Abstract
Neuronal differentiation is a complex process that involves a plethora of regulatory steps. To identify transcription factors that influence neuronal differentiation we developed a high throughput screen using embryonic stem (ES) cells. Seven-hundred human transcription factor clones were stably introduced into mouse ES (mES) cells and screened for their ability to induce neuronal differentiation of mES cells. Twenty-four factors that are capable of inducing neuronal differentiation were identified, including four known effectors of neuronal differentiation, 11 factors with limited evidence of involvement in regulating neuronal differentiation, and nine novel factors. One transcription factor, Oct-2, was studied in detail and found to be a bifunctional regulator: It can either repress or induce neuronal differentiation, depending on the particular isoform. Ectopic expression experiments demonstrate that isoform Oct-2.4 represses neuronal differentiation, whereas Oct-2.2 activates neuron formation. Consistent with a role in neuronal differentiation, Oct-2.2 expression is induced during differentiation, and cells depleted of Oct-2 and its homolog Oct-1 have a reduced capacity to differentiate into neurons. Our results reveal a number of transcription factors potentially important for mammalian neuronal differentiation, and indicate that Oct-2 may serve as a binary switch to repress differentiation in precursor cells and induce neuronal differentiation later during neuronal development.
Keywords: Oct-2, neuronal, embryonic stem cell, differentiation, high throughput
Neuronal development and differentiation are complex processes that involve extracellular factors, local niche, and intracellular factors. Much is known about extracellular factors that mediate neural induction. For example, inhibition of bone morphogenetic protein (BMP) by chordin, noggin (Sasai et al. 1995; Zimmerman et al. 1996), and follistatin (Hemmati-Brivanlou et al. 1994; Fainsod et al. 1997) in cooperation with signaling from bFGF (Delaune et al. 2005) helps generate neural precursors. Following the formation of the neural precursors, differentiation is induced by factors such as sonic hedgehog (Shh) (Hynes et al. 1995; Tanabe et al. 1995) and Notch signaling (Breunig et al. 2007). Likewise, at the transcriptional level, several key players involved in neuronal development and differentiation have been identified. Basic helix–loop–helix (bHLH) family proteins such as Mash1, Math1, NeuroD, and Neurogenin 1–3 (Kageyama et al. 2005) help induce differentiation of neurons and other cell types. Other bHLH family members, Hes1, Hes3, and Hes5, are expressed in neural stem cells and known to inhibit neuronal differentiation (Kageyama et al. 2008). In spite of the large number of transcription factors implicated in neuronal development and differentiation, few systematic studies of these regulators have been performed, and thus it is likely that many other transcription factors regulate neuronal differentiation.
The complete sequencing of the human genome offers new approaches to systematically identify components important for neuronal differentiation. Gray et al. (2004) examined the expression of most mouse transcription factors using in situ hybridization, and found that at least 20% of known transcription factors have spatially restricted expression patterns in developing mouse brains. While this study provided a wealth of information on the potential involvement of factors in neurogenesis, it lacked any functional information. In principle, another approach that could be developed is to inactivate transcription factors on a large scale; however, for many neuronal determinant genes knockouts rarely lead to defects in neurogenesis, which is likely due to functional redundancy (Ma 2006).
To systematically identify transcription factors involved in neuronal differentiation, we developed an overexpression screen in embryonic stem (ES) cells. Previous studies have shown that introduction of single transcription factor could induce the directed differentiation of a precursor cell or transdifferentiation of a mature cell type. Examples include MyoD (myocyte differentiation) (Weintraub et al. 1989), PPARγ2 (adipocyte differentiation) (Tontonoz et al. 1994), and Lmx1a (dopamingeric neuronal differentiation) (Andersson et al. 2006).
We screened 700 human transcription factors for their ability to induce neuronal differentiation in mouse ES (mES) cells and found many known and novel factors that induce differentiation. One factor, Oct-2, was studied in detail. Octamer-binding proteins (Oct) are comprised of a family of five transcription factors (Oct-1, Oct-2, Oct-4, Oct-6, and Oct-11) that recognize the octamer motif (5′-ATGCAAAT-3′). All Oct family members are part of the POU (Pit1, Oct, unc86) domain family of transcription factors that harbor a POU domain connected to a homeodomain by a short linker. Of the mammalian Oct proteins, one, Oct-4 has been found that has several functions with regard to differentiation in ES cells: maintenance of pluripotency (Nichols et al. 1998), differentiation into primitive endoderm (Niwa et al. 2000), as well as promotion of neuronal differentiation (Shimozaki et al. 2003). Other Oct family members have been less well characterized. Oct-1 is a ubiquitously expressed protein whose primary functions are not clear. Oct-1 knockout embryos are defective in erythropoiesis, and Oct-1-deficient B cells show normal development (Wang et al. 2004), yet primary mouse embryonic fibroblasts lacking Oct-1 are deficient in response to stress stimuli (Tantin et al. 2005). Oct-6 is regulated during stem cell differentiation, and is localized to certain regions of the developing brain; however, no specific function has been determined. Oct-2 was initially studied for its role in B-cell development (Veenstra et al. 1997). However, it has been found to be widely expressed throughout the CNS at multiple time points during rat embryonic development, and its expression is restricted to the brain in adult rats (He et al. 1989). Furthermore, treatment of sensory neurons with nerve growth factor results in up-regulation of Oct-2 levels (Wood et al. 1992).
Although neuronal expression of Oct-2 has been noted, little has been reported with regard to its function in neuronal differentiation. Previous work has focused on the ability of various Oct-2 splice forms to regulate specific promoters either through the use of artificial reporter assays (Dent et al. 1991; Wirth et al. 1991) or by examining endogenous promoters (Deans et al. 1995, 1996). However, a direct role for Oct-2 in neuronal differentiation has not been explored.
We discovered that Oct-2 can serve as a bifunctional regulator of neuronal differentiation. One isoform Oct-2.4 has the ability to block neuronal differentiation, whereas a second isoform Oct-2.2 can act as an inducer of neuronal differentiation. Removal of Oct-2 and siRNA knockdown of the Oct-1 homolog inhibits neuronal differentiation. Thus, our results demonstrate a dual function for Oct-2 in neuronal development and assigns a role in neuronal differentiation to Oct family members whose functions have been confined primarily to areas such as B-cell maturation and apoptosis. Additionally, use of our library for rapid functional categorization of transcription factors is expected to be a valuable resource, and may also be applied to the analysis of many other cell lineages.
Results
Generation of a transgenic mES collection and development of a high throughput neurogenesis screen
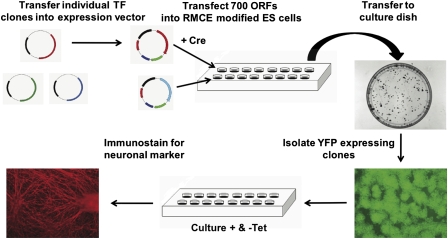
To identify transcription factors that may be involved in neuronal differentiation we established the expression system shown in Figure 1. Briefly, coding regions of interest are cloned into an expression cassette that can be efficiently integrated into a specific genomic location (the ROSA26 locus) in mES cells as reported previously (Masui et al. 2005). The sequence of interest is expressed under an inducible promoter such that transcription is activated by the removal of tetracycline. The expression construct we prepared makes a C-terminal fusion to the transactivation domain of Herpesvirus protein VP16, a potent activator of gene expression (Braselmann et al. 1993), as well as an N-terminal fusion to Hemagluttinin (HA) epitope tag. Upon proper integration a yellow fluorescent protein (YFP) reporter is expressed after induction (Supplemental Fig. S1).
Figure 1.
Overview of the neuronal differentiation screen. Human ORFeome 1.1 clones (Rual et al. 2004) were transferred into the expression vector and were cotransfected with Cre recombinase into EBRTcH3 cells. Drug-resistant colonies were picked from 10-cm plates. Positive clones were selected based on YFP reporter expression. Cell lines were tested for their ability to undergo neuronal differentiation with and without gene induction.
We first tested the system using two transcription factors, mouse Gata6 (mGata6) and human NeuroD1 (hNeuroD1). mGata6 was previously shown to induce differentiation (Masui et al. 2005), and therefore, mGata6 without VP16 was initially used as a control for differentiation into primitive endoderm. It was found that cells in which mGata6 was stably introduced displayed a morphology change characteristic of primitive endoderm within 48 h of inducing mGata6 expression (Supplemental Fig. S1).
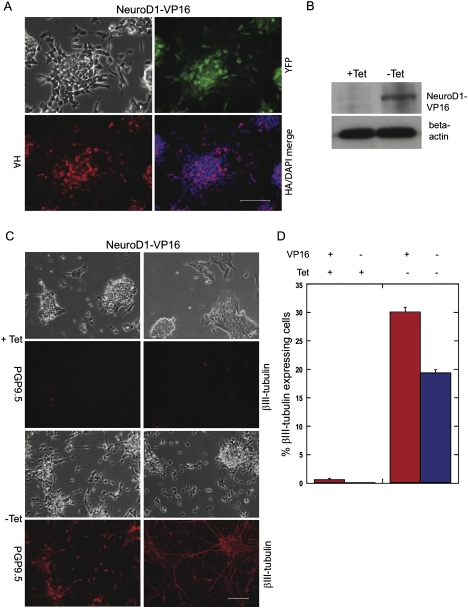
hNeuroD1 has been shown previously to induce neuronal differentiation when stably expressed in mES cells (O'Shea 2001). We generated two stable cell lines, one containing the hNeuroD1-VP16 construct and another lacking VP16, and induced expression in each of these by the removal of tetracycline from the media (Fig. 2B). hNeuroD1-VP16 fusion protein localized properly to the nucleus by immunostaining for the HA epitope (Fig. 2A). Using conditions that do not normally form neurons (Supplemental Fig. S2), we tested for neuronal differentiation by examining cell morphology and then staining for the presence of neuron-specific ubiquitin C-terminal hydrolase (PGP9.5) as well as βIII-tubulin (Fig. 2C). We found that after 72 h many cells formed projections characteristic of neurites and stained positively for both PGP9.5 and βIII-tubulin. This pattern was observed for both hNeuroD1- and hNeuroD1-VP16-expressing cells, but the percentage of cells that differentiated was 30% higher for hNeuroD1-VP16 relative to hNeuroD1 alone (Fig. 2D). Thus, VP16 enhanced the transcriptional activity of NeuroD1 and was included in our screening system. An additional advantage of including the VP16 domain is that it can potentially identify both positive and negative regulators of differentiation (Su et al. 2004).
Figure 2.
Functional confirmation of hNeuroD1-VP16 activity. (A) Induction of transgene expression in cell line Tet-NeuroD1-VP16 resulted in expression of nuclear localized HA-NeuroD1-VP16 fusion protein (bottom right panel) and coexpression of YFP (top right panel). (B) A NeuroD1 fusion protein of proper size is inducibly expressed. (C) Two different markers, PGP9.5 and βIII-tubulin, confirmed that neuronal cell differentiation was induced by NeuroD1-VP16. (D) hNeuroD1 minus VP16 transactivation domain also induced neuronal differentiation. Bars, 100 μm.
Neuronal differentiation from a defined set of neural ORFs
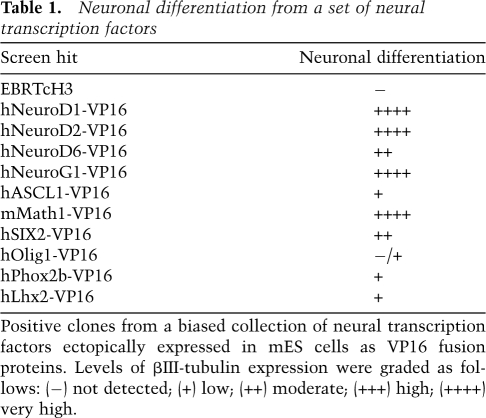
As a pilot screen for factors that induce neuronal cell differentiation, we next examined a small set (23) of human and mouse ORFs known or implicated in the induction of neuronal differentiation for their ability to induce differentiation in the mES expression system as VP16 fusion proteins (Table 1). Several factors, including Mash1, NeuroD2, and Math1, had previously been observed to induce some level of differentiation when overexpressed in embryonic carcinoma cells (Farah et al. 2000). For NeuroD6, an ability to differentiate had previously been demonstrated in rat pheochromocytoma cells (PC12) but not in ES cells (Uittenbogaard and Chiaramello 2002). Two stable lines were chosen and analyzed for each factor and examined 96 h post-induction. Of the 23 factors examined, 10 were found to induce neuronal differentiation as judged by morphology and/or immunofluorescence (summarized in Table 1).
Table 1.
Neuronal differentiation from a set of neural transcription factors
Positive clones from a biased collection of neural transcription factors ectopically expressed in mES cells as VP16 fusion proteins. Levels of βIII-tubulin expression were graded as follows: (−) not detected; (+) low; (++) moderate; (+++) high; (++++) very high.
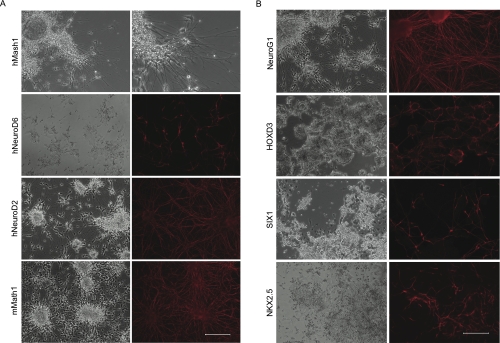
Figure 3A displays the neuronal morphology of several examples. Ectopic expression of Mash1 resulted in a dramatic morphological phenotype indicative of neuronal differentiation in which many long neurites are evident; this phenotype was evident even without the use of immunostaining (Fig. 3A, top panels). Other factors (NeuroD2, Math1, Mash1, and NeuroD1) also strongly induced differentiation, as evidenced by the presence of large numbers of neurites and immunostaining, whereas still others such as NeuroD6, Six2, and Phox2B induced fewer neurites. In general, our results were consistent with the literature.
Figure 3.
Positive clones from neural biased collection and large-scale screen. (A) Cell lines such as Tet-Mash1-VP16 displayed obvious neuronal-like projections at 96 h post-induction. Bar, 200 μm. (B) Positive clones from high throughput screen varied in levels of βIII-tubulin expression and morphology. Bars: NeuroG1, 200 μm; HOXD3, 333 μm; SIX1 and NKX2.5, 100 μm.
There were several unexpected effectors of neuronal differentiation. For example, Olig1 is known to be expressed in motor neuron progenitors, and in the absence of both Olig1 and Olig2 progenitors are diverted to an interneuron and astrocyte fate (Zhou and Anderson 2002). In our assay, Olig1 was able to reproducibly induce neuronal differentiation at a very low level (data not shown), consistent with the VP16 domain, rendering a repressing transcription factor into a dominant positive activator (Immaneni et al. 2000). Of the 13 factors that did not induce some level of neuronal differentiation, none had ever been shown to induce differentiation when ectopically expressed. It is possible that the assay used was too stringent for some low-potency differentiation factors or that the VP16 domain interferes with the native transactivation domains. Nonetheless, these results indicate that multiple factors known to be involved in neural development can induce neuronal differentiation when ectopically expressed in mES cells using our system.
Neuronal differentiation screen of a collection of 700 transgenic mES cell lines
As a result of the high success rate of known factors, we prepared and screened a collection of transcription factors for their ability to induce neuronal differentiation. Approximately 700 human ORFs, mostly encoding transcription factors, were cloned into the inducible expression vector and then transfected into mES cell lines to generate stable cell lines (summarized in Supplemental Table S1). Two cell lines for each ORF were cultured in permissive conditions for a period of 4–5 d before fixation and immunostaining for βIII-tubulin. All positive staining cell lines were retested through the differentiation protocol a second time, but with transgene expression turned both on and off in parallel. Cell lines that produced a clear and reproducible increase in βIII-tubulin-positive cells versus (1) uninduced controls and (2) identically treated parental EBRTcH3 cell lines were deemed true positives. In several cases (<10) cell lines showed a notable increase in the number of neurons produced, but neurons formed independent of induction of expression. Such clones were discounted.
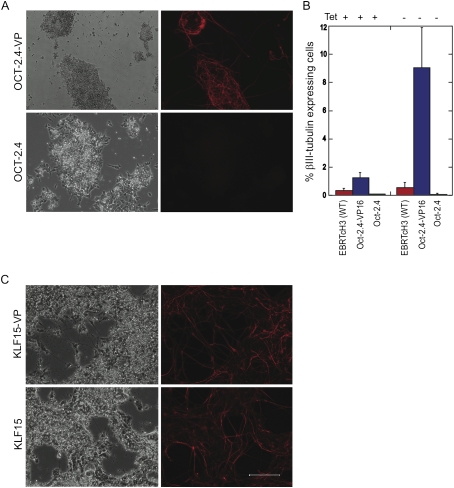
From the collection of 1400 cell lines, 24 neuronal differentiation inducing transcription factors were identified. The morphology, number, and length of the induced neurites varied among the different factors. Well-known factors such as NeuroG1 (Fig. 3B) and NeuroD1 strongly induced large numbers of long neurites. Other neural-inducing genes such as ZNF435 (Supplemental Fig. S3) and ZFP64 (data not shown) were subtle and lacked the long axonal processes observed with NeuroG1. Such weaker factors had a tendency to resemble mES colonies that had not undergone neuronal differentiation, and cells with shortened neuronal-like processes could only be identified with immunostaining. Another category of positives were those in which βIII-tubulin-positive cells were either intertwined in a large clump of cells with disorganized neuronal outgrowths or that had the majority of neuronally differentiated cells on the periphery of colonies. Targets that fit into this latter category included HOXD3 (Fig. 3B), HOXA3 (data not shown), and OCT-2 (Fig. 4A). An overall summary of the different factors is included in Table 2; many of these have been found previously to be expressed in the embryonic CNS (e.g., Hoxd3, Tead2, Klf15, Tcf4) (Gray et al. 2004).
Figure 4.
(A) Positive clones such as OCT-2 demonstrated a decrease or complete loss in neuronal differentiation upon removal of VP16. (B) Quantification of the number of βIII-tubulin-positive cells revealed a dramatic difference in cells expressing OCT-2 versus OCT-2-VP16. Removal of VP16 resulted in a lower production of neurons than the wild-type control. Mouse Oct-2.4 exhibits a similar result as the human isoform. (C) Positive clones such as KLF15 demonstrated strong neuronal differentiation both in the presence and absence of VP16. Bar, 200 μm.
Table 2.
Transcription factors that induce neuronal differentiation
Positive clones from a high throughput neuronal differentiation screen. Twenty-four hits were identified from ∼700 transgenic mES cell lines. Clones that tested positive from the first round of screening were retested without the VP16 fusion. Levels of βIII-tubulin expression were graded as follows: (−) not detected; (+) low; (++) moderate; (+++) high; (++++) very high.
The 24 factors that induced neuronal differentiation as VP16 fusions were next tested for activation in the absence of VP16. Of the 24 positives, nine were able to reproducibly induce neuronal differentiation without VP16. Differentiated NeuroG1- and KLF15-expressing (Fig. 4; data not shown) cell lines were similar in appearance regardless of whether VP16 was present or not, indicating that they are strong activators even in the absence of VP16. Transgenes such as HOXA3, HOXD3, HOXA3, NeuroD6, NKX2.5, SIX1, and ZNF205 that lacked the VP16 transactivation domain also induced βIII-tubulin-positive neurites upon induction of expression, but the number (and in some cases, length) of neurons produced decreased relative to the constructs containing VP16 (data not shown).
The remaining factors that failed to induce differentiation in the absence of VP16 might either be weak activators or repressors of neural differentiation. TEAD2 and DMRTC2 were weak inducers only in the presence of VP16 and might be weak activators. BRAF35 binds to the neural-restrictive silencer (NRS) promoter and is a known repressor (Hakimi et al. 2002), and thus, it is likely to be an activator in the presence of VP16 but a repressor in its absence. As described in the next section, one factor, OCT-2, was found to exhibit a very strong difference in the presence and absence of the VP16 transactivation domain.
OCT-2.4 as a repressor of neuronal differentiation
As a VP16 fusion protein OCT-2 was one of the more potent neuronal differentiation factors, with ∼10% of cells expressing βIII-tubulin when cells were plated on gelatin-coated dishes and allowed to differentiate in standard screen media (Fig. 4A,B). Upon removal of the VP16 transactivation domain the percentage of neurons after 4 d is reduced to less than 0.1%. This figure was lower than the ∼1% of neurons produced by the OCT-2-VP16 transgene when tetracycline was in the media, suggesting that OCT-2 may function as a repressor (Fig. 4B). Analysis of the Tet-OCT-2.4 and Tet-OCT-2.4-VP16 lines revealed that OCT-2.4 was expressed and localized to the nucleus as a wild-type or VP16 fusion protein, suggesting it was functioning properly in each case (Supplemental Fig. S4).
To directly test the ability of OCT-2.4 to act as a repressor of neuronal differentiation we used an embryoid body (EB) differentiation protocol. EBs are multilayered cellular aggregates that form an outer layer of endoderm while the inner cells differentiate toward a fate of primitive ectoderm. After a few days of suspension culture, EBs form fluid filled cysts. When cystic EBs are cultured in the presence of retinoic acid the primitive ectoderm will differentiate toward a neuronal lineage (Bibel et al. 2007).
We placed the Tet-OCT-2.4 cell line and wild-type control cells through the EB/neuronal differentiation protocol. Ectopic expression of OCT-2.4 resulted in a marked reduction of EB-derived neurons as compared with control, as evidenced by immunostaining of anti-βIII-tubulin (Fig. 5C). FACS analysis confirmed that there was an approximately fourfold reduction in βIII-tubulin-expressing cells (Fig. 4C); this figure is likely a large underestimate as the FACS protocol we used shears cells neurites and thereby will miss many positive cells. These data strongly suggest that OCT-2.4 inhibits mES neuronal cell differentiation.
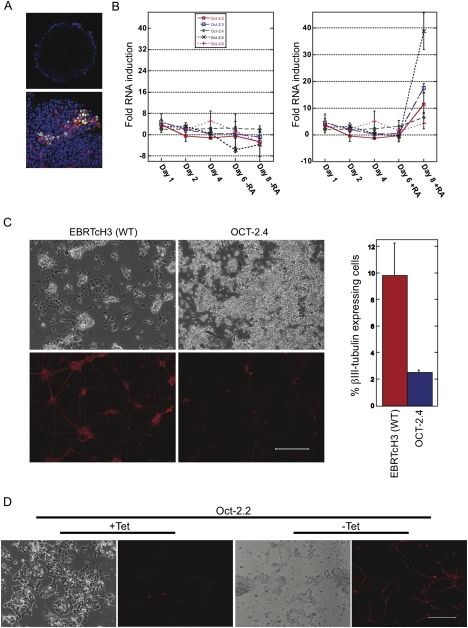
Figure 5.
Different splce isoforms of Oct-2 can either inhibit or induce differentiation. (A) Confocal microscopy of EBs induced to differentiate (bottom panel) and control (top panel). Oct-2 was detected as a nuclear stain (green) in clusters of cells having undergone neuronal differentiation (βIII-tubulin, red). Nucleii were costained with DAPI (blue). (Graph, left) Real-time RT–PCR on EBs revealed Oct-2.4 levels remained relatively unchanged during neuronal differentiation. (Graph, right) Oct-2.2, Oct-2.3, and Oct-2.5 all increased dramatically in expression levels. (B) OCT-2.4 inhibited neuronal differentiation. Ectopic expression of OCT-2.4 blocks retinoic acid-induced differentiation of EBs (bottom panels) relative to vector control (top panels). Quantification of neurons by FACS. (C) Oct-2.2 isoform induced differentiation. Activation domain containing Oct-2.2 was the only isoform capable of inducing ES cells to differentiate into neurons. Bars, 200 μm.
Expression of Oct-2 splice forms during differentiation
A search of annotated human ORFs revealed three entries for OCT-2 (recently renamed POU2F2). Seven entries were present for the mouse oct-2 gene (Supplemental Fig. S5). Of the mouse genes one, Oct-2.4, lacks a transactivation domain. Of the remaining isoforms Oct-2.1, Oct-2.2, Oct-2.3, Oct-2.5, Oct2.6, and Oct-2.7 all contain transactivation domains. Amino acid analysis of human ORF OCT-2 showed that it was nearly identical (97% amino acid identity and 98% amino acid similarity) with mouse Oct-2.4. We therefore refer to our clone of human OCT-2 as OCT-2.4.
To further assess the role of OCT-2 in neuronal differentiation, we analyzed mouse Oct-2 expression by immunostaining of EBs using a monoclonal antibody against Oct-2 (Fig. 5A, top panel). The monoclonal Oct-2 antibody used in this study was raised against amino acids 1–47 of the protein, and therefore, did not distinguish between the various Oct-2 isoforms (Corcoran et al. 2004). The results showed that Oct-2 was absent from undifferentiated mES cells as well as untreated day1 (D1), D2, D4, D6, and D8 EBs (data not shown). D6 and D8 EBs were treated with 2 and 4 d of retinoic acid, respectively. D6 retinoic-treated EBs lacked Oct-2 staining, whereas D8 EBs contained pockets of immunoreactive cells that coincided with the first appearance of βIII-tubulin-positive cell clusters displaying axonal processes (Fig. 5A, bottom panel).
Studies in rodents using RT–PCR have shown that Oct-2 splice forms Oct-2.1, Oct-2.5, and Oct-2.4 are expressed in neuroblastoma lines and primary tissues such as brain. To identify which mouse Oct-2 isoforms were expressed in differentiating EBs TaqMan primers and probes were designed to recognize Oct-2 splice forms Oct-2.2 through Oct-2.6 (Fig. 5B). The Oct-2.4 repressor isoform was present at similar levels throughout the course of differentiation and increased only slightly by the final day of retinoic acid-induced differentiation. The splice form Oct-2.6, which contains a transactivation domain, was also expressed, albeit in very low amounts, throughout all stages of EB formation regardless of the presence or absence of retinoic acid. Interestingly, most isoforms containing known transactivation domains (Oct-2.2, Oct-2.3, and Oct-2.5) showed a sharp increase in RNA fold induction from day 6 to day 8 samples (retinoic acid induced differentiation stages). The fold induction of day 8 retinoic acid-treated EBs was greater than either day 6 or day 8 untreated EBs. The RNA organization of Oct-2.1 did not allow for the simple generation of isoform-specific primers. As a result, a nested primer strategy was used and demonstrated that Oct-2.1 expression increased in D8 EBs treated with retinoic acid, similar to other Oct-2 isoforms containing activation domains. Overall, these results highlight a consistent pattern in which most of the neural-specific Oct-2 isoforms containing a transactivation domain are up-regulated during retinoic acid neuronal differentiation while the single splice form lacking an activation domain is largely unchanged.
Oct-2 and Oct-1 have partially redundant functions in the control of neuronal differentiation
Since Oct-2.1, Oct-2.2, Oct-2.3, and Oct-2.5 were induced during neuronal differentiation; we suspected that some of the isoforms might serve as activators in this process. We therefore tested mouse Oct-2.2 for its ability to induce neuron differentiation in mES cells using the tetracycline-inducible expression system lacking VP16 described above. Overexpression of Oct-2.2 led to an increase in the number of neurons formed relative to uninduced cells (Fig. 5D). This induction was demonstrated to be 20-fold using FACS analysis. Thus, Oct-2.2, unlike Oct-2.4, serves as a potent activator of neuronal differentiation.
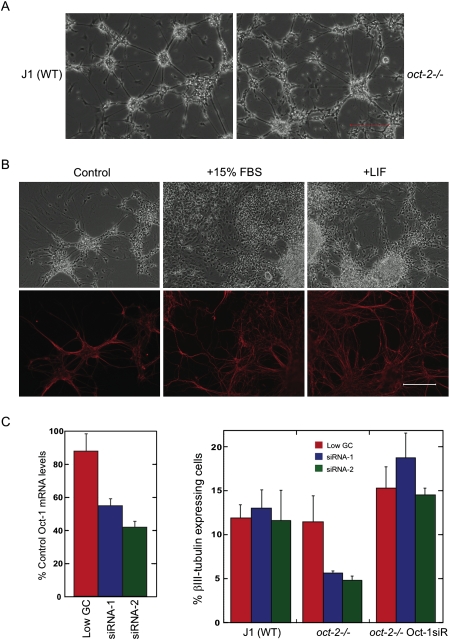
Surprisingly, it was reported that mice null for Oct-2 protein do not display any gross morphological CNS defects, although they do have a slight decrease in serum Ig levels, and they may have synaptic defects (Corcoran et al. 1993). To further examine the role of Oct-2 in neuronal differentiation we examined oct-2−/− mES cells in which none of the Oct-2 splice forms are expressed. oct-2−/− mES cells had previously been generated from blastocysts by mating mice heterozygous for a null allele of Oct-2 (L. Corcoran, unpubl.). Our initial examination of oct-2−/− cells undergoing EB differentiation showed no overt difference compared with wild-type mES cells (Fig. 6A). To more carefully assess the ability of oct-2−/− cells to undergo neuronal differentiation, a stromal cell method of differentiation induction was used. mES cells plated on the cranially derived mesenchymal cell line PA6 (Kawasaki et al. 2000; Kitajima et al. 2005) often differentiate into neurons. As the oct-2−/− cells used in this study were originally derived from mice on the J1 background (Li et al. 1992) differentiation tests were carried out with J1 mES cells. When wild-type J1 mES cells were seeded onto a fixed layer of confluent PA6 cells there were obvious neurite outgrowths from colonies within 4–5 d. By 9 d post-plating the neurite outgrowths had increased in length and number, and βIII-tubulin analysis by in-plate immunostaining revealed a significant increase in the number of neurons in optimized plating conditions, which was confirmed by FACS (Fig. 6B,C).
Figure 6.
Oct-1 functionally compensates for Oct-2 in neuronal differentiation. (A) Oct-2 knockout ES cells lacking all isoforms displayed no overt phenotype versus wild-type cells. Bar, 200 μm. (B) Optimization of stromal cell layer differentiation protocol. (C) Knockdown of Oct-1. Quantification of siRNA knockdown of Oct-1 in wild-type ES cells (left graph). In the background of oct-2 −/− ES cells Oct-1 knockdown led to a reduction in neuronal differentiation. Rescue of neuronal differentiation was achieved with reintroduction of a siRNA-resistant Oct-1 (Oct-1-siR). Bar, 200 μm.
It has previously been suggested that loss of Oct-2 could be functionally compensated by the related POU domain factor Oct-1 because of the high degree of homology in their DNA-binding domains (Wang et al. 2004). Oct-1 animals die several days prior to central neuronal system development, and thus its role in this process cannot be easily deduced in vivo. We tested whether Oct-1 played a redundant role with Oct-2 during neuronal differentiation using the PA6 in vitro system. Transient transfection assays with siRNA oligonucleotides were used because of the high (85%) and consistent transfection rate of mES transfection with labeled oligonucleotides (Supplemental Fig. S6).
Transient transfection of mES cells with siRNAs targeting Oct-1 yielded ∼40%–50% reduction in Oct-1 levels relative to mock transfected cells for two independent sets of oligonucleotides (Fig. 6C, bottom left). mES cells transfected with siRNAs targeting Oct-1 had no effect on neuronal differentiation. In contrast, knockdowns of Oct-1 in cells lacking Oct-2 had 50% fewer neurons relative to control cells (Fig. 6C, bottom right). To verify that the decrease in neuronal phenotype generated by transfection of Oct-1 siRNAs was not a consequence of off-target effects we overexpressed a siRNA-resistant Oct-1 in the knockdown cells. The siRNA-resistant version of Oct-1 (Oct-1-siR) was prepared through synthesis of Oct-1 cDNA, altering codon usage while preserving amino acid identity. Cotransfection experiments with either Oct-1 or Oct-1-siR and Oct-1 siRNAs show that, relative to cells cotransfected with wild-type Oct-1, Oct-1-siR-transfected cells were unaffected (Supplemental Fig. S5).
We next introduced the siRNA-resistant Oct-1-siR construct into the double-knockout lines. Oct-1-siR in a lentiviral vector was used to infect oct-2−/− cells and generate the oct-2−/− Oct-1-siR cell line. FACS analysis of wild-type J1 cells and oct-2 −/− cells transfected with siRNAs targeting Oct-1 resulted in a consistent reduction in the number of βIII-tubulin-positive cells. Oct-1-siR rescued neuronal differentiation in oct-2 −/− cells transfected with Oct-1 siRNAs (Fig. 6C, bottom right), thereby confirming a lack of off target effects from the siRNAs. Furthermore, cells overexpressing Oct-1-siR even had a slightly higher number of neurons than wild-type cells, suggesting that Oct-1 can enhance neuronal formation in this assay. Collectively, these data demonstrate that Oct-1 and Oct-2 together are necessary for wild-type levels of neuronal differentiation in vitro, and that they have functionally redundant roles for this process.
Discussion
Identification of genes important for neuronal differentiation
In this study we performed a screen to identify transcription factors important for neuronal differentiation. From a screen of 700 transcription factors, we identified 24 that induced neuronal differentiation. Some of clones, such as NeuroD1, NeuroD6, NeuroG1, and Nkx2.5, are known to be important for differentiation, and are as valuable controls for our screen. Interestingly, it had been recently reported that overexpression of Nkx2.5 in a muscle cell line led to neuronal transdifferentiation (Riazi et al. 2005); our study demonstrates that it can induce mES cells to form neurons as well.
The majority of positives identified in our screen were comprised of proteins that had either localization or expression studies indicating their potential involvement in neuronal processes, but a direct role in neuronal differentiation had not been demonstrated. For example, transcription factors such as HOXA3 and its paralog HOXD3 have long been known to be expressed during development in hindbrain rhombomeric segments but a connection to neuronal differentiation had yet to be established. Perhaps HOXA3 confers positional identity (e.g., rostral or caudal) on the neurons produced. The screen also identified SIX1 as a neuronal differentiation inducer. SIX1 has been suggested previously to be required for differentiation of neuroblasts during inner ear neurogenesis (Zou et al. 2004). Not surprisingly, targets of both known and candidate activators of neuronal differentiation had the capacity to induce differentiation even after the removal of the VP16 transactivation domain.
In addition to known and candidate inducers of neuronal differentiation we also found eight novel regulators, KLF12, ZNF43, ZNF205 ZNF37A, ZFP64, ZNF435, ZNF408, and WDR34. One of these, OCT-2, is described further below. Of the 24 identified transcription factors, nine maintained their differentiation ability even after removal of the artificial transcriptional transactivation domain. The remainders are likely weak inducers of differentiation or repressors. Some of the factors are consistent with the latter possibility. BRAF35, for example, is known to be part of a complex that binds to the repressor element 1 or NRS (RE1/NRS) (Hakimi et al. 2002). Reduction of expression of ZNF43 by antisense in Ewing Sarcoma causes neuronal differentiation, suggesting that it also might be a repressor in vivo (Gonzalez-Lamuno et al. 2002). Another factor that failed to induce differentiation when lacking VP16 is the homeodomain protein MEOX2. Mice that are null for Meox2 through targeted gene ablation have severe defects in muscle development so that there is an overall decrease in muscle mass and elimination of specific muscles (Mankoo et al. 1999). There is a precedent for transcription factors with the capacity to activate the muscle-specific programs, such as MyoD and Myf5, to actively block neuronal differentiation (Delfini and Duprez 2004). Thus, Meox2 may function in a similar fashion, and mES cells overexpressing Meox2 may exhibit inhibition of neuronal differentiation.
The results obtained from this screen leave available a wide assortment of interesting proteins to be further analyzed for neuron specification and positional identity, both in vitro and in vivo. During the course of this study a report was published by Falk et al. (2007) describing a high throughput transfection screen for neuronal differentiation factors. They identified 15 genes whose expression weakly induced differentiation, none of which overlapped with ours. Their strategy differed from ours by using pooled cDNAs in a transient transfection assay and by screening all types of proteins, whereas we focused on transcription factors and screened them individually using stable integrants; the expression of the factors was under the control of an inducible promoter. As such, we could carefully examine each clone for its ability to induce differentiation in a controlled fashion. Regardless, collectively these results indicate that a large number of progteins are likely to be involved in neuronal differentiation.
Oct-2 as a potential switch for regulating neuronal differentiation
Oct-2.4 was identified as a transcription factor that induces neuronal differentiation when fused to the VP16 transactivation domain but not when lacking it. It was further demonstrated that Oct-2.4 represses differentiation in a well-characterized EB-based differentiation assay. Interestingly, this is the only Oct-2 isoform that lacks a C-terminal glutamate-rich transcriptional transactivation domain.
All Oct-2 isoforms containing transactivation domains except Oct-2.6 were shown to be up-regulated during retinoic acid-induced differentiation of EBs. We found that one of these, Oct-2.2, induces neuronal differentiation when overexpressed. We also tested Oct-2.1, Oct-2.3, and Oct-2.5, which have putative transactivation domains for induction of neuronal differentiation. The results from these three isoforms were not definitive and, at most, modest neuronal formation was observed; perhaps cofactors are required for strong neuronal induction with these isoforms. Nonetheless, the knockout results clearly demonstrate that Oct-2 is required for neuronal differentiation as double mutants lacking Oct-2 and its homolog Oct-1 exhibit in a defect in neuronal differentiation.
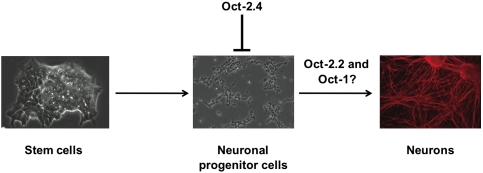
Our results are consistent with a role for Oct-2 as a bifunctional regulator through its different spliced isoforms (Fig. 7). Early in EB formation the Oct-2.4 is one of the more abundant isoforms. This repressive isoform presumably help keep the neuronal differentiation program suppressed. Later during differentiation, the other isoforms containing transcription transactivation domains such as Oct-2.2 become much more abundant and activate neuronal differentiation. In this way a single gene can mediate a switch from repression to activation. Although bifunctional transcriptional regulators have been observed in other contexts (e.g., Max and p53) (Makela et al. 1992; Sauer et al. 2008), we believe this to be the first time that alternatively spliced isoforms of one transcription factor have been shown to have different functional roles in neuronal differentiation of mES cells.
Figure 7.
Oct-2 as a potential bifunctional regulator of neuronal differentiation. We propose that Oct-2.4 is an important isoform in undifferentiated cells that repress neuronal differentiation. Later-activating isoforms of Oct-2 such as Oct-2.2 attain much higher levels and stimulate neuronal differentiation.
Our results are consistent with reporter assays that examined the effects of Oct2 isoforms at individual gene promoters. Oct-2.4 was shown previously to repress transcription of the tyrosine hydroxlyase promoter, whereas Oct-2.1 and Oct-2.5 appear to activate expression of this gene promoter in tissue culture cells (Dawson et al. 1994; Deans et al. 1995). Our study demonstrates that the Oct-2.4 and 2.2 isoforms are repressors and activators of the process of neuronal differentiation, respectively.
Using mES cells to identify functions of human ORFs
Understanding protein function is critical for understanding the many new genes discovered in genome sequencing projects as well the mechanisms controlling basic growth and differentiation and for developing targeted therapies. To date, there have not been many large-scale reagents developed for assessing protein function. Our method using a library of mES cells that are transgenic for a defined set of full-length ORFs is expected to be a powerful and facile approach for assessing protein function. Although human factors were introduced into mouse cells, there is considerable precedent for human proteins that function normally when introduced into mice or mouse cells (e.g., interleukin-6 receptor, A-myb) (Touw et al. 1992; DeRocco et al. 1997). Consistent with this shared function, our results with the human OCT-2.4 clone were identical to those obtained with the mouse Oct-2.4 gene. Because the cell collection is permanent and the cells are easy to grow and maintain, it can be screened for a variety of different assays and will be valuable for many researchers. Of even greater benefit to the research community will be the continuance of the effort begun here to generate transgenic ES lines for all known cDNAs, either in the same expression system or, a similarly designed human ES cell-based system.
Materials and methods
Cell culture and antibiotic selection
Deriviation of oct-2 −/− mES cells and parental wild type J1 cells will be described elsewhere (L. Corcoran, unpubl.). The cell line EBRTcH3 was obtained from the laboratory of Hitoshi Niwa (Laboratory for Pluripotent Cell Studies, RIKEN Center for Developmental Biology, Japan). mES cells were grown in ES + LIF media (DMEM high glucose [Invitrogen, catalog no. 11995-065] supplemented with 15% FBS [Sigma or Inivtrogen] 0.1 mM nonessential amino acids [GIBCO-BRL, catalog no. 11140-050], 0.1 mM 2-mercaptoethanol [Sigma, catalog no. M6250], and 1000 U/mL ESGRO-LIF [Millipore, catalog no. ESG1107]). All stable cell lines derived from EBRTcH3 were grown in ES + LIF + Tet media (ES + LIF supplemented with 2 μg/mL tetracycline). For antibiotic selection of RMCE lines ES + LIF + Tet supplemented with 1.5 μg/mL of puromycin (Sigma, catalog no. P9620) was typically used. An increase to 8.0 μg/mL of puromycin resulted in slower cell growth and was sometimes used to reduce the number of background mES colonies. 293T cells were grown in ES − LIF media (ES + LIF media without LIF).
Antibodies, immunostaining, and Western blotting
Antibody HA.11 was purchased from Covance (catalog no. MMA-101R), antibody TU-20 (anti-βIII-tubulin) and PGP9.5 were purchased from Abcam (TU-20, catalog no. ab7751; PGP9.5, catalog no. ab8189). Secondary antibodies goat anti-mouse Alexa 633, donkey anti-mouse Alexa 555, and donkey anti-rabbit Alexa 555 were purchased from Invitrogen. Immunostaining was carried out as reported previously (Muthuswamy et al. 2001). Briefly, cells were fixed in 4% PFA for 30 min at room temperature, washed twice with PBS, and permeabilized with 0.2% Triton X-100 in PBS. After several PBS washes, 5% goat serum in PBS was used for blocking 1 h at room temperature and then again used for primary antibody dilution. Three 15-min PBS washes were used eliminate unbound and nonspecifically bound primary antibody and a final incubation was done with fluorescence conjugated secondary for 30 min at room temperature. Samples were rinsed with PBS several times before taking pictures.
Western blotting was performed as follows: Protein samples were lysed by suspending cell pellets in 1× RIPA (UpstateBiotechnologies, catalog no. 20-188) containing a 1× Complete protease inhibitor cocktail (Roche, catalog no. 11697498001) and incubating on ice for 45 min. Lysates were mixed and boiled with 2× sample buffer and loaded onto precast 10% polyacrylamide gels (Bio-Rad, catalog no. 131-1155). Following electrophoresis, separated proteins were transferred from polyacrylamide gels to PVDF membrane (Millipore, catalog no. IPVH00010) via semidry transfer. Membranes were blocked with 5% BSA and probed with primary anti-HA (Covance, catalog no. MMS-101R) or β-actin loading control (Sigma, catalog no. A2228).
Generation of transgenic mES cell lines
EBRTcH3 mES cells (Masui et al. 2005) were plated on gelatin coated six-well plates in mES + LIF + Tet media at 1 × 105 cells per well 1 d prior to transfection. For transfection, ∼5–6 μg of each pPthC or pPthC-VP16 ORF containing plasmids were prepared using Qiagen Miniprep columns. In 96 deep-well blocks exchange plasmids were added to 125 μL serum-free media (DMEM with high glucose [GIBCO-BRL, catalog no. 11995-065]) along with 2 μg of pCAGGS-Cre vector. Ten microliters of Lipofectamine 2000 (Invitrogen, catalog no. 11668-019) were added to an equal volume of serum-free media, added to the DNA mixture, and then incubated for 10–15 min at room temperature. Transfection mixes were neutralized with 1.25 mL of mES + LIF media minus antibiotics (DMEM high glucose supplemented with 15% fetal bovine serum and 1000 units/mL ESGRO-LIF) and transferred into aspirated wells containing EBRTcH3 cells. Four hours following transfection cells were washed twice with PBS, trypsinized (0.25% Trypsin-EDTA; Invitrogen, catalog no. 25200-056), and replated in gelatin-coated 10-cm plates containing mES + LIF + Tet media. Cells were switched to selection media (mES media + LIF + Tet + 1.5 μg puromycin) 48 h post-plating, and colonies were allowed to grow out over the next 8–10 d before being picked and trypsinized in 96-well U-bottom plates. Trypsinized cells were neutralized with mES + LIF + Tet media, vigorously pipetted, and then transferred to gelatin-coated 96-well plates where they were allowed to adhere overnight. Cultures were split in half, and one copy of all clones was replated in mES media without LIF and incubated overnight to remove residual tetracycline. Media was changed one more time to mES media without LIF, and after a further 24–48 h transgene-expressing clones were scored by examining levels of Venus YFP expression using fluorescent microscopy.
FACS analysis
The FACS protocol was adopted from that of Sergent-Tanguy et al. (2003). All cells prepared for flow cytometry were dissociated using TryplE Express (Invitrogen, catalog no. 12605-010) and transferred to 50-mL tubes containing PFN (1× PBS, 2% FBS, 0.1% sodium azide) and spun at 1500 rpm for 5 min at 4°C. Cells were fixed in 2% PFA in PBS on ice for 20 min and then washed twice in ice-cold PBS. Washed cells were permeabilized by incubating with 0.5% Saponin in PBS on ice for 20 min. Following two washes in PBS, cells were incubated with primary antibody (Abcam TU-20, 1:500; Covance, HA.11, 1:750) diluted in 0.1% Saponin in PBS, and incubated on ice for 30 min. After two washes with 0.1% Saponin cells were resuspended with goat anti-mouse IgG secondary conjugated with Alexa 633 (Invitrogen) diluted in 0.1% Saponin, and incubated for 30 min on ice. One last wash was done with 0.1% Saponin to remove unincorporated secondary antibody and cells were resuspended in PBS + 0.1% BSA + 1% EM grade methanol-free formaldehyde (Polysciences, Inc., catalog no. 04018). Cell were counted by a hemacytomter and concentration adjusted to 1 × 106 cells per milliliter. Samples were run on a Becton Dickinson FACSCalibur four-color Flow Cytometer and data was analyzed with Becton Dickinson CellQUEST Software version 5.2.1.
Vector design and library construction
DNA fragments generated in this study for plasmid modification were amplified with Platinum Pfx DNA polymerase (Invitrogen, catalog no. 11708013). pPthC-Oct3/4 expression vector (Masui et al. 2005) was modified as follows: Oct3/4 was excised by digesting with XhoI and NotI. The Gateway recombination cassette (Invitrogen) was amplified with forward primer GTGTGTCTCGAGCCACCATGGATTATCCATATGACGTCCCAGACTATGCTACAAGTTTGTACAAAAAAGCTGAAC, which incorporates an HA epitope tag for N-terminal fusions, and reverse primer GTGTGTGCGGCCGCTAAATAGAATAAACCACTTTGTACAAGAAAGCTGA, which incorporates stop codons in all three reading frames immediately downstream from the attR2 recombination site. For VP16 Cterminal fusions a reverse primer with the sequence GTGTGTGCGGCCGCTAAATAGAATAAGCTAGCCACCACTTTGTACAAGAAAGCTGA was used to incorporate an NheI site immediately downstream from the Gateway attR2 site. The VP16 transactivation domain was amplified from plasmid pM3-VP16 AD (Clontech) using the forward and reverse primers GTGTGTACTAGTCCCAAGAAGAAGCGGAAGGTC and GTGTGTGCTAGCCCCACCGTACTCGTCAATTCC. DNAs mOct-1, mOct-1-siR, mOct-2.1, and mOct-2.5 ORFs were synthesized, sequenced, and cloned into pDNR221 by DNA2.0. Human ORFeome 1.1 selected clones were grown in LB supplemented with 100 μg/mL spectinomycin and DNA was extracted using 96-well QIAprep 96 Turbo Miniprep (Qiagen, catalog no. 27191). Shuttling of ORFeome clones into expression vector pPthC-DEST and pPthC-VP16-DEST was done with LR Clonase enzyme (Invitrogen, catalog no. 11791-043). LR reactions were performed in either a 5 μL (96-well format) or 10 μL (individual tube format). Reactions in 10-μL format consisted of 2 μL of 5× LR reaction buffer, 2 μL of 100 ng/μL Entry clone, 2 μL of 100 ng/μL Destination vector, and 4 μL of LR Clonase enzyme.
Lentiviral preparation and infection
Mach7 vector was designed and constructed in collaboration with Patrick Salmon (University of Geneva) and Didier Trono (Laboratory of Virology and Genetics, Ecole Polytechnique Fédérale, Lausanne, Institute of Bioengineering) (E. Theodorou and M. Snyder, in prep.). Lentiviral packaging plasmids ΔR874 and pMD2G were gifts of P. Salmon and D. Trono. For viral packaging 5 × 106 cells were plated in six-well dishes in 2 mL DMEM + 15% FBS minus antibiotics the day before transfection. The day of transfection cells were left without media changing media prior to transfection. Lipofectamine 2000 was used according to manufacturer's specifications, with an optimal L2000 to DBA ratio of ∼2:1. Packaging consisted of tube A [3.3 μg ΔR874 and 0.83 μg pMD2G being added to 83 μL of serum-free media [DMEM minus additives]) and mixed with tube B (10 μL of L2000 added to 83 μL SFM). Transfection mix was added drop-wise to cell cultures and allowed to proceed for ∼5–6 h before media was changed to culture media minus antibiotics. Forty-eight hours post-transfection viral particle-containing media was collected and passed through a 0.45-μm disk filter. Filtered viral supernatant was used immediately for infections or stored for a few days at 4°C or for long-term storage at −80°C. Viral infection with M7-eGFP, M7-Oct-1, and M7-siR-Oct-1 was done by trypsinizing mES cells and resuspending them in standard ES growth media at 1 × 105 cells in a 200-μL volume. To the freshly plated cells, 1.3 mL of undiluted viral supernatant was added along with a final concentration of 6 μg/mL Polybrene (Sigma, catalog no. 107689). Cells were left in viral supernatant overnight before being switched back to standard growth media. Antibiotic selection was applied 48 h post-infection, and cells were maintained in selection media for a minimum of two passages. Transgene expression was confirmed by anti-HA immunostaining of cells having undergone selection.
EB culture
EBs were generated by using culture methods similar to those published previously (Bibel et al. 2007). One day prior to EB formation wild-type and transgenic mES lines were placed in ES + LIF media minus any antibiotics. To form EBs cells were trypsinized and pipeted until they became a primarily single-cell suspension. Single-cell suspensions were plated in 15 mL of ES − LIF media in 10-cm triple-vented petri dishes (Greiner Bio-One, catalog no. 82050-574), and thereafter media was changed every 2 d. For retinoic acid treatment 5 μM all trans-retinoic acid was added to EBs on days 6 and 8 during media changes. On day 8, fully formed EBs were collected and washed twice in PBS by gravity sedimentation and dissociated with freshly made 0.5% trypsin with chymotrypsin inhibitor (TPCK, Worthington Biochemical, catalog no. LS003750) in 0.5% EGTA in PBS at 37°C. TPCK treatment was halted with ES − LIF media and EBs were pipeted vigorously until they were well dissociated. Dissociated EBs were centrifuged for 5 min at 2000 rpm in order to concentrate cells and to remove traces of serum-containing media. Cells were resuspended in N2 media (DMEM:F12 1:1 (GIBCO-BRL, catalog no. 11330-032), 1× N2 supplement (GIBCO-BRL, catalog no. 17502-048), 20 μg/mL insulin (Sigma, catalog no. I0516), 50 μg/mL Fraction V BSA (GIBCO-BRL, catalog no. 15260-037), and 0.25× GlutaMAX (GIBCO-BRL, catalog no. 35050-061), and then added dropwise to a 40-μm cell strainer (BD Falcon, catalog no. 352340) to remove any large cell clumps or debris. Cell suspensions were counted by hemacytomter and plated in poly-ornithine/laminin-coated dishes (BD Biosciences, catalog no. 354658). Prior to plating cells a solution of 5 μg/mL laminin (Roche Diagnostics, catalog no. 11243217001) was added to the precoated plates for 2 h at room temperature to allow for better adherence of neural precursors. N2 suspended cells were plated at a density of 2.5 × 106 cells/well in a six-well plate. N2 media was changed 24 h postplating and the switched to a modified version of Complete media (DMEM:F12 1:1 supplemented with with 1× B27 reagent [GIBCO-BR, catalog no. 17504-044] and 0.25× Glutamax) at 48 h. Four days following the addition of Complete media cells were either directly fixed and stained or prepared for FACS analysis.
Stromal cell induction of neuronal differentiation
PA6 stromal cell (RIKEN) differentiation was performed as published previously (Kawasaki et al. 2000) with slight modification. On the day of mES cells plating, PA6 stromal cells were fixed in 4% paraformaldehyde for 30 min at room temperature, then washed in PBS three times, 15 min each wash. mES cells grown in ES media were trypsinized and then neutralized with ES medium minus LIF, pelleted, and resuspended in ES differentiation media (GMEM [GIBCO-BRL, catalog no. 11710-035] supplemented with 10% KSR [GIBCO-BRL, catalog no. 10828-028], 0.1 mM nonessential amino acids [GIBCO-BRL, catalog no. 11140-050], 1 mM sodium pyruvate [Sigma, catalog no. S-8636], 0.1 mM 2-mercaptoethanol [Sigma, catalog no. M6250]). ESGRO-LIF (1000 U/mL) was added to increase plating efficiency. mES cells were counted and plated at a density of 5 × 104 cells per dish. Twelve hours to 16-h post-plating cells were washed once with PBS and media was changed to ES differentiation media without LIF. Media was changed every 48 h until day 8 when cells were harvested for FACS analysis.
RNA isolation and gene expression analysis
RNA was isolated using the RNeasy Kit (Qiagen, catalog no. 74104) along with Qiashredder columns (Qiagen, catalog no. 79654). RNA concentration was measured using a Nanodrop ND-1000 UV-Vis Spectrophotometer (Nanodrop Technologies). The first strand was generated using Superscript III First-Strand Synthesis Supermix for qRT–PCR (Invitrogen, catalog no. 11752-050) with 1 μg of total RNA. TaqMan assay was performed according to manufacturer's specifications using TaqMan master mix (Applied Biosystems, catalog no. 4369016) and custom primers and probes designed by Applied Biosystems. First-strand samples were typically diluted 1:10. cDNAs spanning the relevant cDNA regions to be amplified were synthesized by DNA2.0 as positive controls for quantifying efficiency of primers. Samples were normalized to actin (Applied Biosystems, catalog no. 4352933E).
Acknowledgments
We thank J. Minshull and C. Gustaffson (DNA 2.0) for DNA synthesis and technical advice, R. West (Van Andel Institute) for FACS analysis, J. Wolenski (Yale University) for confocal microscopy, and C.M. Schwartz (NIH) for technical advice with PA6 stromal assay. Special thanks to H. Im, A. Nath, and E. Miriami for helpful discussions and critical review of this manuscript. This work was supported by grants from the CT Stem Cell initiative and NIH to M.S. and S.W.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1772509.
Supplemental material is available at http://www.genesdev.org.
References
- Andersson E., Tryggvason U., Deng Q., Friling S., Alekseenko Z., Robert B., Perlmann T., Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Bibel M., Richter J., Lacroix E., Barde Y.A. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protocols. 2007;2:1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- Braselmann S., Graninger P., Busslinger M. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins. Proc. Natl. Acad. Sci. 1993;90:1657–1661. doi: 10.1073/pnas.90.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig J.J., Silbereis J., Vaccarino F.M., Sestan N., Rakic P. Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc. Natl. Acad. Sci. 2007;104:20558–20563. doi: 10.1073/pnas.0710156104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran L.M., Karvelas M., Nossal G.J., Ye Z.S., Jacks T., Baltimore D. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes & Dev. 1993;7:570–582. doi: 10.1101/gad.7.4.570. [DOI] [PubMed] [Google Scholar]
- Corcoran L.M., Koentgen F., Dietrich W., Veale M., Humbert P.O. All known in vivo functions of the Oct-2 transcription factor require the C-terminal protein domain. J. Immunol. 2004;172:2962–2969. doi: 10.4049/jimmunol.172.5.2962. [DOI] [PubMed] [Google Scholar]
- Dawson S.J., Yoon S.O., Chikaraishi D.M., Lillycrop K.A., Latchman D.S. The Oct-2 transcription factor represses tyrosine hydroxylase expression via a heptamer TAATGARAT-like motif in the gene promoter. Nucleic Acids Res. 1994;22:1023–1028. doi: 10.1093/nar/22.6.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans Z., Dawson S.J., Buttery L., Polak J.M., Wallace D., Latchman D.S. Direct evidence that the POU family transcription factor Oct-2 represses the cellular tyrosine hydroxylase gene in neuronal cells. J. Mol. Neurosci. 1995;6:159–167. doi: 10.1007/BF02736762. [DOI] [PubMed] [Google Scholar]
- Deans Z., Dawson S.J., Xie J., Young A.P., Wallace D., Latchman D.S. Differential regulation of the two neuronal nitric-oxide synthase gene promoters by the Oct-2 transcription factor. J. Biol. Chem. 1996;271:32153–32158. doi: 10.1074/jbc.271.50.32153. [DOI] [PubMed] [Google Scholar]
- Delaune E., Lemaire P., Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- Delfini M.C., Duprez D. Ectopic Myf5 or MyoD prevents the neuronal differentiation program in addition to inducing skeletal muscle differentiation, in the chick neural tube. Development. 2004;131:713–723. doi: 10.1242/dev.00967. [DOI] [PubMed] [Google Scholar]
- Dent C.L., Lillycrop K.A., Estridge J.K., Thomas N.S., Latchman D.S. The B-cell and neuronal forms of the octamer-binding protein Oct-2 differ in DNA-binding specificity and functional activity. Mol. Cell. Biol. 1991;11:3925–3930. doi: 10.1128/mcb.11.8.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRocco S.E., Iozzo R., Ma X.P., Schwarting R., Peterson D., Calabretta B. Ectopic expression of A-myb in transgenic mice causes follicular hyperplasia and enhanced B lymphocyte proliferation. Proc. Natl. Acad. Sci. 1997;94:3240–3244. doi: 10.1073/pnas.94.7.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fainsod A., Deissler K., Yelin R., Marom K., Epstein M., Pillemer G., Steinbeisser H., Blum M. The dorsalizing and neural inducing gene follistatin is an antagonist of BMP-4. Mech. Dev. 1997;63:39–50. doi: 10.1016/s0925-4773(97)00673-4. [DOI] [PubMed] [Google Scholar]
- Falk A., Karlsson T.E., Kurdija S., Frisen J., Zupicich J. High-throughput identification of genes promoting neuron formation and lineage choice in mouse embryonic stem cells. Stem Cells. 2007;25:1539–1545. doi: 10.1634/stemcells.2006-0485. [DOI] [PubMed] [Google Scholar]
- Farah M.H., Olson J.M., Sucic H.B., Hume R.I., Tapscott S.J., Turner D.L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lamuno D., Loukili N., Garcia-Fuentes M., Thomson T.M. Expression and regulation of the transcriptional repressor ZNF43 in Ewing sarcoma cells. Pediatr. Pathol. Mol. Med. 2002;21:531–540. doi: 10.1080/15227950290112789. [DOI] [PubMed] [Google Scholar]
- Gray P.A., Fu H., Luo P., Zhao Q., Yu J., Ferrari A., Tenzen T., Yuk D.I., Tsung E.F., Cai Z., et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Hakimi M.A., Bochar D.A., Chenoweth J., Lane W.S., Mandel G., Shiekhattar R. A core–BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Treacy M.N., Simmons D.M., Ingraham H.A., Swanson L.W., Rosenfeld M.G. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature. 1989;340:35–41. doi: 10.1038/340035a0. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A., Kelly O.G., Melton D.A. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell. 1994;77:283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Hynes M., Porter J.A., Chiang C., Chang D., Tessier-Lavigne M., Beachy P.A., Rosenthal A. Induction of midbrain dopaminergic neurons by Sonic hedgehog. Neuron. 1995;15:35–44. doi: 10.1016/0896-6273(95)90062-4. [DOI] [PubMed] [Google Scholar]
- Immaneni A., Lawinger P., Zhao Z., Lu W., Rastelli L., Morris J.H., Majumder S. REST-VP16 activates multiple neuronal differentiation genes in human NT2 cells. Nucleic Acids Res. 2000;28:3403–3410. doi: 10.1093/nar/28.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Hatakeyama J., Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. Roles of Hes genes in neural development. Dev. Growth Differ. 2008;50:S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- Kawasaki H., Mizuseki K., Nishikawa S., Kaneko S., Kuwana Y., Nakanishi S., Nishikawa S.I., Sasai Y. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- Kitajima H., Yoshimura S., Kokuzawa J., Kato M., Iwama T., Motohashi T., Kunisada T., Sakai N. Culture method for the induction of neurospheres from mouse embryonic stem cells by coculture with PA6 stromal cells. J. Neurosci. Res. 2005;80:467–474. doi: 10.1002/jnr.20469. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Ma Q. Transcriptional regulation of neuronal phenotype in mammals. J. Physiol. 2006;575:379–387. doi: 10.1113/jphysiol.2006.113449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela T.P., Koskinen P.J., Vastrik I., Alitalo K. Alternative forms of Max as enhancers or suppressors of Myc-ras cotransformation. Science. 1992;256:373–377. doi: 10.1126/science.256.5055.373. [DOI] [PubMed] [Google Scholar]
- Mankoo B.S., Collins N.S., Ashby P., Grigorieva E., Pevny L.H., Candia A., Wright C.V., Rigby P.W., Pachnis V. Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature. 1999;400:69–73. doi: 10.1038/21892. [DOI] [PubMed] [Google Scholar]
- Masui S., Shimosato D., Toyooka Y., Yagi R., Takahashi K., Niwa H. An efficient system to establish multiple embryonic stem cell lines carrying an inducible expression unit. Nucleic Acids Res. 2005;33:e43. doi: 10.1093/nar/gni043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy S.K., Li D., Lelievre S., Bissell M.J., Brugge J.S. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat. Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Scholer H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- O'Shea K.S. Neuronal differentiation of mouse embryonic stem cells: Lineage selection and forced differentiation paradigms. Blood Cells Mol. Dis. 2001;27:705–712. doi: 10.1006/bcmd.2001.0435. [DOI] [PubMed] [Google Scholar]
- Riazi A.M., Lee H., Hsu C., Van Arsdell G. CSX/Nkx2.5 modulates differentiation of skeletal myoblasts and promotes differentiation into neuronal cells in vitro. J. Biol. Chem. 2005;280:10716–10720. doi: 10.1074/jbc.M500028200. [DOI] [PubMed] [Google Scholar]
- Rual J.F., Hirozane-Kishikawa T., Hao T., Bertin N., Li S., Dricot A., Li N., Rosenberg J., Lamesch P., Vidalain P.O., et al. Human ORFeome version 1.1: A platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y., Lu B., Steinbeisser H., De Robertis E.M. Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature. 1995;376:333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- Sauer M., Bretz A.C., Beinoraviciute-Kellner R., Beitzinger M., Burek C., Rosenwald A., Harms G.S., Stiewe T. C-terminal diversity within the p53 family accounts for differences in DNA binding and transcriptional activity. Nucleic Acids Res. 2008;36:1900–1912. doi: 10.1093/nar/gkn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergent-Tanguy S., Chagneau C., Neveu I., Naveilhan P. Fluorescent activated cell sorting (FACS): A rapid and reliable method to estimate the number of neurons in a mixed population. J. Neurosci. Methods. 2003;129:73–79. doi: 10.1016/s0165-0270(03)00210-3. [DOI] [PubMed] [Google Scholar]
- Shimozaki K., Nakashima K., Niwa H., Taga T. Involvement of Oct3/4 in the enhancement of neuronal differentiation of ES cells in neurogenesis-inducing cultures. Development. 2003;130:2505–2512. doi: 10.1242/dev.00476. [DOI] [PubMed] [Google Scholar]
- Su X., Kameoka S., Lentz S., Majumder S. Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation. Mol. Cell. Biol. 2004;24:8018–8025. doi: 10.1128/MCB.24.18.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y., Roelink H., Jessell T.M. Induction of motor neurons by Sonic hedgehog is independent of floor plate differentiation. Curr. Biol. 1995;5:651–658. doi: 10.1016/s0960-9822(95)00130-8. [DOI] [PubMed] [Google Scholar]
- Tantin D., Schild-Poulter C., Wang V., Hache R.J., Sharp P.A. The octamer binding transcription factor Oct-1 is a stress sensor. Cancer Res. 2005;65:10750–10758. doi: 10.1158/0008-5472.CAN-05-2399. [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Spiegelman B.M. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Touw I., van Gurp R., Schipper P., van Agthoven T., Lowenberg B. Introduction of the human interleukin-6 (IL-6) receptor in murine IL-3-dependent hematopoietic cells restores responsiveness to IL-6. Blood. 1992;79:2867–2872. [PubMed] [Google Scholar]
- Uittenbogaard M., Chiaramello A. Constitutive overexpression of the basic helix–loop–helix Nex1/MATH-2 transcription factor promotes neuronal differentiation of PC12 cells and neurite regeneration. J. Neurosci. Res. 2002;67:235–245. doi: 10.1002/jnr.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra G.J., van der Vliet P.C., Destree O.H. POU domain transcription factors in embryonic development. Mol. Biol. Rep. 1997;24:139–155. doi: 10.1023/a:1006855632268. [DOI] [PubMed] [Google Scholar]
- Wang V.E., Schmidt T., Chen J., Sharp P.A., Tantin D. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol. Cell. Biol. 2004;24:1022–1032. doi: 10.1128/MCB.24.3.1022-1032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Tapscott S.J., Davis R.L., Thayer M.J., Adam M.A., Lassar A.B., Miller A.D. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Priess A., Annweiler A., Zwilling S., Oeler B. Multiple Oct2 isoforms are generated by alternative splicing. Nucleic Acids Res. 1991;19:43–51. doi: 10.1093/nar/19.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.N., Lillycrop K.A., Dent C.L., Ninkina N.N., Beech M.M., Willoughby J.J., Winter J., Latchman D.S. Regulation of expression of the neuronal POU protein Oct-2 by nerve growth factor. J. Biol. Chem. 1992;267:17787–17791. [PubMed] [Google Scholar]
- Zhou Q., Anderson D.J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zimmerman L.B., De Jesus-Escobar J.M., Harland R.M. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- Zou D., Silvius D., Fritzsch B., Xu P.X. Eya1 and Six1 are essential for early steps of sensory neurogenesis in mammalian cranial placodes. Development. 2004;131:5561–5572. doi: 10.1242/dev.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]