Abstract
The nucleolus is a dynamic structure that has roles in various processes, from ribosome biogenesis to regulation of the cell cycle and the cellular stress response. Such functions are frequently mediated by the sequestration or release of nucleolar proteins. Our understanding of protein targeting to the nucleolus is much less complete than our knowledge of membrane-spanning translocation systems—such as those involved in nuclear targeting—and the experimental evidence reveals that few parallels exist with these better-characterized systems. Here, we discuss the current understanding of nucleolar targeting, explore the types of sequence that control the localization of a protein to the nucleolus, and speculate that certain subsets of nucleolar proteins might act as hub proteins that are able to bind to multiple protein targets. In parallel to other subnuclear structures, such as PML bodies, the proteins that are involved in the formation and maintenance of the nucleolus are inexorably linked to nucleolar trafficking.
Keywords: hub protein, nucleolar localization, nucleolin, nucleolus, nucleophosmin
Glossary
ARF alternative reading frame product of the CDKN2A locus
dsRNA double-stranded RNA
EGFP enhanced green-fluorescent protein
FGF2 fibroblast growth factor 2
HIF hypoxia-inducible factor
HIV-1 human immunodeficiency virus type 1
HSV-1 herpes simplex virus type 1
HVS herpesvirus saimiri
MIZ1 Myc-associated zinc-finger protein
NF-κB nuclear factor-κB
NOM1 nucleolar protein with MIF4G domain 1
NPM nucleophosmin, also known as B23.1 (there is also an alternative splice variant called B23.2)
NRF nuclear factor-κB-repressing factor
ORF open reading frame
PML promyelocytic leukaemia protein
PP1 protein phosphatase 1 (a serine/threonine phosphatase)
PRRSV porcine reproductive and respiratory syndrome virus
rDNA ribosomal DNA
RelA nuclear factor-κB p65/Rel A
rRNA ribosomal RNA
SUMO small ubiquitin-like modifier
UBF upstream binding factor
UL24 unique long 24
US11 unique short 11
VHL von Hippel–Lindau tumour-suppressor protein
Introduction
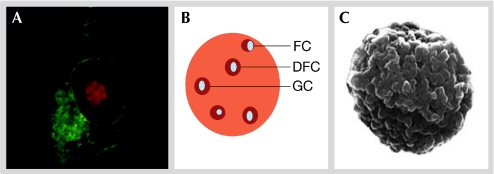
The nucleus is a highly ordered structure that contains non-membrane-bound subcompartments—including PML bodies, splicing speckles, Cajal bodies and the nucleolus—that have specific functions. The nucleolus is the largest subnuclear structure (Fig 1) and is easily visible under the light microscope owing to its high refractive index. It is centred on rDNA repeats within the chromosomes and is traditionally associated with ribosome biogenesis. In mammalian cells, the number and activity of nucleoli vary during the cell cycle according to differing metabolic conditions and cell types.
Figure 1.
Structure of the nucleolus. (A) Live-cell laser-scanning confocal microscope image showing the localization of a fluorescently tagged nucleolar marker protein (red) with a fluorescently tagged cytoplasmic marker protein (green). The nucleolus constitutes a significant proportion of the nucleus and contains defined features. (B) Diagrammatic representation of the mammalian nucleolus showing the positions of the FC, DFC and GC. (C) Scanning electron micrograph of a nucleolus purified from HeLa cells. The surface corresponds to a shell of highly condensed heterochromatin that surrounds the nucleolus in vivo (image courtesy of Angus Lamond, University of Dundee, UK). DFC, dense fibrillar component; FC, fibrillar centre; GC, granular component.
The mammalian nucleolus can be morphologically divided into several discrete regions—the fibrillar centre (FC), the dense fibrillar centre (DFC) and the granular component (GC)—that have roles in the various steps of rRNA synthesis. The FC contains the transcription factor UBF and is rich in RNA polymerase I. The DFCs are associated with, and surround, the FCs and contain fibrillarin, an RNA methyltransferase and nucleolin—a protein that has multiple roles in nucleolar and cellular biology (Mongelard & Bouvet, 2007). Surrounding both the FC and the DFC is the GC, which is the site of the partial maturation and assembly of pre-ribosomes, accumulates NPM, and is enriched with ribosomal proteins and assembly factors. The GC might also contain regions that comprise protein complexes that are devoid of RNA (Politz et al, 2005).
Our understanding of nucleolar function changed markedly with the formulation of the plurifunctional nucleolus hypothesis (Pederson, 1998), which has now been underpinned by proteomic analysis (Boisvert et al, 2007), and proposes that the nucleolus has multiple functions in health and disease (Matthews & Olson, 2006; Stark & Taliansky, 2009). For example, there is extensive evidence that the nucleolus is involved in the response to cellular stress (Mayer & Grummt, 2005; Rubbi & Milner, 2003), and in the regulation of the cell cycle and cell growth (Martindill & Riley, 2008). The nucleolus can also be a target for virus infection (Hiscox, 2007) and perturbations to the nucleolus have been observed in a wide range of cellular diseases, from auto-immunity to cancer (Montanaro et al, 2008).
The regulation of many nucleolar roles is mediated by the sequestration or release of specific proteins, thereby providing insight into one of the mechanisms by which the nucleolus operates. This is illustrated by the sequestration of NF-κB (RelA) to the nucleolus in colorectal cancer cell lines in the presence of non-steroidal anti-inflammatory drugs such as aspirin, which induces apoptosis (Thoms et al, 2007). Another implication of continuous protein sequestration and release is that the nucleolar proteome is continuously changing in response to metabolic conditions and driving metabolic change (Lam et al, 2007). For example, the ribosomal—and part nucleolar—protein L23 is retained in the nucleolus by NPM, thereby enhancing MIZ1-mediated cell-cycle arrest (Wanzel et al, 2008). NPM also sequesters the ribosomal protein S9 in nucleoli and directly affects ribosome biogenesis (Lindstrom & Zhang, 2008). The localization and stability of nucleolin—one of the most abundant nucleolar proteins—can also change in response to differing metabolic conditions such as osmotic changes (Yang et al, 2008) and developmental stages (Bicknell et al, 2005). Similarly, alterations in cellular pH—for example, owing to hypoxia or acidosis—have been shown to cause the nucleolar sequestration of the VHL protein (Mekhail et al, 2004), one function of which is to degrade HIF, which activates genes involved in tumour vascularization and oxygen homeostasis. Therefore, the sequestration of VHL to the nucleolus during hypoxia or acidosis—which is mediated by a H+-responsive nucleolar localization signal (NoLS)—allows the activation of HIF until normal cellular conditions are reached, which results in the release of VHL from the nucleolus and the subsequent inhibition of HIF function (Mekhail et al, 2004).
The continuous exchange of nucleolar proteins with the nucleoplasm and cytoplasm in response to fluctuating cellular conditions indicates that nucleolar proteins might have specific signals or pathways that determine nucleolar sequestration or release.
Hub proteins in the nucleolus?
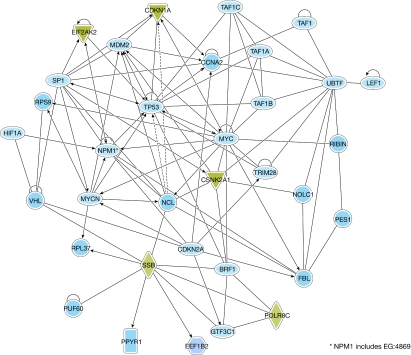
The nucleolus consists of complex protein–protein and protein–nucleic acid interactions, and, although it is imaged as a steady-state structure, high-throughput proteomic analysis of purified nucleoli coupled with live-cell imaging of fluorescently labelled nucleolar proteins has revealed a dynamic nuclear compartment, the components of which undergo continuous exchange with the nucleoplasm (Andersen et al, 2005; Hernandez-Verdun, 2006). The current human nucleolar proteome database—which has an estimated 80% coverage—consists of more than 4,500 proteins (Ahmad et al, 2009). Many nucleolar proteins interact with one another, and with other nuclear and cytoplasmic proteins depending on trafficking and the cell-cycle stage (Fig 2), and perturbations to certain proteins in the nucleolus—either enrichment or ablation—have a knock-on effect on partner proteins and, hence, function. The nucleolus is assembled in mitosis, when nucleolin, NPM, fibrillarin and other factors form nucleolus-derived foci (Hernandez-Verdun et al, 2002). Therefore, members of a subset of the nucleolar proteome act as building blocks of the nucleolus around rDNA repeats, and it is formed in an incremental manner (Dundr et al, 2000). For assembly to occur in this way, these blocks must bind to multiple partner molecules, thereby probably functioning as so-called hub proteins.
Figure 2.
Pathway analysis of direct interactions between selected nucleolar proteins, and other nuclear and cytoplasmic proteins. This interaction map was generated using Ingenuity Pathway Analysis software (Ingenuity Systems, Inc., Redwood City, CA, USA), and shows the complexity of the interactions—many of which are interlinked—and how perturbations to one protein might affect the function of another. Examples of proteins that can traffic to the nucleolus and that are discussed in the text, such as VHL, are also shown. The full names of all of the proteins that are shown on the map can be found in supplementary Table 1 online. FBL, fibrillarin; NCL, nucleolin; NPM1, nucleophosmin; VHL, von Hippel–Lindau tumour-suppressor protein.
A common characteristic of hub proteins is the ability to bind to 10 or more distinct proteins (Krasowski et al, 2008). Hub proteins have been extensively studied in Saccharomyces cerevisiae, in which two types have been identified (Ekman et al, 2006). Dynamic hub proteins are proposed to bind to different partner molecules at different times or subcellular locations, whereas static hubs interact with most partners simultaneously. Traditionally, hub proteins have been thought to be important components of protein–protein interaction networks; however, the possibility that their capacity to interact with large numbers of target proteins could also have a structural role has so far been neglected. The nucleolus and other nuclear structures without membranes are dynamic, with their constituent proteins in constant flux. Therefore, if a hub protein is present in one compartment in sufficient quantity, we speculate that sequential transient interactions between the targeted protein and multiple hub proteins could be sufficient to induce a period of residence within the compartment. Hub proteins are known to facilitate interactions with a wide range of partner proteins (Kim et al, 2008), while retaining the ability to distinguish between suitable partners. In this case, localization works through the recognition of structural or sequence-based motifs within a protein, which make possible its identification as a protein that must be retained within a specific subcellular localization.
Hub proteins might be responsible for much of the nucleolar localization that occurs in the absence of protein–RNA interactions. Each hub protein could have different recognition requirements and/or multiple recognition sites, thereby explaining the existence of different NoLSs. This hypothesis is supported by the observation that many of the NoLSs for which binding partners are known interact with the same small subset of nucleolar proteins, thereby pointing to hub protein candidates, which include nucleolin and NPM. Whether such proteins are dynamic or static hubs is difficult to elucidate, and they might share characteristics of the two types. For example, both proteins can form multiple stable interactions throughout the cell cycle (Fig 2), which is a characteristic of static hubs; however, given the transitory nature of a subset of the nucleolar proteome—including ribosomal proteins, which are exported to the cytoplasm (Andersen et al, 2005)—they also show characteristics of a dynamic hub protein.
Nucleolin and NPM are predicted to contain disordered domains in an important part of the overall protein structure—corresponding to 55% and 47%, respectively (Fig 3)—which is a characteristic of hub proteins. Disordered proteins tend to have larger surface-to-volume ratios than ordered ones (Gunasekaran et al, 2003), and extensive surface areas are advantageous for binding to different ligands. Disordered proteins have been shown to interact with multiple protein or nucleic-acid partners, while maintaining an overall size that is two to three times smaller than ordered proteins (Gunasekaran et al, 2003); this characteristic is also advantageous for proteins that act as scaffolds for dense compartments such as the nucleolus, which would otherwise be considerably larger. Furthermore, disordered domains have been proposed to be involved in nucleolar protein–RNA interactions (Yiu et al, 2006). The known importance of nucleolin and NPM for the formation of nucleoli also supports their roles as hub proteins. As nucleoli breakdown and reform during M phase, the idea that such disruption could be controlled by affecting a small number of proteins that are crucial for maintaining the structural integrity of nucleoli has obvious advantages (Ma et al, 2007). The ablation of either nucleolin or NPM using RNA interference is sufficient to disrupt the nucleolar structure, although the exact nature of the disruption varies between the two proteins (Amin et al, 2008; Ma et al, 2007; Ugrinova et al, 2007). The depletion of NPM results in an accumulation of SUMOylated proteins within nucleoli (Yun et al, 2008), whereas the depletion of nucleolin results in the accumulation of cells in the G2 phase of the cell cycle, apoptosis, centrosome-control deficiencies and nuclear deformities, including multiple nuclei, micronuclei and large nuclei (Ugrinova et al, 2007). These findings illustrate the concept of the centrality–lethality rule, whereby the deletion of a hub protein is more likely to be lethal than the deletion of a non-hub protein (He & Zhang, 2006). Indeed, transgenic-knockout studies have shown that NPM is essential in the maintenance of genomic stability and embryonic development (Grisendi et al, 2005). Similarly, genetic mutations that cause the cytoplasmic localization of NPM and result in the stabilization of c-Myc are the most frequent alterations observed in acute myelogenous leukaemia (Bonetti et al, 2008). Conversely, overexpression of NPM can also contribute to the transforming ability of c-Myc (Li et al, 2008). In addition, changes in nucleolar structure can result from virus infection (Dove et al, 2006); nucleolin is dispersed by the UL24 protein of HSV-1, resulting in the alteration of nucleolar structure (Bertrand & Pearson, 2008; Lymberopoulos & Pearson, 2007).
Figure 3.
Diagrammatic illustration of nucleolin and nucleophosmin. Various protein domains are highlighted, as well as the consensus results of a disorder analysis that was performed using several disorder predictors (see supplementary Fig 1 and supplementary Table 2 online for details of this analysis). NLS, nuclear localization signal.
Targeting proteins to and from the nucleolus
The localization of proteins to membrane-bound compartments such as the nucleus is mediated by dedicated targeting systems that recognize specific protein motifs and vary depending on protein size, hydrodynamic radius and concentration. Many proteins that traffic to and from the nucleus by the nuclear-pore complex contain one or more nuclear localization signals (NLSs) and, if necessary, also nuclear export signals (NESs). These motifs are well characterized and moderately conserved, and can therefore be predicted by suitable algorithms (Cokol et al, 2000; la Cour et al, 2004). However, the motifs that are involved in regulating nucleolar localization are not well defined and it is believed that nucleolar localization of a protein results from either direct or indirect interaction with one of the nucleolar building blocks—that is, with rDNA, its transcripts or protein components (Carmo-Fonseca et al, 2000). Therefore, rather than a targeting signal that acts as a recognition motif for the binding to import machinery, such signals would be responsible for high-affinity interactions with proteins or nucleic acids that reside within the nucleolus and thereby impart localization. Initial support for this hypothesis came from early studies of nucleolar proteins (Schmidt-Zachmann & Nigg, 1993); for example, although nucleolin was shown to contain a NLS, no defined NoLS could be identified, and it was therefore postulated that RNA-binding domains that were present within nucleolin were responsible for nucleolar accumulation. Similarly, other proteins that localize to the nucleolus might do so by binding directly to dsRNA, such as the NRF (Niedick et al, 2004).
Nucleolin has been shown to associate with NPM—which contains a NoLS—and therefore probably transports nucleolin to the nucleolus (Li et al, 1996). NPM can also associate with the ARF tumour-suppressor protein and retarget it to the nucleolus, where its function is inhibited (Korgaonkar et al, 2005). Another example of a protein that can localize to the nucleolus in the absence of a specific NoLS is PP1. In this case, nucleolar localization is driven by the NoLS-containing nucleolar protein NOM1 (Gunawardena et al, 2008). The association with NPM and other nucleolar proteins is also necessary for the trafficking and function of several viral proteins (Hiscox, 2007) such as the ORF3 protein of the plant umbravirus groundnut rosette virus, which associates with fibrillarin for the formation of viral ribonucleoprotein particles, the nucleolar translocation of which is essential for infection (Kim et al, 2007).
In some cases, the NoLSs are part of the NLSs, making their identification problematic and again pointing to the lack of consensus. FGF2—which regulates cell proliferation—contains two NLSs and the carboxy-terminal one also acts as a NoLS (Sheng et al, 2004). The ORF57 protein of HSV contains two NLSs, both of which are involved in nucleolar localization (Boyne & Whitehouse, 2006). Parafibromin—which is a tumour-suppressor protein—contains a bipartite NLS and three distinct NoLSs, all of which contribute to the efficiency of the nucleolar localization of this protein (Hahn & Marsh, 2007).
Several motifs have been identified that can be both necessary and sufficient—that is, they will direct an exogenous protein—to target a protein to the nucleolus; these are composed mostly of Arg or Lys residues. Such motifs can range in size from short sequences of seven or eight amino acids to approximately 30 residues (Table 1; Birbach et al, 2004; Reed et al, 2006). For example, studies of the NoLS of the human La protein and sequence alignment with other motifs indicated that the (R/K)(R/K)X(R/K) motif appeared once or as multiple copies in peptides that were targeted to the nucleolus (Horke et al, 2004); such motifs had been known since the discovery of NoLSs in HIV-1 and cellular proteins (Hatanaka, 1990; Dang & Lee, 1989). The conjugation of defined NoLSs to fluorescent proteins such as EGFP indicates that the motifs responsible for the function of a protein within the nucleolus can be distinct from those involved in nucleolar localization. For example, the nucleocapsid protein of the PRRSV—which binds to viral RNA—travels between the nucleolus and the cytoplasm (Rowland et al, 1999; You et al, 2008), and has separate motifs that are responsible for NoLS activity and fibrillarin binding, the association to which might cause defects in rRNA production (Rowland et al, 2003; Yoo et al, 2003). Presumably, nucleolar proteins that are above the size exclusion limit of the nuclear pore complex might also require a NLS for active transport through the nuclear envelope. This might explain why many NLSs and NoLSs contain shared motifs, as genetic stability and the availability of interfaces that are accessible for interaction with the trafficking machinery would drive the selection for proximal motifs.
Table 1.
Nucleolar localization sequences
| Protein | NoLS sequence | Position | Accession number | References |
|---|---|---|---|---|
| IBV N protein | WRRQARFK | 71–78 | AAA46214 | Reed et al, 2006 |
| ApLLP | MAKSIRSKHRRQMRMMKRE | 1–19 | ABY66901 | Kim et al, 2003 |
| HIV-1 TAT | RKKRRQRRRAHQ | 48–61 | AAC82591 | Siomi et al, 1990 |
| GGNNVα | RRRANNRRR | 23–31 | ACB21052 | Guo et al, 2003 |
| Angiogenin | IMRRRGL | 53–59 | AAA51678 | Lixin et al, 2001 |
| HSV type 1 γ(1)34.5 | MARRRRHRGPRRPRPP | 1–16 | P08353 | Cheng et al, 2002 |
| HIV-1 REV | RRNRRRRWRERQRQI | 38–52 | CAA41586 | Cochrane et al, 1990 |
| FGF2 | RSRKYTSWYVALKR | 249–262 | NP_001997 | Sheng et al, 2004 |
| MDM2 | KKLKKRNK | 466–473 | Q00987 | Lohrum et al, 2000 |
| NIK | RKKRKKK | 143–149 | Q99558 | Birbach et al, 2004 |
| Nuclear VCP-like protein | KRKGKLKNKGSKRKK | 112–126 | NP_996671 | Nagahama et al, 2004 |
| p120 | SKRLSSRARKRAAKRRLG | 40–57 | P46087 | Valdez et al, 1994 |
| HIC p40 | GRCRRLANFGPRKRRRRRR | 44–62 | Q9P1T7 | Thebault et al, 2000 |
| MDV MEQ protein | RRRKRNRDARRRRRKQ | 62–78 | AAB48631 | Liu et al, 1997 |
| HVS ORF57 | KRPR-RRPSRPFRKP (a bipartite NoLS; 25 residues separate the two halves in wild type, but it retains its functionality when they are joined) | 91–94, 119–128 | NP_040259 | Boyne & Whitehouse, 2006 |
| LIMK2 | KKRTLRKNDRKKR | 491–503 | CAG30399 | Goyal et al, 2006 |
| PRRSV N protein | PGKKNKKKNPEKPHFP LATEDDVRHHFTPSER | 41–72 | AAG13733 | Rowland et al, 1999; Rowland et al, 2003 |
ApLLP, Aplysia LAPS18-like protein; FGF2, fibroblast growth factor 2; GGNNVα, betanodavirus greasy grouper (Epinephelus tauvina) nervous necrosis virus protein α; HIC p40, human I-mfa domain-containing protein, p40; HIV-1 Rev, human immunodeficiency virus-1 regulator of virion protein; HIV-1 TAT, human immunodeficiency virus-1 transactivator of transcription protein; HVS, herpesvirus saimiri; HSV type 1 γ(1) 34.5, herpes simplex virus type 1 γ(1) 34.5 protein; IBV N, infectious bronchitis virus nucleocapsid; LIMK2, LIM kinases 2; MDM2, murine double minute 2 protein; MDV, Marek disease virus; MEQ, MDV Eco Q; NIK, nuclear factor-κB inducing kinase; NoLS, nucleolar localization signal; ORF57, open reading frame 57; PRRSV N, porcine reproductive and respiratory syndrome virus nucleocapsid; VCP, nuclear valosin-containing protein.
NoLSs might be targets for other nucleolar proteins or RNAs. In this regard, NPM has been shown to interact with an artificial NoLS to direct exogenous proteins to specific compartments within the nucleolus (Lechertier et al, 2007). Indeed, the fusion of the same artificial NoLS with another nucleolar protein, fibrillarin—which contains its own NoLS—showed that it could be retargeted from the DFC to the GC by conferring on fibrillarin high affinity for NPM (Lechertier et al, 2007). This suggests that NoLSs might have a hierarchal dominance in terms of directing proteins to the nucleolus and its subcompartments. Swapping NoLSs between viral proteins that have different nucleolar localization and trafficking rates has certainly illustrated that the NoLS can determine both of these properties, although different NoLS sequences might confer the same biological activity (Emmott et al, 2008). For example, abolishing the NoLS of the ORF57 protein of HVS prevented the trafficking of intronless HVS RNA from the nucleolus to the cytoplasm, yet this activity was rescued by the addition of the NoLS of the HIV-1 Rev protein (Boyne & Whitehouse, 2006).
Several factors might be responsible for determining the activity of NoLSs. Similar to the positioning of NESs, NoLSs might be situated on the exterior of the parent protein, making them available for interaction with other molecules that are thereby also targeted to the nucleolus (Horke et al, 2004). The activity of NoLSs might also be regulated by post-translational modifications; a notable example of this is found in the NoLS and NES of the HSV-1 US11 protein, which comprises a single shared proline-rich motif the activity of which—to mediate nucleolar localization or nuclear export—is regulated by phosphorylation (Catez et al, 2002). This finding also emphasizes the fact that Arg and Lys residues are not the sole functional residues of NoLSs, as is the case with NPM, for which Trp residues were reported to be important (Nishimura et al, 2002). The subcellular localization of NPM is also controlled by SUMOylation, which occurs in close proximity to the NoLS motif (Liu et al, 2007) and can affect the role of NPM in 28S rRNA maturation (Haindl et al, 2008).
Conclusion
The use of proteomics and fluorescent live-cell imaging has rapidly expanded the repertoire of known nucleolar proteins and our understanding of their functions. These proteins might be resident in the nucleolus throughout interphase or their localization might be dependent on the metabolic state of the cell. The localization of proteins to the nucleolus is undoubtedly dynamic, and requires suitable signalling and trafficking cascades. The challenge in understanding nucleolar trafficking is separating these cascades in order to build a complex model that illustrates nucleolar dynamics, from the beginning of nucleolar assembly at the end of mitosis to the subsequent nucleolar disassembly at the end of cell division. The stoichiometry of proteins in the nucleolus is crucial for its successful functioning, as the release or sequestration of proteins to the nucleolus has profound biological consequences. The study of the nucleolar proteome has revealed families of proteins that localize in the nucleolus, thereby increasing the chances of discovering potential classes of NoLSs and their interacting partners. The nucleolus might be built around hub proteins that allow the binding of multiple protein partners, which could be orchestrated in a position-dependent and time-dependent manner, in that both localization within the nucleolus and the trafficking rate could be determined by the NoLS. These NoLSs might be recognized by the hub nucleolar proteins, which are centred around the rDNA (see sidebar A). There are clearly various classes of NoLSs, and their identification, characterization and classification will undoubtedly increase our understanding of nucleolar targeting and formation.
Sidebar A | In need of answers.
The generation of a dynamic interactome map of the nucleolus would aid the prediction of loss and/or gain of function through the sequestration and release of nucleolar proteins as a result of disease.
How can nucleolar localization signals (NoLSs) be identified using bioinformatics-based approaches? Several studies are now beginning to address this problem (Boden & Teasdale, 2008).
Complete structural information on native nucleolar proteins with and without appropriate post-translational modifications would help to determine how they might signal or control the activity of the NoLS.
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Supplementary Material
Regions of predicted disorder within nucleolin and nucleophosmin using online resources (1–6).
List of abbreviations accompanying the interactome map presented in Fig 2.
Regions of predicted disorder within nucleolin and nucleophosmin using online resources, to accompany Sup Fig 1.

Edward Emmott & Julian A. Hiscox
Acknowledgments
Our laboratory is supported by the Biotechnology and Biological Sciences Research Council (BBSRC; UK), the Health Protection Agency (UK), the Medical Research Council (UK) and the National Pork Board (USA). E.E. is supported by a BBSRC Doctoral Training Grant awarded to the Astbury Centre for Structural Molecular Biology, University of Leeds, UK.
References
- Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI (2009) NOPdb: Nucleolar Proteome Database: 2008 update. Nucleic Acid Res 37: D181–D184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin MA, Matsunaga S, Uchiyama S, Fukui K (2008) Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem J 415: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M (2005) Nucleolar proteome dynamics. Nature 433: 77–83 [DOI] [PubMed] [Google Scholar]
- Bertrand L, Pearson A (2008) The conserved N-terminal domain of herpes simplex virus 1 UL24 protein is sufficient to induce the spatial redistribution of nucleolin. J Gen Virol 89: 1142–1151 [DOI] [PubMed] [Google Scholar]
- Bicknell KA, Brooks G, Kaiser P, Chen H, Dove BK, Hiscox JA (2005) Nucleolin is regulated both at the level of transcription and translation. Biochem Biophys Res Comm 332: 817–822 [DOI] [PubMed] [Google Scholar]
- Birbach A, Bailey ST, Ghosh S, Schmid JA (2004) Cytosolic, nuclear and nucleolar localization signals determine subcellular distribution and activity of the NF-kappaB inducing kinase NIK. J Cell Sci 117: 3615–3624 [DOI] [PubMed] [Google Scholar]
- Boden M, Teasdale RD (2008) Determining nucleolar association from sequence by leveraging protein–protein interactions. J Comput Biol 15: 291–304 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Bonetti P, Davoli T, Sironi C, Amati B, Pelicci PG, Colombo E (2008) Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. J Cell Biol 182: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne JR, Whitehouse A (2006) Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc Natl Acad Sci USA 103: 15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Mendes-Soares L, Campos I (2000) To be or not to be in the nucleolus. Nat Cell Biol 2: E107–E112 [DOI] [PubMed] [Google Scholar]
- Catez F, Erard M, Schaerer-Uthurralt N, Kindbeiter K, Madjar JJ, Diaz JJ (2002) Unique motif for nucleolar retention and nuclear export regulated by phosphorylation. Mol Cell Biol 22: 1126–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Brett ME, He B (2002) Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the gamma(1)34.5 protein of herpes simplex virus type 1. J Virol 76: 9434–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane AW, Perkins A, Rosen CA (1990) Identification of sequences important in the nucleolar localization of human immunodeficiency virus Rev: relevance of nucleolar localization to function. J Virol 64: 881–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol M, Nair R, Rost B (2000) Finding nuclear localization signals. EMBO Rep 1: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Lee WM (1989) Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV tat proteins. J Biol Chem 264: 18019–18023 [PubMed] [Google Scholar]
- Dove BK, You JH, Reed ML, Emmett SR, Brooks G, Hiscox JA (2006) Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cell Microbiol 8: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Misteli T, Olson MO (2000) The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol 150: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman D, Light S, Bjorklund AK, Elofsson A (2006) What properties characterize the hub proteins of the protein–protein interaction network of Saccharomyces cerevisiae? Genome Biol 7: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E, Dove BK, Howell G, Chappell LA, Reed ML, Boyne JR, You JH, Brooks G, Whitehouse A, Hiscox JA (2008) Viral nucleolar localisation signals determine dynamic trafficking within the nucleolus. Virology 380: 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal P, Pandey D, Siess W (2006) Phosphorylation-dependent regulation of unique nuclear and nucleolar localization signals of LIM kinase 2 in endothelial cells. J Biol Chem 281: 25223–25230 [DOI] [PubMed] [Google Scholar]
- Grisendi S, Bernardi R, Rossi M, Cheng K, Khandker L, Manova K, Pandolfi PP (2005) Role of nucleophosmin in embryonic development and tumorigenesis. Nature 437: 147–153 [DOI] [PubMed] [Google Scholar]
- Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R (2003) Extended disordered proteins: targeting function with less scaffold. Trends Biochem Sci 28: 81–85 [DOI] [PubMed] [Google Scholar]
- Gunawardena SR, Ruis BL, Meyer JA, Kapoor M, Conklin KF (2008) NOM1 targets protein phosphatase I to the nucleolus. J Biol Chem 283: 398–404 [DOI] [PubMed] [Google Scholar]
- Guo YX, Dallmann K, Kwang J (2003) Identification of nucleolus localization signal of betanodavirus GGNNV protein alpha. Virology 306: 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Marsh DJ (2007) Nucleolar localization of parafibromin is mediated by three nucleolar localization signals. FEBS Lett 581: 5070–5074 [DOI] [PubMed] [Google Scholar]
- Haindl M, Harasim T, Eick D, Muller S (2008) The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep 9: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka M (1990) Discovery of the nucleolar targeting signal. Bioessays 12: 143–148 [DOI] [PubMed] [Google Scholar]
- He X, Zhang J (2006) Why do hubs tend to be essential in protein networks? PLoS Genet 2: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D (2006) Nucleolus: from structure to dynamics. Histochem Cell Biol 125: 127–137 [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Roussel P, Gebrane-Younes J (2002) Emerging concepts of nucleolar assembly. J Cell Sci 115: 2265–2270 [DOI] [PubMed] [Google Scholar]
- Hiscox JA (2007) RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol 5: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horke S, Reumann K, Schweizer M, Will H, Heise T (2004) Nuclear trafficking of La protein depends on a newly identified nucleolar localization signal and the ability to bind RNA. J Biol Chem 279: 26563–26570 [DOI] [PubMed] [Google Scholar]
- Kim H, Chang DJ, Lee JA, Lee YS, Kaang BK (2003) Identification of nuclear/nucleolar localization signal in Aplysia learning associated protein of slug with a molecular mass of 18 kDa homologous protein. Neurosci Lett 343: 134–138 [DOI] [PubMed] [Google Scholar]
- Kim PM, Sboner A, Xia Y, Gerstein M (2008) The role of disorder in interaction networks: a structural analysis. Mol Syst Biol 4:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Macfarlane S, Kalinina NO, Rakitina DV, Ryabov EV, Gillespie T, Haupt S, Brown JW, Taliansky M (2007) Interaction of a plant virus-encoded protein with the major nucleolar protein fibrillarin is required for systemic virus infection. Proc Natl Acad SciUSA 104: 11115–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE (2005) Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol 25: 1258–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowski MD, Reschly EJ, Ekins S (2008) Intrinsic disorder in nuclear hormone receptors. J Proteome Res 7: 4359–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17: 527–536 [DOI] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechertier T, Sirri V, Hernandez-Verdun D, Roussel P (2007) A B23-interacting sequence as a tool to visualize protein interactions in a cellular context. J Cell Sci 120: 265–275 [DOI] [PubMed] [Google Scholar]
- Li YP, Busch RK, Valdez BC, Busch H (1996) C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Euro J Biochem 237: 153–158 [DOI] [PubMed] [Google Scholar]
- Li Z, Boone D, Hann SR (2008) Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc Natl Acad SciUSA 105: 18794–18799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS, Zhang Y (2008) Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J Biol Chem 283: 15568–15576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Lee LF, Ye Y, Qian Z, Kung HJ (1997) Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J Virol 71: 3188–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Liu Z, Jang SW, Ma Z, Shinmura K, Kang S, Dong S, Chen J, Fukasawa K, Ye K (2007) Sumoylation of nucleophosmin/B23 regulates its subcellular localization, mediating cell proliferation and survival. Proc Natl Acad SciUSA 104: 9679–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lixin R, Efthymiadis A, Henderson B, Jans DA (2001) Novel properties of the nucleolar targeting signal of human angiogenin. Biochem Biophys Res Commun 284: 185–193 [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH (2000) Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol 2: 179–181 [DOI] [PubMed] [Google Scholar]
- Lymberopoulos MH, Pearson A (2007) Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology 363: 397–409 [DOI] [PubMed] [Google Scholar]
- Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K (2007) Nucleolin functions in nucleolus formation and chromosome congression. J Cell Sci 120: 2091–2105 [DOI] [PubMed] [Google Scholar]
- Martindill DM, Riley PR (2008) Cell cycle switch to endocycle: the nucleolus lends a hand. Cell Cycle 7: 17–23 [DOI] [PubMed] [Google Scholar]
- Matthews DA, Olson MO (2006) What is new in the nucleolus? Workshop on the nucleolus: new perspectives. EMBO Rep 7: 870–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I (2005) Cellular stress and nucleolar function. Cell Cycle 4: 1036–1038 [DOI] [PubMed] [Google Scholar]
- Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S (2004) HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol 6: 642–647 [DOI] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P (2007) Nucleolin: a multiFACeTed protein. Trends Cell Biol 17: 80–86 [DOI] [PubMed] [Google Scholar]
- Montanaro L, Trere D, Derenzini M (2008) Nucleolus, ribosomes, and cancer. Am J Pathol 173: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama M, Hara Y, Seki A, Yamazoe T, Kawate Y, Shinohara T, Hatsuzawa K, Tani K, Tagaya M (2004) NVL2 is a nucleolar AAA-ATPase that interacts with ribosomal protein L5 through its nucleolar localization sequence. Mol Biol Cell 15: 5712–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedick I, Froese N, Oumard A, Mueller PP, Nourbakhsh M, Hauser H, Koster M (2004) Nucleolar localization and mobility analysis of the NF-kappaB repressing factor NRF. J Cell Sci 117: 3447–3458 [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Ohkubo T, Furuichi Y, Umekawa H (2002) Tryptophans 286 and 288 in the C-terminal region of protein B23.1 are important for its nucleolar localization. Biosci Biotechnol Biochem 66: 2239–2242 [DOI] [PubMed] [Google Scholar]
- Pederson T (1998) The plurifunctional nucleolus. Nucleic Acid Res 26: 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Polena I, Trask I, Bazett-Jones DP, Pederson T (2005) A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell 16: 3401–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed ML, Dove BK, Jackson RM, Collins R, Brooks G, Hiscox JA (2006) Delineation and modelling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic 7: 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland RR, Kerwin R, Kuckleburg C, Sperlich A, Benfield DA (1999) The localisation of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res 64: 1–12 [DOI] [PubMed] [Google Scholar]
- Rowland RRR, Schneider P, Fang Y, Wootton S, Yoo D, Benfield DA (2003) Peptide domains involved in the localization of the porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus. Virology 316: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22: 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Nigg EA (1993) Protein localization to the nucleolus: a search for targeting domains in nucleolin. J Cell Sci 105: 799–806 [DOI] [PubMed] [Google Scholar]
- Sheng Z, Lewis JA, Chirico WJ (2004) Nuclear and nucleolar localization of 18-kDa fibroblast growth factor-2 is controlled by C-terminal signals. J Biol Chem 279: 40153–40160 [DOI] [PubMed] [Google Scholar]
- Siomi H, Shida H, Maki M, Hatanaka M (1990) Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol 64: 1803–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark LA, Taliansky M (2009) Old and new faces of the nucleolus. Workshop on the Nucleolus and Disease. EMBO Rep 10: 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thebault S, Basbous J, Gay B, Devaux C, Mesnard JM (2000) Sequence requirement for the nucleolar localization of human I-mfa domain-containing protein (HIC p40). Eur J Cell Biol 79: 834–838 [DOI] [PubMed] [Google Scholar]
- Thoms HC, Dunlop MG, Stark LA (2007) CDK4 inhibitors and apoptosis: a novel mechanism requiring nucleolar targeting of RelA. Cell Cycle 6: 1293–1297 [DOI] [PubMed] [Google Scholar]
- Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P (2007) Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol 8: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez BC, Perlaky L, Henning D, Saijo Y, Chan PK, Busch H (1994) Identification of the nuclear and nucleolar localization signals of the protein p120. Interaction with translocation protein B23. J Biol Chem 269: 23776–23783 [PubMed] [Google Scholar]
- Wanzel M, Russ AC, Kleine-Kohlbrecher D, Colombo E, Pelicci PG, Eilers M (2008) A ribosomal protein L23-nucleophosmin circuit coordinates Miz1 function with cell growth. Nat Cell Biol 10: 1051–1061 [DOI] [PubMed] [Google Scholar]
- Yang L, Reece JM, Cho J, Bortner CD, Shears SB (2008) The nucleolus exhibits an osmotically regulated gatekeeping activity that controls the spatial dynamics and functions of nucleolin. J Biol Chem 283: 11823–11831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu CP, Beavil RL, Chan HY (2006) Biophysical characterisation reveals structural disorder in the nucleolar protein, Dribble. Biochem Biophys Res Commun 343: 311–318 [DOI] [PubMed] [Google Scholar]
- Yoo D, Wootton SK, Li G, Song C, Rowland RR (2003) Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J Virol 77: 12173–12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JH, Howell G, Pattnaik AK, Osorio FA, Hiscox JA (2008) A model for the dynamic nuclear/nucleolar/cytoplasmic trafficking of the porcine reproductive and respiratory syndrome virus (PRRSV) nucleocapsid protein based on live cell imaging. Virology 378: 34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Wang Y, Mukhopadhyay D, Backlund P, Kolli N, Yergey A, Wilkinson KD, Dasso M (2008) Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J Cell Biol 183: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Regions of predicted disorder within nucleolin and nucleophosmin using online resources (1–6).
List of abbreviations accompanying the interactome map presented in Fig 2.
Regions of predicted disorder within nucleolin and nucleophosmin using online resources, to accompany Sup Fig 1.