. AGO7 expression is restricted to the pith region and vasculature below the SAM (Supplemental Fig. S7A), such that the weak tasiR-ARF activity detected within the incipient leaf (Fig. 1A–C) likely results from small RNA mobility (Fig. 5D).
Similarly, intercellular movement might be a possible explanation for the disparity between the accumulation patterns of miR390 and its precursors in the meristem region. Although expression of artificial miRNAs from a vascular-specific promoter suggests phloem-derived miRNAs act cell-autonomously in mature leaves (Tretter et al. 2008), it is interesting to note that the ARGONAUTE family member PINHEAD/ZWILLE is required in the vasculature below the shoot apex to non-cell-autonomously maintain pluripotency of the overlaying stem cells (Tucker et al. 2008). The shoot apex may provide a unique developmental context that permits the intercellular movement of miRNAs. At least one other miRNA, miR391, shows a similar discrepancy between the localization pattern of the mature miRNA and the transcriptional activity of its precursor (data not shown). Unlike miR390, miR391 possesses a 5′ U and is loaded into AGO1 (Montgomery et al. 2008), suggesting that the possible movement of miR390 would not be an outcome of its unique recruitment into an AGO7 complex, but may be a general property of miRNAs expressed beneath the SAM.
Our study reveals two novel RNAi-based developmental patterning mechanisms. Although miR390 accumulates in a seemingly nonspecific pattern throughout the leaf, its activity becomes polarized by the restricted expression of AGO7 and TAS3A (Fig. 5A,B). Likewise, the activities of other miRNAs with similarly broad localization patterns as miR390 may become refined by the restricted spatial expression of effector complexes. Although the activity of AGO1, through which most plant miRNAs act, is found throughout developing leaves, expression of PINHEAD/ZWILLE is adaxially restricted (Lynn et al. 1999). It will be interesting to see what other subspecialized mechanisms exist at the miRNA effector level to permit the discrete functioning of miRNAs during development.
The mobility of tasiR-ARFs illustrates a second RNAi-based patterning mechanism and identifies the first positional signal in adaxial–abaxial leaf polarity. Movement of this low abundant small RNA from a defined source of biogenesis creates a gradient of accumulation (Figs. 2C, 5C). Mathematical modeling predicts that such small RNA gradients can sharply define the expression boundary of target genes (Levine et al. 2007). This scenario is empirically supported by this study, as the gradient of tasiR-ARF accumulation across the adaxial-abaxial axis creates a sharp boundary of ARF3 expression (Figs. 1H, 2C). The idea of stably dividing a field of cells into adaxial and abaxial fates through a gradient of mobile small RNAs is elegant, and reminiscent of morphogenetic activity. Although the precise localization patterns of developmentally important small RNAs (Wienholds et al. 2005) argues against their long-distance movement in animals, mobility over finite cellular distances and in specific developmental contexts, as is likely the case in plants, should be considered. Given the scope of small RNA-regulated networks during development, the prospect of small RNA movement has important ramifications with respect to their potential to act as inductive signals and generators of pattern.
Materials and methods
GUS analysis
Seedlings grown on a 16 h light/8 h dark schedule for 10 d at 22°C on Murashige and Skoog medium supplemented with either 50 μg/mL hygromycin or 50 μg/mL kanamycin were harvested quickly into ice-cold acetone and prefixed for 20 min at room temperature. Seedlings were washed with 100 mM sodium phosphate (pH 7.0), 10 mM EDTA, 0.1% Triton-X, 6 mM ferrocyanide, and 6 mM ferricyanide buffer and allowed to sit for 5 min on ice. The same buffer supplemented with 0.05% X-Gluc was then added and the seedlings were vacuum-infiltrated for 30 min at 600 mm Hg. Seedlings were incubated for 6 h (MIR390 reporter lines), 12 h (tasiR-ARF sensor lines), or 96 h (extended staining) at 37°C and subsequently dehydrated to 50% ethanol, fixed in FAA (50% ethanol, 5% formaldehyde, 10% acetic acid), and embedded and sectioned as described (Kidner and Timmermans 2006). At least six independently transformed lines were characterized for each reporter construct.
In situ hybridization
Riboprobes to detect the MIR390 and TAS3A precursors and mature miR390 are described in the Supplemental Material. Histological methods, probe preparation, and tissue hybridizations were performed as described (Kidner and Timmermans 2006). All probes were hybridized at 50°C except the tasiR-ARF LNA probe, which was hybridized at 37°C.
Plant materials
ARF3 promoter sequence, consisting of 3024 base pairs of 5′ regulatory sequence, was PCR-amplified from genomic DNA, cloned into pENTR/DTOPO (Invitrogen), and recombined into the plant transformation vector pMDC163 (Curtis and Grossniklaus 2003). Transgenic plants were selected on MS medium supplemented with cefotaxime (100 μg/mL) and appropriate antibiotic before being transferred to soil. pARF3:ARF3-GUS, pARF3:ARF3-GUS rdr6-15, pARF3:ARF3m-GUS, pMIR390A:GUS, pMIR390B:GUS, pAGO7:GUS, and the TAS3A gene trap are previously described lines (Fahlgren et al. 2006; Garcia et al. 2006; Montgomery et al. 2008). rdr6-15, sgs3-11, and se-2 are previously described alleles (Allen et al. 2005; Grigg et al. 2005; Hunter et al. 2006).
Acknowledgments
We thank C. Fernandez-Marco and A. Husbands for technical assistance; T. Mulligan for plant care; R. Schwab for discussions; and M. Scanlon, G. Hannon, C. Kuhlemeier, and members of the Timmermans and Carrington Laboratories for critique of the manuscript. D.C. was an NSF Graduate Research Fellow and is a George A. and Marjorie H. Anderson Fellow. This work is supported by grants to M.T. from the USDA (2006-03420) and the NSF (IOB-0615721), and to J.C. from the NSF (MCB-0618433), the NIH (AI43288), and the USDA (2006-35301-17420).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1770009.
Supplemental material is available at http://www.genesdev.org.
References
- Aboobaker A., Tomancak P., Patel N., Rubin G., Lai E. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl. Acad. Sci. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C., Eshoo T., Bartel D., Axtell M. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouche N., Gasciolli V., Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A., Carrington J. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Alvarez J., Pekker I., Goldshmidt A., Blum E., Amsellem Z., Eshed Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell. 2006;18:1134–1151. doi: 10.1105/tpc.105.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M., Jan C., Rajagopalan R., Bartel D. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Curtis M., Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T., Howell M., Allen E., Dvorak S., Alexander A., Carrington J. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Garcia D., Collier S., Byrne M., Martienssen R. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Grigg S., Canales C., Hay A., Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- Hunter C., Willmann M., Wu G., Yoshikawa M., dela Luz Guierrez-Nava M., Poethig S. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez M., Kui J., Thomas J., Heller B., Timmermans M. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- Kidner C., Timmermans M. In situ hybridization as a tool to study the role of microRNAs in plant development. Methods Mol. Biol. 2006;342:159–179. doi: 10.1385/1-59745-123-1:159. [DOI] [PubMed] [Google Scholar]
- Levine E., McHale P., Levine H. Small regulatory RNAs may sharpen spatial expression patterns. PLoS Comput. Biol. 2007;3:e233. doi: 10.1371/journal.pcbi.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K., Fernandez A., Aida M., Sedbrook J., Tasaka M., Masson P., Barton M. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Montgomery T., Howell M., Cuperus J., Li D., Hansen J., Alexander A., Chapman E., Fahlgren N., Allen E., Carrington J. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- Nogueira F., Madi S., Chitwood D., Juarez M., Timmermans M. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes & Dev. 2007;21:750–755. doi: 10.1101/gad.1528607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira F., Chitwood D., Madi S., Ohtsu K., Schnable P., Scanlon M., Timmermans M. Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet. 2009;5:e1000320. doi: 10.1371/journal.pgen.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizotto E., Dunoyer P., Rahm N., Himber C., Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes & Dev. 2004;18:2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekker I., Alvarez J., Eshed Y. AUXIN RESPONSE FACTORS mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H., Poethig R. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes & Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter E., Alvarez J., Eshed Y., Bowman J. Activity range of Arabidopsis small RNAs derived from different biogenesis pathways. Plant Physiol. 2008;147:58–62. doi: 10.1104/pp.108.117119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M.R., Hinze A., Tucker E.J., Takada S., Jurgens G., Laux T. Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development. 2008;135:2839–2843. doi: 10.1242/dev.023648. [DOI] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A., Hilbert J., Bartel D., Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Viswanathan S., Daley G., Gregory R. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W., Miska E., Alvarez-Saavedra E., Berezikov E., deBruijn E., Horvitz H.R., Kauppinen S., Plasterk R. microRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]