Abstract
Trans-acting small interfering RNAs (tasiRNAs) comprise a class of endogenous small RNAs that are generated from TAS gene-derived transcripts after these are cleaved at a microRNA (miRNA) target site. Arabidopsis thaliana has four families of TAS genes: miR173 triggers tasiRNA production from TAS1 and TAS2, miR390 from TAS3 and miR828 from TAS4. The two-hit trigger model postulates that dual target sites in the same transcript are often sufficient to initiate tasiRNA production, but two hits are not always required for tasiRNA formation. Here, we characterize the function of miR173 in the formation of tasiRNAs from TAS1 transcripts, as well as the importance of the TAS1 and TAS3 transcript sequences outside the miRNA-targeting sites for tasiRNA production. We show that tasiRNAs can be produced from heterologous transcripts containing miR173 or miR390 target sites, indicating that these trigger sequences are the only cis sequences essential for tasiRNA formation.
Keywords: microRNA, trans-acting small RNA, tasiRNA, transitivity, Arabidopsis
Introduction
Trans-acting small interfering RNAs (tasiRNAs) are a specialized class of small RNAs (sRNAs) that originate from TAS gene transcripts and, similar to microRNAs (miRNAs), they act in trans to regulate messenger RNAs (mRNAs) at the post-transcriptional level (Vazquez et al, 2004). The generation of tasiRNAs itself is triggered by an miRNA that targets the TAS transcript, resulting in the production of 21 nucleotide sRNAs that are phased with respect to the miRNA cleavage site. This process depends on several proteins, including SUPPRESSOR OF GENE SILENCING 3 (SGS3), RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) and DICER-LIKE 4 (DCL4; Peragine et al, 2004; Vazquez et al, 2004; Allen et al, 2005; Gasciolli et al, 2005; Xie et al, 2005; Yoshikawa et al, 2005).
Four families of TAS genes have been identified in Arabidopsis thaliana. TAS1 and TAS2 transcripts are recognized by miR173, which triggers the production of phased tasiRNAs downstream from the cleavage site (Allen et al, 2005). A similar pattern is seen for TAS4, which is targeted by miR828 (Rajagopalan et al, 2006). By contrast, miR390 triggers the production of tasiRNAs from TAS3 transcripts upstream from the miR390-guided cleavage site (Allen et al, 2005).
One of the main questions about tasiRNA generation is why TAS transcripts, but not the vast majority of other miRNA-targeted transcripts, form siRNAs. Axtell et al (2006) proposed a two-hit trigger model in which tasiRNAs are often spawned when transcripts are targeted at two positions by one or more sRNAs. This model was based on the observation that TAS3 transcripts in Physcomitrella patens and Pinus taeda have a second, cleavable miR390 target site, with most tasiRNAs being formed between the two miR390 target motifs. A second, upstream miR390 complementary motif is also present in A. thaliana TAS3, but owing to additional mismatches, it is not cleaved. Nonetheless, the production of tasiRNAs from A. thaliana TAS3 is dependent on the presence of both sites (Axtell et al, 2006).
Replacing the downstream, cleavable miR390 target site with another miRNA complementary motif does not affect the generation of tasiRNAs, as long as the new site is recognized and cleaved through the activity of the alternative miRNA. By contrast, the upstream, non-cleavable miR390 target site in TAS3a is essential for the production of tasiRNAs. When this site is replaced with another miRNA-targeting motif, tasiRNAs are no longer formed, even if the mismatches in the alternative site are engineered to resemble the original site. Notably, miR390 is unique compared with other miRNAs and is preferentially loaded into ARGONAUTE 7 (AGO7; Montgomery et al, 2008).
In the case of TAS1, TAS2 and TAS4, which seem to have only single miRNA target motifs, the specific functions of miR173 and miR828 in tasiRNA production are not yet clear. Here, we show that the miR173 target site in TAS1 transcripts is not only necessary but also sufficient to trigger the formation of tasiRNAs. Similarly, the two miR390 target sites from TAS3 transcripts are shown to be sufficient for tasiRNA production.
Results
miR173 is necessary for tasiRNA formation from TAS1
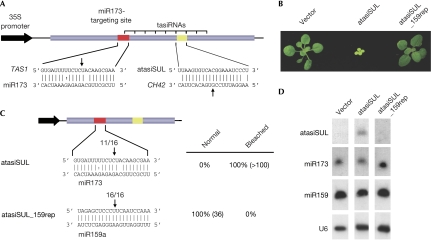
The formation of TAS1- and TAS2-derived tasiRNAs is initiated by miR173. To investigate in more detail the function of miR173 in this process, we took advantage of one of the properties of tasiRNAs––that the phase of production is determined by the miRNA-guided cleavage site. The phasing allows the prediction of the sRNAs that will be spawned from a TAS gene, which can be exploited to generate artificial (synthetic) tasiRNAs (atasiRNAs/syn-tasiRNAs; Montgomery et al, 2008). Rules developed for artificial miRNAs (Schwab et al, 2006) were used to design a sRNA, artificial tasiRNA-SULFUR (atasiSUL), that should specifically cause cleavage of the mRNA of CHLORINA 42 (CH42), the A. thaliana homologue of tobacco SULFUR (Koncz et al, 1990; Ossowski et al, 2008). CH42 encodes a magnesium chelatase involved in the biosynthesis of chlorophyll, and its inactivation causes bleaching of green tissue. The siR255 sequence in TAS1a was replaced with the atasiSUL sequence (Fig 1A; supplementary Fig 1 online). Plants expressing the TAS1-atasiSUL chimaera under the control of the strong constitutive cauliflower mosaic virus (CaMV) 35S promoter were very pale and much smaller than wild type (Fig 1B).
Figure 1.
A target site for miR173, but not miR159, triggers the generation of trans-acting small interfering RNAs. (A) Diagram of TAS1a-derived TAS1-atasiSUL construct. The tasiRNA-spawning region is indicated by brackets, the miR173 target site is shown in red and the atasiSUL sequence in yellow. Arrows indicate expected miRNA-guided cleavage in the TAS1-atasiSUL transcript or atasiSUL-guided cleavage in its CH42 target. (B) Phenotype of atasiSUL-expressing plants. (C) Constructs testing miR173 and miR159 target site functionality. The numbers above arrows refer to the fraction of 5′ RACE products terminating at the canonical miRNA target site; the numbers of analysed plants are given in parentheses in the table. (D) Small RNA blot analysis; U6 was used as a loading control. atasiSUL, artificial (synthetic) tasiRNA-SULFUR; CH42, CHLORINA 42; miRNA, microRNA; RACE, rapid amplification of cloned ends; tasiRNA, trans-acting small interfering RNA.
To test the importance of miR173-guided cleavage for the production of tasiRNAs from the TAS1a transcript, we replaced the miR173 complementary motif in TAS1-atasiSUL with an miR159 target. miR159 is among the most abundant miRNAs in A. thaliana, it is broadly expressed, it is very effective in causing target cleavage in seedlings (Fig 1D; http://asrp.cgrb.oregonstate.edu/db; Allen et al, 2007; Palatnik et al, 2007) and has been used for studying the generation of TAS previously (Montgomery et al, 2008). Although the miR159 target site in the RNA transcribed from this construct, TAS1-atasiSUL_159rep, was cleaved at the expected position (Fig 1C), TAS1-atasiSUL_159rep did not seem to produce any atasiRNA against CH42 (Fig 1D) and the plants were not bleached (Fig 1B). We conclude that the miR173 target site in TAS1 transcripts is essential and that the normal function of miR173-containing effector complexes extends beyond transcript cleavage.
miR173 target site sufficient for tasiRNA production
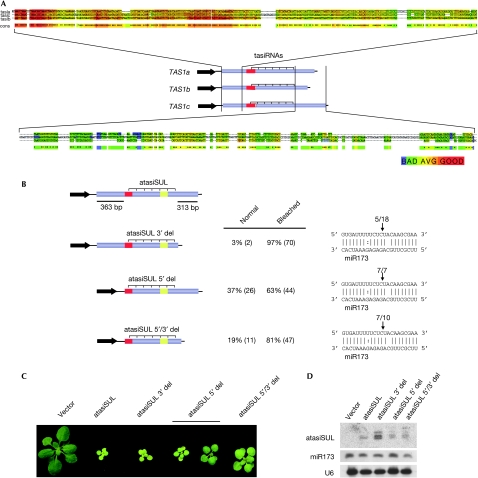
The fact that two miR390 complementary motifs are necessary for tasiRNA production from TAS3, and that several other TAS transcripts spawn secondary sRNAs only from a portion of the transcripts, led Axtell et al (2006) to propose a two-hit trigger mechanism for tasiRNA production. As TAS1 and TAS2 lack obvious second target sites for known A. thaliana miRNAs, we aligned TAS1 and TAS2 family member sequences to identify conserved regions that might participate in the generation of tasiRNAs. In TAS1 and TAS2, the miR173-targeting motif is upstream from the region that gives rise to tasiRNAs. According to the two-hit trigger model, one might therefore expect sequences downstream from the tasiRNA-spawning region to be involved in tasiRNA biogenesis; however, there was little sequence conservation downstream from the tasiRNA-generating region (Fig 2A; supplementary Figs 2,3 online). By contrast, the TAS1 and TAS2 loci had considerable sequence similarity in the region upstream from the miR173 target site.
Figure 2.
Formation of artificial (synthetic) trans-acting small interfering RNAs from a TAS1 derivative. (A) Alignment of TAS1 family transcripts. The alignment of regions flanking the tasiRNA-producing region is shown; colours are based on the alignment score generated by the CORE function of T-Coffee (Llave et al, 2002). (B) TAS1-atasiSUL constructs with 5′ and 3′ deletions still generate tasiRNAs. The full-length construct is shown at the top (see Fig 1A). Numbers in parentheses indicate the number of plants analysed. (C) Plants expressing the various TAS1 derivatives. (D) Small RNA blots of the various TAS1 derivatives. atasiRNA, artificial (synthetic) trans-acting small interfering RNA.
To test whether this upstream conserved sequence or the sequences downstream from the tasiRNA-producing region are important for the generation of tasiRNAs, we deleted these sequences individually and in combination in the TAS1-atasiSUL construct. For the upstream region, we removed all sequences 5′ to the miR173 target site, whereas the downstream deletion started a few nucleotides after the last tasiRNA with a predicted target (Axtell et al, 2006; supplementary Fig 1 online). The deleted downstream region does spawn a few sRNAs without known targets (http://asrp.cgrb.oregonstate.edu/db/). On the basis of the production of sRNAs and the characteristic bleaching phenotype, we concluded that neither of these sequences has an essential role in the biogenesis of tasiRNAs (Fig 2B–D). Taken together, our observations suggest that the only sequence that is essential for the production of tasiRNAs in TAS1a is the miR173 target site.
Triggering transitivity in non-TAS transcripts with miR173
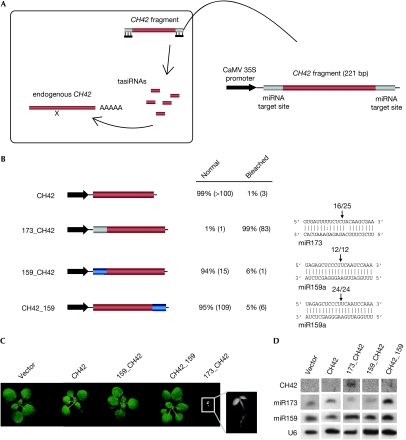
Although our results so far indicated that TAS1a did not contain additional sequences necessary for the production of tasiRNAs, it was still unclear whether miR173 cleavage is the only necessary trigger for tasiRNA biogenesis in TAS1. Indeed, it has been suggested that perhaps a second binding element recruits RDR6 in a sRNA-independent manner (Yoshikawa et al, 2005). To test the sufficiency of miR173, we developed a CH42 silencing reporter. In this reporter, a CH42 fragment is flanked by an miRNA-binding site of choice (Fig 3A). If no secondary sRNAs are formed, expression of such a construct in transgenic plants should be innocuous. The advantage of this reporter is that, apart from ease of scoring the bleaching resulting from the inactivation of CH42, perfect phasing is not required for causing a phenotype, resulting in very sensitive detection of secondary sRNA production. In addition, the 221 nucleotide fragment of the CH42 gene is the same size as the fragment separating the two miR390 target sites in TAS3, thus allowing for an appropriate comparison with endogenous TAS3 (see below).
Figure 3.
Sufficiency of the miR173 target site for trans-acting small interfering RNA formation. (A) Diagram of CH42 silencing reporter system. (B) Construct diagrams (left), fraction of transgenic plants with bleaching phenotype (middle) and 5′ RACE results (right). CH42 sequences are indicated in red and miRNA target sites in grey or blue. Numbers in parentheses indicate the number of plants analysed. (C) Plants expressing the various CH42 silencing reporters. (D) Small RNA blots; for CH42, the fragment present in the silencing reporter was used. CaMV, cauliflower mosaic virus; CH42, CHLORINA 42; miRNA, microRNA; RACE, rapid amplification of cloned ends.
We introduced an miR173 and an miR159 target site separately upstream from a fragment of CH42, and expressed these constructs, 35S:173_CH42 and 35S:159_CH42, in plants. Most of the 35S:173_CH42 plants presented a marked bleaching phenotype, arresting at the seedling stage and finally dying (Fig 3C). By contrast, most of the 35S:159_CH42 plants were normal and did not produce secondary sRNAs (Fig 3B–D). To determine whether miR159 could trigger the production of sRNA upstream from the cleavage site, we also analysed plants expressing a construct in which the miR159 target motif had been placed downstream from the CH42 fragment (35S:CH42_159). These plants were also phenotypically normal and did not produce detectable secondary sRNAs. We conclude that an miR173 target site is not only necessary but also sufficient for triggering the formation of tasiRNAs.
Triggering transitivity in non-TAS transcripts with miR390
Unlike other TAS transcripts, TAS3 transcripts contain two miR390 complementary motifs flanking the tasiRNA-spawning region. Both sites are necessary for the production of tasiRNAs, but the downstream motif can be replaced with other miRNA target sites that result in transcript cleavage (Axtell et al, 2006). Nonetheless, it is not clear whether additional sequences in TAS3 transcripts have a function in triggering the production of tasiRNAs. By using the silencing reporter described above, we generated transgenic plants expressing a CH42 fragment flanked by the miR390 target motifs found in TAS3 (35S:390_CH42_390). As a control, the same CH42 fragment was placed between two genuine miR159 complementary motifs (35S:159_CH42_159). Most of the 35S:159_CH42_159 plants were normal, whereas the 35S:390_CH42_390 plants presented a pale phenotype (Fig 4). Interestingly, bleaching was strongest close to the veins, similar to that already described for atasiRNAs expressed from the TAS3 backbone and targeting another gene required for the biosynthesis of chlorophyll (Montgomery et al, 2008). Our results suggest that the biosynthesis of tasiRNAs from TAS3 transcripts involves no other specific sequences outside the miR390 target motifs.
Figure 4.
Dual targeting by miR390, but not miR159, can trigger the formation of small interfering RNA. (A) Construct diagrams (left), fraction of transgenic plants with bleaching phenotype (middle) and 5′ RACE results (right). CH42 sequences are indicated in red and miRNA target sites in green or blue. Cleavage at the downstream miR390 target site could only be mapped using a modified RACE procedure (see Methods). (B) Plants expressing the various CH42 silencing reporters. (C) Small RNA analysis of plants expressing CH42 reporters. CH42, CHLORINA 42; RACE, rapid amplification of cloned ends.
Discussion
We have shown that the miR173 target site is sufficient for the production of tasiRNAs at the TAS1a locus. miR173 cannot apparently be replaced by an arbitrarily chosen miRNA, and the miR173 effector complex perhaps has unique properties for triggering the formation of tasiRNAs. Our results also indicate that other sequences in the TAS1 backbone have only a minor function in the biogenesis of tasiRNAs, and that they do not contain any essential feature necessary for the production of tasiRNAs. Similarly, the TAS3 backbone apparently has only a minor role in triggering the formation of tasiRNAs, based on the fact that miR390 dual targeting is sufficient to initiate the production of secondary sRNAs, a process also known as transitivity. Finally, our data support the idea that transcripts can be routed to tasiRNA production, once certain TAS criteria are satisfied.
The two-hit trigger model postulates that a given transcript, once targeted twice by sRNAs, is predisposed for the production of secondary sRNAs (Montgomery et al, 2008). A two-hit trigger has also been invoked to explain transitivity at an overexpressed alien transcript containing a single target motifs that is perfectly complementary to an endogenous miRNA (Axtell et al, 2006). However, from genome-wide analyses, it is clear that the presence of two sRNA complementary motifs is not always associated with the production of tasiRNAs (Parizotto et al, 2004; Axtell et al, 2006). A trivial explanation for this observation could be that the potential triggers are not co-expressed, and that therefore the two-hit trigger situation does not apply. To test explicitly whether two triggers are sufficient for the formation of tasiRNAs, we tested a construct in which two miR159 target sites flanked a fragment of CH42. This chimaeric transgene, 35S:159_CH42_159, was much less effective in causing bleaching than the 35S:390_CH42_390 transgene. This is unlikely to be due to insufficient activity of miR159 per se, as substituting the downstream miR390 complementary motif in TAS3 with an miR159 target site does not affect TAS3 function (Howell et al, 2007).
Our results, together with those of Montgomery et al (2008), support two conclusions: (1) dual targeting is not sufficient for the formation of tasiRNAs, and (2) only some sRNAs, such as miR173 and miR390, are efficient triggers of transitivity. This raises the question of what makes these miRNAs unique. In the case of TAS3, the exclusive interaction of miR390 with AGO7 is the crucial factor that allows the generation of tasiRNAs (Montgomery et al, 2008). A. thaliana has 10 different AGOs, but their preference for different sRNAs is only known for a subset (Baumberger & Baulcombe, 2005; Qi et al, 2005, 2006; Montgomery et al, 2008; Takeda et al, 2008). What is known is that miR173 does not associate with AGO7, but with AGO1, probably the Arabidopsis AGO with the broadest role in sRNA-guided slicing, and to a lesser extent with AGO5, an AGO of unknown function (Mi et al, 2008; Montgomery et al, 2008). One possibility is that interaction of miR173 with one of the AGOs not yet studied results in an miR390/AGO7-like interaction. However, as AGO1 is required for the formation of tasiRNAs from the TAS1 locus, it seems more likely that an miR173/AGO1 complex has a special capacity to recruit another factor required for the production of tasiRNAs. Finally, as we found miR173 to be sufficient to trigger transitivity, our conclusions probably also apply to the miR173-targeted TAS2 (Baumberger & Baulcombe, 2005).
Another important question is the similarity of the mechanisms of tasiRNA production triggered by miR390/AGO7 and miR173/AGO1. The initial steps are clearly different, as reflected in the different requirements for miRNA targeting. However, both pathways, for TAS1/2/4 and TAS3, converge on the recruitment of SGS3 and RDR6, and subsequent processing by DCL4 (Peragine et al, 2004; Allen et al, 2005; Gasciolli et al, 2005; Xie et al, 2005; Yoshikawa et al, 2005). That only a subset of AGO1/miRNA complexes, such as AGO1/miR173, can trigger the formation of tasiRNAs might be explained by a change of AGO1 conformation induced by miR173, which would then mimic AGO7/miR390. Structural studies should shed light on these questions.
Finally, we found that constitutive expression of a CH42 gene fragment linked to an miR173 target site caused a severe CH42 loss-of-function phenotype. This system could thus present yet another effective approach to gene silencing. It could, for example, be used to create dominant knockouts in non-model systems, by transforming plants with a cassette that expresses miR173 and at the same time contains an miR173 target site next to an outward facing promoter.
Methods
Generation of transgenic lines. Overlapping PCR was used to replace the siR255 sequence in TAS1a (At2g27400) with the atasiSUL sequence, and the miR173 target site in atasiSUL constructs with an miR159 target site. Deletion derivatives were also generated by PCR amplification. The CH42 (At4g18480) silencing reporters were generated by PCR using oligonucleotide primers that introduced an miRNA target site. For expression in plants behind the CaMV 35S promoter, the pGreen binary vector (Vazquez et al, 2004) was used. Transgenes were introduced into accession Col-0 by Agrobacterium-mediated transformation (Hellens et al, 2000).
RNA analysis. Total RNA was extracted using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). miRNA-guided cleavage site was detected by 5′ RACE (rapid amplification of cloned ends) using modifications of a published protocol (Weigel & Glazebrook, 2002). To detect cleavage at the 3′ miR390 target site in the 390_CH42_390 transcript, a modified RNA adaptor-nested primer was used to amplify specifically products resulting from cleavage at the expected miR390 target motif. For small RNA blots, total RNA was resolved on a 17% PAGE gel under denaturing conditions (7 M urea) and hybridized with DNA probes that had been radioactively labelled, either with 32P-dATP and OptiKinase™ (USB, Cleveland, OH, USA, in the case of oligonucleotide probes) or 32P-dCTP and Prime-a-gene® labelling kit (Promega, Madison, WI, USA, in the case of a DNA fragment probe).
Supplementary information is available at EMBO reports online (http://www.emboreports.org)
Supplementary Material
Supplementary Information
Acknowledgments
We thank Jim Carrington and Sascha Laubinger for discussion and a critical reading of the paper. This study was supported by a Deutscher Akademischer Austauschdienst fellowship to F.F.F., European Community FP6 IP SIROCCO (Silencing RNAs: Organisers and Coordinators of Complexity in eukaryotic Organisms; contract LSHG-CT-2006-037900) and a Gottfried Wilhelm Leibniz Award to D.W., and the Max Planck Society.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Allen RS, Li J, Stahle MI, Dubroue A, Gubler F, Millar AA (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104: 16371–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC (2007) Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J (1990) Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J 9: 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056 [DOI] [PubMed] [Google Scholar]
- Mi S et al. (2008) Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC (2008) Specificity of ARGONAUTE7–miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Palatnik JF et al. (2007) Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell 13: 115–125 [DOI] [PubMed] [Google Scholar]
- Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O (2004) In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev 18: 2237–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ (2006) Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443: 1008–1012 [DOI] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y (2008) The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol 49: 493–500 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information