Abstract
The 13 Fanconi anemia (FA) proteins cooperate in a common DNA repair pathway. Eight of these proteins are assembled into a multisubunit E3 ligase called the FA core complex. During S phase, the FA core complex is loaded by the FANCM protein into chromatin where it monoubiquitinates its substrates. In mitosis, the FA core complex is released from FANCM by an unknown mechanism. Here we show that FANCM is hyperphosphorylated and degraded during mitosis. β-TRCP and Plk1 are the key regulators of FANCM degradation. Nondegradable mutant forms of FANCM retain the FA core complex in the chromatin and disrupt the FA pathway. Our data provide a novel mechanism for the cell cycle-dependent regulation of the FA pathway.
Keywords: Fanconi anemia, FANCM, mitosis, β-TRCP, Plk1, ubiquitin
Fanconi anemia (FA) is a genetic disease characterized by chromosomal instability and increased susceptibility to cancer (D'Andrea and Grompe 2003). There are 13 complementation groups (FANC-A, -B, -C, -D1, -D2, -E, -F, -G, -I, -J, -L, -M, and -N) and the gene products cooperate in a cellular repair mechanism that removes detrimental DNA cross-links. Deficiency in any of the 13 FA proteins, renders cells hypersensitive to DNA cross-linking agents, such as Mitomycin C (MMC). Among the 13 FA proteins, eight are assembled into a nuclear complex (A, B, C, E, F, G, L, and M), termed the FA core complex (Kennedy and D'Andrea 2005). Of the eight subunits of the complex, only two have recognizable enzymatic domains, with FANCL possessing a PHD E3 ubiquitin ligase domain and FANCM possessing a helicase and a degenerate nuclease domain (Meetei et al. 2003, 2005; Mosedale et al. 2005; Gurtan et al. 2006). The remaining subunits are believed to provide mainly a scaffolding role (Kennedy and D'Andrea 2005).
A major function of the FA core complex is to monoubiquitinate its two substrates, FANCD2 and FANCI (Garcia-Higuera et al. 2001; Dorsman et al. 2007; Sims et al. 2007; Smogorzewska et al. 2007). The monoubiquitinated FANCD2 subsequently forms DNA damage-induced nuclear foci that contain other key DNA damage response proteins, including γ-H2AX, BRCA1, BRCA2, and RAD51 (Taniguchi et al. 2002; Wang et al. 2004; Taniguchi and D'Andrea 2006; Bogliolo et al. 2007). Failure to monoubiquitinate FANCD2 and FANCI results in decreased efficiency of DNA cross-link repair and homologous recombination (HR) (Kennedy and D'Andrea 2005; Nakanishi et al. 2005; Smogorzewska et al. 2007). Several studies suggested that the FA core complex has additional functions independent of monoubiquitinating FANCD2 and FANCI (Matsushita et al. 2005; Medhurst et al. 2006; Mirchandani et al. 2008), supporting the notion that the FA core complex coordinates various DNA repair mechanisms.
Monoubiquitination of FANCD2 is dynamically regulated throughout cell cycle. During S phase, FANCD2 is actively monoubiquitinated, while during M phase, the monoubiquitinated form rapidly disappears. There are at least two possible underlying mechanisms for the cell cycle-specific regulation. First, the FANCD2-specific deubiquitinating enzyme complex USP1/UAF1 (Cohn et al. 2007) may be highly active at G2/M phases, while the activity decreases during S phases. Second, the E3 ubiquitin ligase activity of the FA core complex may be up-regulated during S phase and down-regulated at G2/M phases. Little is known regarding the temporal and spatial regulation of the FA core complex. However, a series of recent data suggest a model in which FANCM, in complex with its binding partner FAAP24, serves to recruit the FA core complex to chromatin in order to activate FANCD2 monoubiquitination during S phase (Ciccia et al. 2007; Niedernhofer 2007; Kim et al. 2008). In accordance with this observation, FANCM appears to be the only subunit in the FA core complex that binds directly to DNA (Gari et al. 2008). How FANCM signals the association and dissociation of the FA core complex with chromatin is an important unanswered question.
The ubiquitin and proteasome system is a major regulatory mechanism for many cellular pathways, including the DNA damage response and the cell cycle. Proteins targeted by the ubiquitin/proteasome system are polyubiquitinated by a serial cascade of E1, E2, and E3 enzymes (Pickart 2001). Among these enzymes, the E3 ligases confer specificity to the system, by selectively interacting with their cognate substrates. The two best characterized E3 ligases that regulate the stability of checkpoint and cell cycle regulatory proteins are SCF (Skp1/Cullin/F-box) and APC (anaphase-promoting complex) E3 ubiquitin ligase complexes (Petroski and Deshaies 2005; Peters 2006). Particularly, the SCF E3 ligases target a number of proteins in different phases of cell cycle. Target specificity depends on the substrate interacting modules, such as F-box proteins (Skowyra et al. 1997; Winston et al. 1999a) and BTB proteins (Xu et al. 2003). One of the best characterized F-box proteins is β-TRCP (transducin repeat-containing protein), which targets several proteins involved in the cell cycle progression and checkpoint signalings, such as Emi1, Cdc25A, Wee1, Claspin, Ikβ, and REST (Winston et al. 1999b; Busino et al. 2003; Jin et al. 2003; Margottin-Goguet et al. 2003; Watanabe et al. 2004; Mailand et al. 2006; Mamely et al. 2006; Peschiaroli et al. 2006; Westbrook et al. 2008). In most cases, β-TRCP targets substrates by recognizing phosphorylated DSGxxS motifs, or variants, present within the substrates. Here we report that FANCM undergoes degradation during mitosis, regulated by β-TRCP and the Polo-like kinase 1 (Plk1). Consistent with reports that FANCM is required for chromatin loading of the FA core complex (Mosedale et al. 2005; Kim et al. 2008), we show that the degradation of FANCM controls the dissociation of the FA core complex from chromatin. Degradation provides a cellular mechanism for assuring the timely inactivation of the DNA repair machinery during mitosis.
Results and Discussion
FANCM is degraded during mitosis
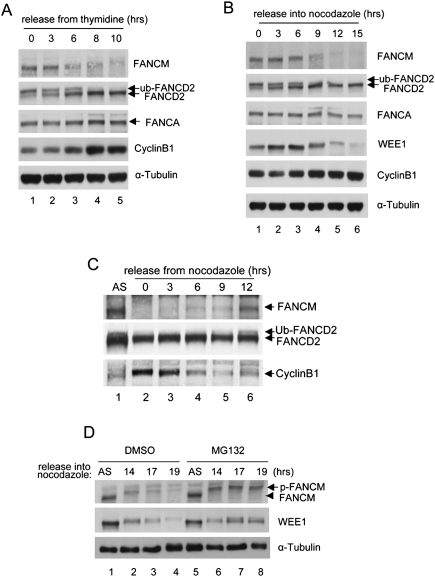
FANCM is highly phosphorylated during mitosis (Kim et al. 2008), and this hyperphosphorylation is temporally correlated with dissociation of the FA core complex from chromatin and loss of monoubiquitination of FANCD2 (Fig. 1A). When we examined the status of FANCM in cell populations highly enriched for mitotic cells (i.e., cells prepared by mitotic shake-off after nocodazole-mediated arrest), we noted the disappearance of FANCM in mitotic-arrested cells (Fig. 1B). The timing of disappearance is similar to that of WEE1, a kinase known to be degraded at G2/M phases. When cells were released from mitotic arrest, the FANCM level was restored by the time the cells entered G1/S phase (Fig. 1C, lane 6). Furthermore, a significant fraction of FANCM was restored following exposure of the arrested cells to a proteasome inhibitor MG132, suggesting that the proteasome degrades FANCM in mitosis (Fig. 1D, lanes 5–8).
Figure 1.
FANCM is degraded at G2/M phase. (A) HeLa cells were synchronized by double thymidine block and released into regular media and samples were harvested at indicated time points. (B) HeLa cells were synchronized by the same procedure as A, but released into nocodazole-containing media. Note the disappearance of phosphorylated FANCM. (C) Nocodazole-arrested HeLa cells were released into fresh medium and the status of FANCM was monitored at indicated time points. (D) HeLa cells were arrested using nocodazole for indicated times and 10 μM MG132 was added during the last 4 h of the nocodazole treatment before the cells were harvested.
β-TRCP component of the SCF E3 ligase mediates degradation of FANCM
Since the FA core complex contains an E3 ubiquitin ligase activity (Meetei et al. 2003; Gurtan et al. 2006) and FANCM is an integral part of the core complex, one possibility is that the FA core complex might polyubiquitinate and trigger degradation of FANCM, a mechanism analogous to the CSA-mediated degradation of CSB helicase in the transcription-coupled repair (Groisman et al. 2006). To test this hypothesis, we compared the status of FANCM in a patient-derived FA-A cell line that was either complemented with vector or FANCA cDNA. Loss of the FANCA protein level leads to destabilization of other subunits of the core complex (Garcia-Higuera et al. 2000; Kennedy and D'Andrea 2005). A similar level of FANCM degradation was observed in these two isogenic cell lines (Supplemental Fig. S1), suggesting that the degradation of FANCM is independent of the E3 ligase activity of the FA core complex. Furthermore, the level of FANCA remained unchanged during mitotic arrest, suggesting that mitotic degradation is specific to FANCM, and not other subunits of the FA core complex (Fig. 1B).
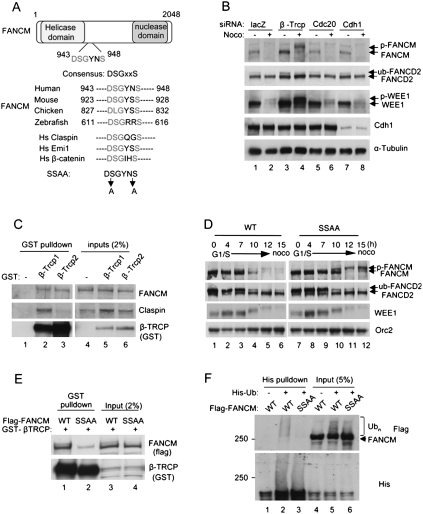
Two major E3 ubiquitin ligase complexes are known to be active during mitosis, the SCF E3 ligase complex and the APC (Peters 2006; Pines 2006). We hypothesized that one of these complexes may be responsible for the mitotic degradation of FANCM. Inspection of the primary amino acid sequence of FANCM revealed the consensus DSGxxS sequence, known to direct SCFβ-TRCP-mediated degradation (Fig. 2A), suggesting that the E3 ligase might mediate the degradation of FANCM. We used siRNA knockdown of the β-TRCP components of the SCF complex or of substrate specificity components of the APC (Cdc20 or Cdh1) (Peters 2006) to test whether depletion of any of these factors stabilizes FANCM. We used previously validated siRNA sequence targeting β-TRCP (Peschiaroli et al. 2006), and Cdc20 and Cdh1 (Brummelkamp et al. 2002). Knockdown of β-TRCP was confirmed by expressing exogenous protein (Supplemental Fig. S2), due to the inefficient detection of endogenous β-TRCP using our antibody. Stabilization of FANCM was observed when cells were treated with the siRNA against β-TRCP, but not against Cdc20 or Cdh1, suggesting that β-TRCP is required for degradation of FANCM (Fig. 2B, lanes 3,4). Consistent with previous reports, WEE1, a known target of β-TRCP (Watanabe et al. 2004), was also stabilized by depletion of β-TRCP. Interestingly, the phosphorylated form of FANCM was stabilized by depletion of β-TRCP, further supporting the hypothesis that phosphorylation precedes the degradation of FANCM. To test whether FANCM is a direct target of β-TRCP, GST-β-TRCP1 and β-TRCP2, two β-TRCP isoforms with indistinguishable biochemical function (Jin et al. 2003), were overexpressed in 293T cells, and pull-down experiments were performed to test the physical interactions (Fig. 2C). A fraction of endogenous FANCM was detected in both pull-down fractions. Claspin, another known substrate of SCFβ-TRCP ligase, was also detected in the pull-down fractions, consistent with previous reports that Claspin binds β-TRCP (Mailand et al. 2006; Mamely et al. 2006; Peschiaroli et al. 2006). Taken together, these results suggest that the β-TRCP component of SCF E3 complex mediates the degradation of FANCM.
Figure 2.
β-TRCP mediates degradation of FANCM. (A) A schematic for FANCM protein and alignment of amino acids corresponding to the DSGxxS sequence with FANCM orthologs and other β-TRCP substrates. (B) HeLa cells were treated with indicated siRNAs before being arrested using nocodazole. The higher migrated bands of FANCM and WEE1 indicate phosphorylated forms. (C) GST-β-TRCP proteins were overexpressed in 293T cells and pull-down assay was performed using glutathione sepharose. (D) Flag-FANCM wild type or SSAA mutant were expressed in HeLa cells that were synchronized by double thymidine block followed by release into nocodazole-containing media for indicated times. (E) GST-β-TRCP1 was coexpressed with either Flag-FANCM wild type or SSAA mutant in 293T cells and pull-down assay was performed using glutathione sepharose. (F) 293T cells were cotransfected with 6XHis-Ub plasmid and Flag-FANCM wild type or SSAA plasmids, followed by nocodazole and MG132 treatment. Cells lysates were subjected to pull-down using Ni-NTA beads and the eluates were analyzed by Western blots.
Degradation of FANCM requires the DSGxxS phosphodegron
To test the importance of the DSGYNS sequence of FANCM, we generated a cDNA encoding a full length mutant form of FANCM with the sequence DAGYNA (indicated as SSAA) (Fig. 2A). We expressed Flag-tagged FANCM wild type or the mutant (SSAA) in HeLa cells. Interestingly, the mutant protein did not undergo degradation during mitosis but remained stable as high-molecular-weight forms (Fig. 2D, lanes 11,12), under the condition where wild-type FANCM proteins were destabilized. High-molecular-weight species of SSAA mutants indeed were phosphorylated forms as they disappeared by phosphatase treatment (Supplemental Fig. S3). These results are consistent with the siRNA experiment, where depletion of β-TRCP also stabilized phosphorylated forms of FANCM (see Fig. 2B), and further suggest that the phosphorylation events precede the degradation of FANCM. To further test the significance of the DSGYNS sequence, we performed pull-down experiments by coexpressing GST-β-TRCP with either FANCM wild type or SSAA mutant (Fig. 2E). The interaction of the SSAA mutant with GST-β-TRCP was significantly reduced compared with wild type, suggesting that the serine residues are critical for recognition by β-TRCP. Furthermore, intracellular FANCM protein was polyubiquitinated, while the nondegradable FANCM protein (SSAA mutant) had significantly reduced polyubiquitination (Fig. 2F). Taken together, these results demonstrate that the DSGYNS sequence is a key determinant for the degradation of FANCM during mitosis.
Plk1 induces phosphorylation and degradation of FANCM
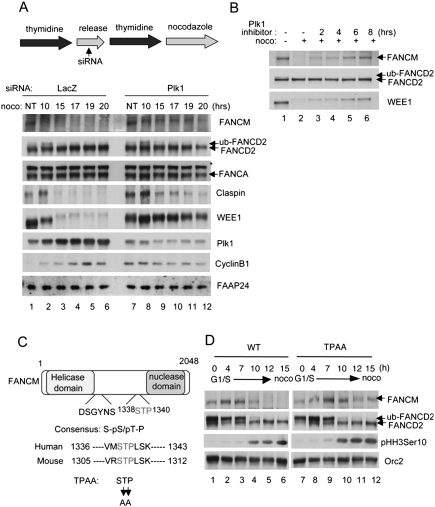
Since the mitotic phosphorylation of FANCM precedes the degradation of FANCM, we reasoned that depletion of a kinase might also stabilize FANCM. Plk1, a critical kinase regulating mitotic progression and cell cycle checkpoints, was shown previously to regulate several β-TRCP targets, such as Emi1, WEE1, and Claspin (Hansen et al. 2004; Watanabe et al. 2004; Mailand et al. 2006; Mamely et al. 2006; Peschiaroli et al. 2006). Therefore, we hypothesized that degradation of FANCM might also be regulated by Plk1. To test this hypothesis, we synchronized HeLa cells by double thymidine block, depleted Plk1 using siRNAs, and monitored the level of FANCM at different time points (Fig. 3A). To minimize potentially deleterious effects of Plk1 depletion to the cells, we used a moderate depletion of Plk1. SiRNAs to Plk1 partially inhibited the degradation of its known substrates Claspin and WEE1 (Fig. 3A, lanes 7–12). Phosphorylation and degradation of FANCM was also significantly inhibited, suggesting that Plk1 is the key kinase triggering mitotic phosphorylation and degradation of FANCM. Plk1 knockdown had no effect on FANCA and FAAP24 level and relatively little effect on FANCD2 monoubiquitination. These results further support a model in which Plk1-mediated phosphorylation of FANCM is a prerequisite for degradation. To rule out potentially indirect effects of Plk1 siRNA treatment, we used a small molecule inhibitor of Plk1 (Lenart et al. 2007). Acute treatment of the Plk1 inhibitor BI2536 stabilized FANCM during the mitotic arrest (Fig. 3B), but had little effect on cell cycle distribution (Supplemental Fig. S4). Treatment with an Aurora-B kinase inhibitor Hesperadin (Hauf et al. 2003) did not alter FANCM stability (data not shown). Previous studies suggested that the interactions between Plk1 with its substrates are largely mediated by recognition of Ser–pSer/pThr–Pro (S–pS/pT–P) motifs within the substrates by the Polo-box domain (PBD) of Plk1 (Elia et al. 2003). Since FANCM contains a conserved STP sequence (Fig. 3C), we tested whether this motif is a genuine recognition sequence of Plk1. Indeed, when we mutated the threonine and proline residues to alanines (indicated as TPAA) (Figure 3D), stabilization of FANCM was observed, suggesting that the STP motif is critical for the degradation induced by Plk1. Interestingly, the high-molecular-weight phosphorylated forms observed in the SSAA mutant of FANCM were significantly reduced (Fig. 3D, lanes 11,12), suggesting that Plk1-mediated phosphorylation precedes degradation of FANCM. Taken together, these results suggest that Plk1, along with β-TRCP, is a critical regulator triggering degradation of FANCM during mitosis.
Figure 3.
Plk1 mediates degradation of FANCM (A, top panel). A schematic for the synchronization assay. siRNAs were treated during a release period after the first thymidine block. (A, bottom panel) HeLa cells were synchronized at G1/S, and released into nocodazole-containing media. Samples were harvested at indicated time points and analyzed by Western blots. Asterisks indicate cross-reacting bands. (B) Nocodazole-arrested HeLa cells were treated with Plk1-specific inhibitor BI2536 for indicated times and the samples were analyzed by Western blots. (C) A schematic for FANCM protein and alignment of amino acids corresponding to the STP sequence with mouse Fancm ortholog. (D) Flag-FANCM wild type or TPAA mutant were expressed in HeLa cells that were synchronized by double thymidine block followed by release into nocodazole-containing media for indicated times.
FANCM degradation is required for release of the FA core complex during mitosis and for genomic stability.
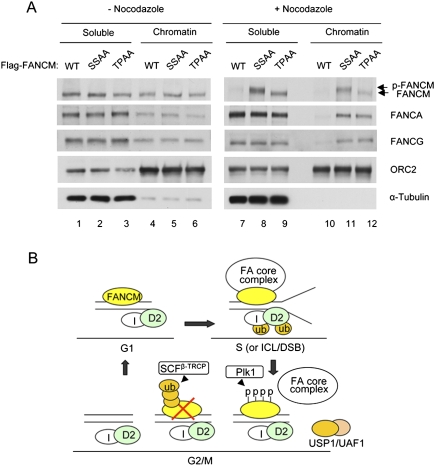
We showed previously that FANCM is required for the DNA damage-inducible or S-phase-specific recruitment of the FA core complex to the chromatin (Kim et al. 2008). How the FA core complex is released from chromatin during G2 and M phases remains unknown. A plausible mechanism is that phosphorylation and degradation of FANCM during mitosis serve as a signal for the release of the core complex from chromatin. To test this hypothesis, we tested whether localization of the FA core complex is disrupted in cells expressing the nondegradable forms of FANCM (SSAA and TPAA) (Fig. 4A). Indeed, the FANCA and FANCG subunits of the core complex was significantly enriched in the chromatin fraction under mitotic arrest in the cells expressing either of the mutant forms of FANCM, compared with the cells expressing wild-type FANCM (Fig. 4A, cf. lanes 11,12 and 10). This strongly suggests that phosphorylation and degradation of FANCM is necessary for releasing the FA core complex from chromatin during mitosis.
Figure 4.
Degradation of FANCM regulates localization of the FA core complex (A) Flag-FANCM wild type, SSAA, or TPAA mutant were expressed in HeLa cells that were synchronized by double thymidine block followed by release into nocodazole-containing media. At 15 h from nocodazole treatment, mitotic cells were collected and fractionated to nucleo/cytoplasmic soluble fractions (S) and chromatin-enriched fractions (P). (B) Model. During G1/S phase, the FA core complex is recruited to chromatin by FANCM, where FANCD2 is monoubiquitinated. During G2/M phase, FANCM is degraded and subsequently the FA core complex is released, leading to inactivation of the FA pathway.
We next determined whether degradation of FANCM is required for cellular function of the FA pathway. The hallmark of FA cells is their hypersensitivity to DNA cross-linking agents, such as Mitomycin C (MMC). Interestingly, heterologous expression of nondegradable FANCM proteins (SSAA and TPAA mutant proteins) resulted in increased MMC-induced chromosomal radial formation (Supplemental Fig. S5), suggesting that FANCM depletion is an important functional event in the FA pathway.
Our results are summarized in the model described in Figure 4B. During S phase, or when DNA is damaged, the FA core complex is recruited to the chromatin via FANCM, where it monoubiquitinates its substrates. As cells enter G2 and M phases, FANCM becomes hyperphosphorylated and degraded, thus releasing the FA core complex from chromatin. FANCD2 and FANCI become deubiquitinated, possibly due to the high activity of the USP1/UAF1 deubiquitinating enzyme complex (Cohn et al. 2007). Consistent with the observation that β-TRCP and Plk1 mediate timely degradation of FANCM, depletion of either protein resulted in increased chromosomal radials following cellular exposure to MMC (Supplemental Fig. S6).
Recently, it was reported that FANCM and FAAP24 form a complex with ATR and HCLK2, and this complex is required for an S-phase checkpoint (Collis et al. 2008). We therefore tested whether there is a functional relationship between the formation of the checkpoint complex and FANCM degradation. FANCM was degraded normally, even when ATR was depleted by siRNA (Supplemental Fig. S7). Consistent with this result, we observed a normal level of DNA damage-induced Chk1 phosphorylation when either wild-type or the nondegradable forms of FANCM were expressed (data not shown). These results suggest that degradation of FANCM is not functionally linked to its role in the S-phase checkpoint.
Previous studies indicate that DNA repair processes are highly coordinated during the cell cycle (Branzei and Foiani 2008). For example, nonhomologous end-joining (NHEJ) tends to occur in the G1 phase, while HR repair is primarily an S-phase process. How the cell cycle coordinates these DNA repair processes is largely unknown. Our results highlight the importance of cross-talk between the cell cycle and DNA repair machinery. By regulating the stability of FANCM, cells may efficiently terminate the FA pathway, while they progress into G2/M phases. It is important to note that β-TRCP and Plk1 play critical roles in regulating the stability of several important cell cycle regulators such as Wee1, Emi1, and Claspin. Degradation of these factors is essential for faithful cell cycle progression through G2 and M. We now show that FANCM joins the list of proteins that are degraded in a β-TRCP and Plk1-dependent manner. We speculate that FANCM may have additional unknown roles in regulating the cell cycle progression, particularly during transition to G2 and M phases, which are independent of DNA repair.
Materials and methods
Cell lines
HeLa and 293T cells were cultured using Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) supplemented with 15% fetal calf serum (FCS). The patient-derived FA-A cell line (GM6914) complemented with pMMP vector and pMMP-FANCA were as described previously (Garcia-Higuera et al. 2001).
Cell synchronization
Synchronization of HeLa cells were described previously (Kim et al. 2008). For the preparation of cellular extracts from mitotic shake-off, cells were treated with nocodazole-containing (100 ng/mL) medium for 18–20 h, the rounded cells were gently disrupted from dishes using pipettes, and the cells were washed with PBS buffer, before finally being harvested at 2500 rpm for 5 min. Cells were resuspended with 1× SDS-PAGE loading buffer and boiled for the SDS-PAGE analyses. Information regarding antibodies, siRNAs, and protein techniques can be found in the Supplemental Material.
Acknowledgments
We thank Wade Harper for the β-TRCP plasmids, Jan-Michael Peters for the Plk1 inhibitor, Stephan Jentsch for the His-ubiquitin-expressing plasmid, and Lisa Moreau for chromosomal breakage analysis. We thank members of the D'Andrea laboratory for helpful discussions. This work was supported by NIH grants RO1-HL52725, RO1-DK43889, and PO1-HL54785 to A.D. Y.K. is a recipient of a Leukemia and Lymphoma Society Fellowship.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1761309.
Supplemental material is available at http://www.genesdev.org.
References
- Bogliolo M., Lyakhovich A., Callen E., Castella M., Cappelli E., Ramirez M.J., Creus A., Marcos R., Kalb R., Neveling K., et al. Histone H2AX and Fanconi anemia FANCD2 function in the same pathway to maintain chromosome stability. EMBO J. 2007;26:1340–1351. doi: 10.1038/sj.emboj.7601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Busino L., Donzelli M., Chiesa M., Guardavaccaro D., Ganoth D., Dorrello N.V., Hershko A., Pagano M., Draetta G.F. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- Ciccia A., Ling C., Coulthard R., Yan Z., Xue Y., Meetei A.R., Laghmani el H., Joenje H., McDonald N., de Winter J.P., Wang W., West S.C. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Cohn M.A., Kowal P., Yang K., Haas W., Huang T.T., Gygi S.P., D'Andrea A.D. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 2007;28:786–797. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Collis S.J., Ciccia A., Deans A.J., Horejsi Z., Martin J.S., Maslen S.L., Skehel J.M., Elledge S.J., West S.C., Boulton S.J. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol. Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- D'Andrea A.D., Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- Dorsman J.C., Levitus M., Rockx D., Rooimans M.A., Oostra A.B., Haitjema A., Bakker S.T., Steltenpool J., Schuler D., Mohan S., et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell. Oncol. 2007;29:211–218. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A.E., Cantley L.C., Yaffe M.B. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I., Kuang Y., Denham J., D'Andrea A.D. The fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood. 2000;96:3224–3230. [PubMed] [Google Scholar]
- Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., D'Andrea A.D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Gari K., Decaillet C., Stasiak A.Z., Stasiak A., Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Groisman R., Kuraoka I., Chevallier O., Gaye N., Magnaldo T., Tanaka K., Kisselev A.F., Harel-Bellan A., Nakatani Y. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes & Dev. 2006;20:1429–1434. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtan A.M., Stuckert P., D'Andrea A.D. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J. Biol. Chem. 2006;281:10896–10905. doi: 10.1074/jbc.M511411200. [DOI] [PubMed] [Google Scholar]
- Hansen D.V., Loktev A.V., Ban K.H., Jackson P.K. Plk1 regulates activation of the anaphase promoting complex by phosphorylating and triggering SCFβTrCP-dependent destruction of the APC Inhibitor Emi1. Mol. Biol. Cell. 2004;15:5623–5634. doi: 10.1091/mbc.E04-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S., Cole R.W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C.L., Peters J.M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore–microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Shirogane T., Xu L., Nalepa G., Qin J., Elledge S.J., Harper J.W. SCFβ-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes & Dev. 2003;17:3062–3074. doi: 10.1101/gad.1157503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy R.D., D'Andrea A.D. The Fanconi anemia/BRCA pathway: New faces in the crowd. Genes & Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- Kim J.M., Kee Y., Gurtan A., D'Andrea A.D. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J.J., Hoffmann M., Rettig W.J., Kraut N., Peters J.M. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Bartek J., Lukas J. Destruction of Claspin by SCFβTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell. 2006;23:307–318. doi: 10.1016/j.molcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Mamely I., van Vugt M.A., Smits V.A., Semple J.I., Lemmens B., Perrakis A., Medema R.H., Freire R. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 2006;16:1950–1955. doi: 10.1016/j.cub.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Margottin-Goguet F., Hsu J.Y., Loktev A., Hsieh H.M., Reimann J.D., Jackson P.K. Prophase destruction of Emi1 by the SCF(βTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell. 2003;4:813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- Matsushita N., Kitao H., Ishiai M., Nagashima N., Hirano S., Okawa K., Ohta T., Yu D.S., McHugh P.J., Hickson I.D., et al. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol. Cell. 2005;19:841–847. doi: 10.1016/j.molcel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Medhurst A.L., Laghmani H., Steltenpool J., Ferrer M., Fontaine C., de Groot J., Rooimans M.A., Scheper R.J., Meetei A.R., Wang W., et al. Evidence for subcomplexes in the Fanconi anemia pathway. Blood. 2006;108:2072–2080. doi: 10.1182/blood-2005-11-008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei A.R., de Winter J.P., Medhurst A.L., Wallisch M., Waisfisz Q., van de Vrugt H.J., Oostra A.B., Yan Z., Ling C., Bishop C.E., et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Meetei A.R., Medhurst A.L., Ling C., Xue Y., Singh T.R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C.G., et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirchandani K.D., McCaffrey R.M., D'Andrea A.D. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst.) 2008;7:902–911. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale G., Niedzwiedz W., Alpi A., Perrina F., Pereira-Leal J.B., Johnson M., Langevin F., Pace P., Patel K.J. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- Nakanishi K., Yang Y.G., Pierce A.J., Taniguchi T., Digweed M., D'Andrea A.D., Wang Z.Q., Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L.J. The Fanconi anemia signalosome anchor. Mol. Cell. 2007;25:487–490. doi: 10.1016/j.molcel.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Peschiaroli A., Dorrello N.V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N.E., Pagano M. SCFβTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Peters J.M. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Petroski M.D., Deshaies R.J. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pines J. Mitosis: A matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Sims A.E., Spiteri E., Sims R.J., III, Arita A.G., Lach F.P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Craig K.L., Tyers M., Elledge S.J., Harper J.W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., III, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D'Andrea A.D., et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., D'Andrea A.D. Molecular pathogenesis of Fanconi anemia: Recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Garcia-Higuera I., Andreassen P.R., Gregory R.C., Grompe M., D'Andrea A.D. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- Wang X., Andreassen P.R., D'Andrea A.D. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol. Cell. Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Arai H., Nishihara Y., Taniguchi M., Watanabe N., Hunter T., Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFβ-TrCP. Proc. Natl. Acad. Sci. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook T.F., Hu G., Ang X.L., Mulligan P., Pavlova N.N., Liang A., Leng Y., Maehr R., Shi Y., Harper J.W., et al. SCFβ-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston J.T., Koepp D.M., Zhu C., Elledge S.J., Harper J.W. A family of mammalian F-box proteins. Curr. Biol. 1999a;9:1180–1182. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Strack P., Beer-Romero P., Chu C.Y., Elledge S.J., Harper J.W. The SCFβ-TRCP–ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes & Dev. 1999b;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wei Y., Reboul J., Vaglio P., Shin T.H., Vidal M., Elledge S.J., Harper J.W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]