Abstract
PTIP regulates gene transcription by controlling the methylation of histone H3, and also has important roles in cellular responses to DNA damage or to perturbed DNA replication. The available data suggest that the functions of PTIP in transcription and preserving genome stability might be independent and mediated by functionally distinct cellular pools of PTIP. Although considerable progress has been made in understanding how PTIP influences transcription, a coherent picture of how it protects cells from DNA damage at the molecular level has yet to emerge. Here, we describe recent progress made in understanding the cellular roles of PTIP and the relevance of PTIP-interacting proteins, as well as the questions that have yet to be answered.
Keywords: 53BP1, ATM, DNA damage, histone methylation, MLL, PTIP
Glossary
53BP1 p53-binding protein 1
ALR ALL1-related gene product
ASH2 (absent, small or homeotic)-like 2
ATM ataxia telangiectasia mutated
ATR ataxia telangiectasia mutated and radiation sensitive 3-related
BRCA1 breast cancer gene 1
BRCT protein domain present at the BRCA1 C terminus
CBP CREB-binding protein
CHK checkpoint kinase
COM1 candidate of metastasis 1
DOT1L disruptor of telomeric silencing 1-like
DSB DNA double-strand break
H2AX member X of the H2A histone family
H3K lysine residue of histone H3
HALR homologue of ALL1-related gene product
HDAC1 histone deacetylase 1
HMTase histone methyltransferase
ING inhibitor of growth
IRIF ionizing radiation induced foci
MDC1 mediator of DNA damage checkpoint 1
MLL mixed-lineage leukaemia
MRN MRE11/RAD50/NBS1
PA1 PTIP-associated protein 1
PAX paired box
PCNA proliferating cell nuclear antigen
PHD plant homeodomain
pS/T-Q phospho-Ser/Thr-Gln
PTIP PAX-transactivation domain-interacting protein
RBQ3 retinoblastoma-binding protein clone Q3
RNF8 RING finger protein 8
SET Su(Var)3-9, E(z), Trx
TGF-β transforming growth factor-β
UBC13 ubiquitin-conjugating enzyme 13
USP28 ubiquitin-specific protease 28
WDR5 WD-repeat-associated repeat protein 5
Introduction
Pax genes encode developmentally regulated transcription factors that have crucial roles in embryogenesis (Dahl et al, 1997). Mutations in Pax2 cause eye and kidney abnormalities (Sanyanusin et al, 1995; Devriendt et al, 1998), and Pax2-null mice are viable but lack kidneys and genital tracts (Torres et al, 1995). PTIP was initially discovered in a yeast two-hybrid screen for factors that interact with mouse Pax2 (Lechner et al, 2000), and it was subsequently shown that the transcription of several Pax2-regulated genes depends on PTIP (Patel et al, 2007). In Xenopus laevis embryos, Ptip—also known as Swift—interacts with Smad2, which is part of the Smad2–Smad4 complex that binds to gene promoters and activates transcription in response to activin or TGF-β (Massague & Gomis, 2006). When overexpressed, Ptip stimulates Smad2-dependent transcription, whereas a mutant form of Ptip, which is unable to interact with Smad2, inhibits Smad2-dependent transcription and mesoderm development (Shimizu et al, 2001). These results indicate that PTIP stimulates PAX2-dependent and Smad2-dependent gene expression, and the subsequent observation that PTIP can regulate histone methylation has begun to explain the underlying mechanisms.
PTIP and histone methylation
Histone methylation is an important epigenetic mechanism that can regulate transcription; the methylation of H3K4, for example, is associated with transcriptional activation (Kouzarides, 2007). The evolutionarily conserved SET domain is a characteristic of the seven families of proteins with lysine methyltransferase activity, which are exemplified by the SET1 multiprotein complex. In humans, several SET1-like complexes have been identified, each containing one or two SET domain proteins—such as SET1, MLL1, MLL2/ALR, MLL3/HALR and MLL4–5—that are responsible for the enzymatic activity of the complex (Ruthenburg et al, 2007). All MLL complexes share the core subunits ASH2, RBQ3 and WDR5, which contribute to the structural scaffolding of SET1-like complexes and influence their substrate specificity (Ruthenburg et al, 2007). Importantly, PTIP has also been found to be an integral component of several such HMTase complexes (Fig 1) that differ in the catalytic subunits they contain—MLL2 (Issaeva et al, 2007), MLL3 plus MLL4 (Cho et al, 2007) or MLL2 plus MLL3 (Patel et al, 2007). Consistent with these observations, anti-PTIP immunoprecipitates contain H3K4—and H3K9—methyltransferase activity (Cho et al, 2007; Issaeva et al, 2007; Patel et al, 2007). The differences in the catalytic components of the PTIP complexes identified in these studies might indicate that PTIP associates with a core subunit that is common to all MLL complexes, or might reflect the differential expression or differences in the abundance of the MLL catalytic subunits in various cell types.
Figure 1.
PTIP targets HMTase complexes to gene promoters. PAX2 binds to PREs in the promoters of a range of genes that are required for the development of various organs. PTIP/Swift is brought to promoters by PAX2, and recruits SET1-like lysine methyltransferase complexes that methylate H3K4, thereby activating gene transcription. PTIP was found to be an integral component of several SET domain-containing HMTase complexes that differ in their catalytic subunits, which can be MLL2 (Issaeva et al, 2007), MLL3 plus MLL4 (Cho et al, 2007) or MLL2 plus MLL3 (Patel et al, 2007). The differences in the catalytic components of the PTIP complexes identified in these studies might indicate that PTIP associates with a core subunit that is common to all MLL complexes, or might reflect the different HMTase complexes that exist in the various cell types used in these studies. H3K4, lysine 4 of histone H3; HMTase, histone methyltransferase; Me, methyl; MLL, mixed-lineage leukaemia; ORF, open reading frame; PAX2, paired box gene 2; PREs, PAX2-response elements; PTIP, PAX-transactivation domain-interacting protein; SET, Su(Var)3-9, E(z), Trx.
Although PTIP associates with MLL-containing HMTase complexes, it is not required for their enzymatic activity (Cho et al, 2007). Instead, PTIP is believed to recruit HMTase complexes to gene promoters by interacting with promoter-bound transcription factors (Fig 1). Chromatin-immunoprecipitation analyses have shown that PTIP associates with gene promoters that are known to be regulated by MLL2 (Issaeva et al, 2007). PAX2 recruits PTIP to PAX2-response elements in gene promoters and PTIP, in turn, recruits an MLL2-containing complex to these promoters (Patel et al, 2007). Furthermore, the depletion of PTIP from human cells prevents PAX2-dependent gene transcription, and the conditional ablation of Ptip from the developing spinal cord of PtipLoxP/LoxP embryos results in a global decrease in the levels of methylated H3K4 (Patel et al, 2007). In addition, the conditional deletion of Ptip in the renal inner medulla of mice results in renal function defects similar to those associated with defects in Pax2 function (Kim et al, 2007). Therefore, the available data strongly support a model in which PTIP interacts with transcription factors that bind directly to promoters—such as PAX2—and facilitates the recruitment of HMTase complexes, thereby activating transcription (Fig 1). PTIP has been reported to interact with p8/COM1, which, in turn, interacts with the p300/CBP histone acetyl transferase (Hoffmeister et al, 2002). Therefore, it is possible that PTIP regulates histone acetylation as well as methylation, although this remains to be tested.
PTIP and genome stability
Disruption of murine Ptip has shown that it has a crucial role in the preservation of genome stability. The homozygous disruption of Ptip in mice is lethal at embryonic day 9.5 (Cho et al, 2003). At embryonic day 7.5, the cells have entered S phase and are replicating DNA although a high number of DSBs can be detected. These DSBs do not appear to be a consequence of apoptosis because the nuclei are not pyknotic (Cho et al, 2003). By embryonic day 8.5, the cells are arrested in G2 and contain a high number of DSBs, and they die before embryonic day 9.5. Therefore, chromosomes appear to fragment during DNA replication in Ptip−/− cells, causing G2 arrest and death (Cho et al, 2003). This phenotype is reminiscent of embryos from mice lacking the ATR protein kinase (Brown & Baltimore, 2000; de Klein et al, 2000). When replisomes stall at obstacles such as DNA damage, ATR stabilizes them and prevents their disassembly. PTIP might also be required to protect replisomes that stall, and the failure to stabilize stalled replisomes in its absence would cause S phase-specific DSBs (Cimprich & Cortez, 2008). Replication forks collapse, occasionally forming DSBs, which are rescued and repaired by homologous recombination (HR; Sancar et al, 2004). An interesting possibility is that PTIP could regulate HR, in which case the accumulation of DSBs in S phase observed in Ptip-null embryos might reflect collapsed forks that are not rescued by HR. It will be important to evaluate this possibility and whether PTIP is also required to prevent DSB formation during S phase in adult cells, as the appearance of high numbers of DSBs in PTIP-null embryos might reflect the rapid rates of embryonic cell division. In fact DSBs initially appear in the embryonic ectoderm, which seems to be particularly sensitive to DNA damage. It has yet to be established whether PTIP is required for S-phase progression in adult tissues; however, its localization at the sites of DNA replication indicates that PTIP might have an important role in S phase in adult cells (Issaeva et al, 2007).
Human cells that are depleted of PTIP, or cells from Ptip-null mice, are hypersensitive to ionizing radiation (IR), which causes DSBs (Cho et al, 2003; Gong et al, 2009; Jowsey et al, 2004). There are two main mechanisms for the repair of DSBs: non-homologous end joining (NHEJ), which occurs primarily in G1 and involves the re-ligation of DNA ends; and HR, which is used primarily in S and G2 phases, and requires an intact sister chromatid or homologous chromosome to direct the repair of the DSB (Sonoda et al, 2006). HR is important not only for DSB repair but also for the rescue of blocked or collapsed replication forks. Therefore, it is tempting to speculate that the appearance of DSBs in Ptip-null embryos and the IR hypersensitivity of cells lacking PTIP could be explained by a common role for PTIP in regulating HR. It will be important to measure how well agents that cause DSBs or block replisomes can induce HR in cells lacking PTIP. PTIP interacts with the MRN complex and with the Bloom syndrome protein (Cho et al, 2007; Patel et al, 2007), both of which are known to be crucial for DNA-damage signalling and HR; however, the significance of these interactions is not yet clear.
The stalling of replisomes at DNA lesions leads to Lys 63-linked polyubiquitination of PCNA—a processivity factor for DNA polymerases—that facilitates the bypass of these lesions (Ulrich, 2005). The depletion of PTIP from Xenopus extracts or from human cells markedly reduces the level of PCNA ubiquitination induced by replication blockage and attenuates the recruitment of lesion-bypass factors to chromatin (Gohler et al, 2008). Could defective DNA-lesion bypass explain the high level of DSBs seen in PTIP-null embryos (Cho et al, 2003)? Defects in lesion bypass cause early embryonic lethality in mice (Wittschieben et al, 2000); however, more work is needed to determine whether this is the cause of DSBs in embryos lacking PTIP. The molecular mechanisms whereby PTIP promotes PCNA ubiquitination are not yet understood, but could involve the recruitment of PCNA ubiquitin ligases to sites of blocked DNA replication. The identity of these ligases is not yet clear, although two candidates have been proposed (Motegi et al, 2008; Unk et al, 2008). However, given the connection between PTIP and chromatin modifiers, it is also possible that PTIP controls PCNA ubiquitination indirectly by creating a chromatin environment that favours the access of PCNA-modifying factors. An important question is therefore whether histone methylation influences PCNA modification.
PTIP and DNA-damage signalling
DNA damage poses a potentially serious threat to genome stability. DNA lesions activate the ATM and ATR protein kinases, which, in turn, activate the CHK1 and CHK2 kinases (Cimprich & Cortez, 2008). Together, these kinases orchestrate many protective cellular responses, including the slowing or arrest of cell-cycle progression, the activation of DNA repair and apoptosis (Sancar et al, 2004). ATM and ATR phosphorylate a wide range of substrates on pS/T-Q motifs (Sancar et al, 2004), although they require adaptor proteins—such as 53BP1—to enable the phosphorylation of different subsets of downstream substrates, such as the tumour suppressor p53, and the CHK1 and CHK2 kinases. 53BP1 is important not only to facilitate the phosphorylation of some substrates by ATM/ATR, but also for DNA repair and the activation of cell-cycle checkpoints (Zgheib et al, 2005).
PTIP seems to be an adaptor protein for ATM. In response to DSBs, ATM and CHK2 phosphorylate the p53 tumour suppressor at Ser 15 and Ser 20, respectively. Depletion of PTIP from human cells does not affect the DSB-induced stabilization of p53, but causes a reduction of the ATM-dependent phosphorylation of p53 at Ser 15 (Jowsey et al, 2004), which normally promotes p53-dependent transcription, thereby leading to the upregulation of proteins that block cell-cycle progression or cause apoptosis (Shiloh, 2006). Consequently, cells that lack PTIP have defects in the induction of p53-responsive genes such as p21 (Jowsey et al, 2004). How PTIP and the other adaptor proteins function at the molecular level to facilitate the phosphorylation of distinct subsets of ATM/ATR targets is an essential, but poorly understood, issue.
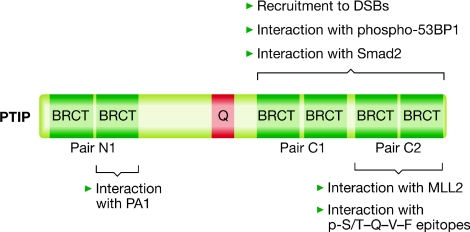
BRCT domains are small modular domains that are often, but not always, found in pairs in proteins that regulate responses to DNA damage, such as 53BP1, MDC1 and BRCA1 (Bork et al, 1997). Some BRCT domain pairs are able to bind to phosphorylated serine or threonine residues (Manke et al, 2003; Yu et al, 2003). PTIP has three pairs of BRCT domains: one at the amino terminus (pair N1) and two at the carboxyl terminus (pairs C1 and C2; Fig 2), which points to a role in DNA-damage signalling. Yaffe and colleagues identified a fragment of human PTIP in a screen for cDNAs that, when translated, bound to a library of degenerate phosphopeptides containing pS/T-Q motifs phosphorylated by ATM/ATR. Binding of the PTIP fragment to pS/T-Q-motifs was mediated by BRCT pair C2 (Fig 2), and the optimal binding motif was pS/T-Q-V-F. BRCT pair C2 was also able to retrieve 53BP1 from extracts of cells, but only after exposure of the cells to IR (Manke et al, 2003). These results suggested that when DNA damage occurs in cells, PTIP BRCT pair C2 binds to a pS/T-Q-V-F motif in 53BP1 phosphorylated by ATM. Shortly thereafter, endogenous PTIP was shown to bind to 53BP1 after exposure of the cells to IR in an ATM-dependent manner (Jowsey et al, 2004), and the mutation of a single ATM phosphorylation site—Ser 25 (DiTullio et al, 2002)—in 53BP1 was shown to prevent its interaction with PTIP after DNA damage in vivo (Munoz et al, 2007). However, Ser 25 of 53BP1 does not conform to the pS/T-Q-V-F consensus (Manke et al, 2003). Furthermore, BRCT pair C2 of PTIP does not seem to be sufficient to interact with the phospho-Ser 25 of 53BP1 in vivo or in vitro. Instead, pairs C1 and C2 both seem to be required (Gong et al, 2009; Munoz et al, 2007). The need for two BRCT domain pairs in PTIP to bind to 53BP1 Ser 25 is not yet understood, especially given the fact that pair C2 alone can bind to other pS/T-Q-V-F phosphopeptides (Manke et al, 2003). In any case, the mutation of 53BP1 Ser 25 causes defects in the DNA-damage response, as do mutations in the PTIP C-terminal BRCT domains that prevent its interaction with 53BP1 phospho-Ser 25 (Munoz et al, 2007). Therefore, it is likely that the interaction between PTIP and 53BP1 phospho-Ser 25 is an important, although poorly understood, event for an intact DNA-damage response. 53BP1 is important for cellular resistance to IR owing to its role in NHEJ (Riballo et al, 2004). PTIP could also regulate NHEJ, although, as mentioned earlier, it might also have a role in HR.
Figure 2.
Organization and known roles of the PTIP BRCT domains. Schematic diagram of the domain organization of PTIP and known cellular functions. Both pairs of PTIP carboxy-terminal BRCT domains are required for binding to sites of DNA damage, and for interacting with Smad2 and with phospho-53BP1, and neither pair alone can carry out these functions. By contrast, the extreme C-terminal pair can interact with MLL2 and with pS-Q-V-F phospho-epitopes. PA1 interacts with the second BRCT domain of pair N1. BRCT, protein domain present at the BRCA1 C terminus; C-terminal, carboxy-terminal; DSB, DNA double-strand break; MLL, mixed-lineage leukaemia; PA1, PTIP-associated protein 1; 53BP1, p53-binding protein 1; pS-Q-V-F, phospho-Ser/Thr-Gln; PTIP, PAX-transactivation domain-interacting protein; Q, polyglutamine-tract region; Smad2, SMA- and MAD-related protein 2.
The PA1–PTIP complex
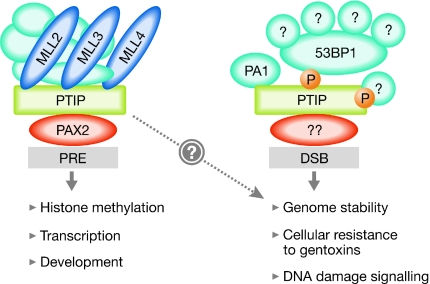
Gel-filtration assays have recently revealed that there are two pools of PTIP in cells (Cho et al, 2007; Gong et al, 2009): a higher molecular weight pool (of >2 MDa) containing the HMTase complexes, and a lower molecular weight pool (of ∼200 kDa) that contains the newly identified protein PA1 (Fig 3). PA1 associates directly with PTIP (Cho et al, 2007) and, importantly, depletion of PA1 renders cells hypersensitive to IR (Gong et al, 2009), as is the case for PTIP. The second BRCT domain of the N1 pair of PTIP is required for its interaction with PA1 (Fig 2) and a mutant form of PTIP lacking this domain cannot rescue the IR hypersensitivity of PTIP-null cells (Gong et al, 2009), suggesting that the interaction of PTIP with PA1 is crucial for the preservation of genome stability. PA1 depends on PTIP to form IRIF, whereas the binding of PTIP at or near DSBs does not require PA1 (Gong et al, 2009). Taken together, these data suggest that the pool of PTIP containing PA1 is important for DNA-damage responses. The molecular mass of the PTIP–PA1 complex in non-irradiated cells is not much larger than the combined masses of PTIP and PA1, suggesting that there might be no other proteins in this complex. However, it is likely that extra proteins are recruited to the PTIP–PA1 complex after DNA damage in the same manner that 53BP1 binds to PTIP, and it would be interesting to perform gel-filtration experiments after the exposure of cells to IR in order to verify this possibility. In the gel-filtration experiments described above, 53BP1 eluted in a high-molecular weight complex (>2 MDa) of unknown composition, which probably contained USP28 (Zhang et al, 2006) and was distinct from the larger PTIP-containing complex. We predict that, after DNA damage, 53BP1 is phosphorylated by ATM and the entire 53BP1 complex becomes associated with the PTIP–PA1 complex. Therefore, identifying all the components of each complex is important to obtain a complete picture of how 53BP1 and PTIP act to protect genome stability. The consequences of the genetic ablation of PA1 are unknown and it will be important to see how closely the phenotype of PA1-null organisms recapitulates that of PTIP-null mice.
Figure 3.
PTIP exists in at least two functionally distinct complexes. Gel filtration has shown that there are two pools of PTIP in cells: a higher molecular weight pool that contains the known PTIP-associated HMTases (Fig 1) and a smaller complex that also contains PA1. It is clear that PA1–PTIP is involved in DNA damage responses, but it is not yet clear whether PTIP-associated HMTases participate in these responses. 53BP1 is part of a large (non-PTIP-containing) protein complex before DNA damage; after DNA damage, ATM-phosphorylated 53BP1 binds to PTIP, presumably to the pool associated with PA1. It is likely that PTIP also associates with other proteins in a phosphorylation-dependent manner after DNA damage. 53BP1, p53-binding protein 1; ATM, ataxia telangiectasia mutated; DSB, DNA double-strand break; HMTase, histone methyltransferase; MLL, mixed-lineage leukaemia; PA1, PTIP-associated protein 1; PAX, paired box; PRE, PAX2-response element; PTIP, PAX-transactivation domain-interacting protein.
PTIP binds at or near sites of DNA damage
PTIP binds to sites of DNA damage (Issaeva et al, 2007; Jowsey et al, 2004; Manke et al, 2003); however, the underlying molecular mechanisms of its recruitment are only beginning to be understood. The histone variant H2AX is phosphorylated in response to DNA damage on Ser 139, thereby allowing its direct interaction with a pair of BRCT domains in the adaptor protein MDC1 (Stucki et al, 2005). MDC1 subsequently recruits the MRN complex (Stucki et al, 2005) and the E3 ubiquitin ligase RNF8—which catalyses the ubiquitination of proteins at sites of DNA damage (Bin & Elledge, 2007; Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007). MDC1, RNF8 and the E2 ubiquitin-conjugating enzyme UBC13 are all required for the binding of 53BP1 to sites of DNA damage, possibly because 53BP1 binds to proteins that are ubiquitinated by UBC13–RNF8 at DSBs. UBC13-dependent and RNF8-dependent ubiquitination events have also recently been shown to be required for the formation of PTIP IRIF after PTIP overexpression (Gong et al, 2009). Although RNF8 is also required for 53BP1 recruitment, and although PTIP and 53BP1 associate in an ATM-dependent manner after DNA damage, the recruitment of PTIP to IRIF is independent of 53BP1 (Gong et al, 2009; Jowsey et al, 2004). PTIP does not seem to be capable of binding to ubiquitin directly (Gong et al, 2009) and has no obvious ubiquitin binding motifs; therefore, it is possible that a PTIP-associated protein mediates the binding of PTIP to ubiquitinated proteins near the sites of DNA damage. This protein is unlikely to be a component of the MLL HMTase complexes, as these proteins do not form IRIF (Issaeva et al, 2007), and MLL3, for example, is not required for PTIP to bind at sites of DNA damage (Gong et al, 2009). By contrast, it is possible that UBC13-catalysed and RNF8-catalysed ubiquitination alters the activity of one or more proteins or enzymes at DSBs, which in turn leads to PTIP recruitment. In any case, the C-terminal BRCT domains of PTIP are required for its retention at DSBs (Fig 2). It is likely that the identification of ligands of these domains other than those already known—that is, 53BP1, MLL2 and Smad2—will be important for resolving this issue.
PTIP in lower eukaryotes?
Esc4 is a multi-BRCT domain-containing protein from Saccharomyces cerevisiae with a relatively high degree of sequence similarity to PTIP (Rouse, 2004)—especially in the BRCT domains—that was implicated in the response to DNA-damaging agents in several yeast genetic screens (Chang et al, 2002; Hanway et al, 2002). Esc4 is required to overcome blocks to DNA replication and is an important target of Mec1, the yeast orthologue of ATR kinase (Roberts et al, 2006; Rouse, 2004; Zappulla et al, 2006). Although Esc4 and PTIP are both required to preserve genome stability during S phase (Cho et al, 2003; Roberts et al, 2006; Rouse, 2004; Zappulla et al, 2006), there are some notable differences between the two proteins: PTIP has one pair of BRCT domains at the N terminus and two at the C terminus, whereas Esc4 has two pairs at the N terminus and one at the C terminus; PTIP is essential in mice, whereas Esc4 is not essential in yeast (Chang et al, 2002; Hanway et al, 2002); PTIP is required for cellular responses to DSBs, but Esc4 is not (Roberts et al, 2006; Rouse, 2004; Zappulla et al, 2006); and none of the reported Esc4-interacting proteins are HMTases (Chin et al, 2006). Therefore, in functional terms, Esc4 and PTIP have more differences than similarities and it is unclear whether PTIP is a genuine orthologue of Esc4 that has evolved additional functions in higher eukaryotes or whether the sequence similarity between Esc4 and PTIP is simply due to the conservation of the BRCT domains.
Dual roles for PTIP?
It is clear that PTIP controls transcription by targeting HMTases to gene promoters, and this function is relatively well understood in mechanistic terms (Fig 1). PTIP also controls aspects of the cellular response to DNA damage, although our understanding of this process is still at an early stage. Are these roles separate or related? H3K4 methylation is known to be important for the maintenance of genome stability. For example, the PHD domain of the ING2 tumour suppressor—a subunit of the HDAC1 histone deacetylase complex—binds to methylated H3K4 after DNA damage. This stabilizes the association of the HDAC1 complex with promoters that are repressed in response to DNA damage (Shi et al, 2006). Chd1, which is a protein that also binds to methylated H3K4, regulates S-phase progression in yeast (Biswas et al, 2008). In addition, the ING1 tumour suppressor must also bind to methylated H3K4 to promote DNA repair and/or apoptosis after DNA damage (Pena et al, 2008). Therefore, defects in H3K4 methylation could, in principle, contribute to the formation of the S-phase DSBs observed in PTIP-null embryos. The methylation of histone H3 at other lysine residues is also important for DNA-damage responses, as it promotes the assembly of signalling complexes at sites of DNA damage. For example, the methylation of H3K79 by DOT1L enables the binding of the Tudor domains of 53BP1, which contributes to 53BP1 recruitment to DSBs (Huyen et al, 2004)—a process in which the methylation of H4K20 also has a role (Botuyan et al, 2006). PR-Set7/SET8 methylates H4K20, localizes at sites of DNA replication (Tardat et al, 2007) and its depletion causes defects that are notably similar to those seen in cells lacking PTIP, such as the induction of DSBs in S phase and an accumulation of cells in G2 (Houston et al, 2008; Jorgensen et al, 2007; Tardat et al, 2007). This phenotype is notably reminiscent of the phenotype of PTIP-null embryos (Cho et al, 2003) and it is tantalizing to speculate that PTIP regulates H4K20 methylation in some manner. However, there is currently no evidence that PTIP regulates H4 methylation and immunoprecipitates of cells in which PTIP has been overexpressed contain no detectable H4 HMTase activity (Cho et al, 2007).
Histone methylation is important for DNA-damage responses, but are the PTIP-associated HMTases involved? Although PTIP and PA1 form IRIF, the components of the MLL HMTase complexes do not (Gong et al, 2009; Issaeva et al, 2007), and the knockdown of MLL3 does not affect PTIP IRIF (Gong et al, 2009). Therefore, it has been argued that this type of complex does not have a role in preserving genome integrity but rather regulates transcription. Our impression is that this is the case, although excluding a role for PTIP HMTases in DNA-damage responses based on IRIF analysis might not be valid without testing the effect that depleting MLL HMTases has on cellular sensitivity to DNA damage. Not all proteins that are involved in DNA-damage responses form foci and, even when they do, focus formation is not always functionally important (Ball et al, 2005). Therefore, it will also be important to address definitively whether the PTIP-associated HMTases are required for a DNA-damage response. In any case, the most important task is to determine the full composition and mode of operation of the PTIP complexes that are known to be DNA-damage responsive before and after DNA damage (Fig 3).
Conclusion and future challenges
There are at least two PTIP-associated complexes that most of the available data suggest are functionally distinct (Fig 3), although further experiments are needed to confirm this. We need to know what controls the distribution of PTIP among these complexes. Does the depletion of MLL2/MLL3/MLL4 HMTases cause genotoxin hypersensitivity, or defects in DNA repair or signalling? By what mechanism does the PTIP–PA1 complex promote cell survival after DNA damage? Are there other proteins apart from 53BP1 that associate with PTIP exclusively after DNA damage or in S phase? PTIP is recruited by transcription factors to gene promoters, where it recruits H3K4 methyltransferases to modulate gene expression. It also binds to sites of DNA damage in a manner that seems to require protein ubiquitination. How is this achieved? Does PTIP regulate HR? Why does PTIP interact with 53BP1? Clearly, much more information is needed to understand how PTIP regulates genome stability. It will be fundamentally important to fill in these gaps and to reconcile the functions of PTIP in histone modification, transcription and DNA damage (Sidebar A).
Sidebar A | In need of answers.
Are the transcriptional and DNA-damage associated functions of PTIP mechanistically separate?
Why is protein ubiquitination required for the recruitment of PTIP to sites of DNA damage?
What are the molecular mechanisms whereby PTIP and PTIP-PA1 control genome stability and resistance to DNA damage?
How does PTIP promote PCNA ubiquitination?
What is the role of PA1 at the molecular level?
How is PTIP distributed among several different complexes?

Ivan M. Muñoz

John Rouse
Acknowledgments
We thank J. Blow and members of the Rouse laboratory, especially G. Toh, for critical reading of the manuscript. Work in the J.R. laboratory is supported by the Medical Research Council UK, the Association for International Cancer Research (I.M.M.) and GlaxoSmithKline/Research Councils UK. J.R. is a European Molecular Biology Organization Young Investigator. We apologize to those scientists whose work we were unable to cite owing to space limitations.
References
- Ball HL, Myers JS, Cortez D (2005) ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell 16: 2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin W, Elledge SJ (2007) Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Takahata S, Xin H, Dutta-Biswas R, Yu YX, Formosat T, Stillman DJ (2008) A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics 178: 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV (1997) A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J 11: 68–76 [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen JJ, Mer G (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D (2000) ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 14: 397–402 [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Boone C, Brown GW (2002) A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci USA 99: 16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JK, Bashkirov VI, Heyer WD, Romesberg FE (2006) ESC4/RTT107 and the control of recombination during replication. DNA Repair 5: 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EA, Prindle MJ, Dressler GR, Lechner MS, Levitan I (2003) BRCT domain-containing protein PTIP is essential for progression through mitosis. Mol Cell Biol 23: 1666–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW et al. (2007) PTIP associates with MLL3-and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl E, Koseki H, Balling R (1997) Pax genes and organogenesis. Bioessays 19: 755–765 [DOI] [PubMed] [Google Scholar]
- de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH (2000) Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol 10: 479–482 [DOI] [PubMed] [Google Scholar]
- Devriendt K, Matthijs G, Van Damme B, Caesbroeck D, Eccles M, Vanrenterghem Y, Fryns JP, Leys A (1998) Missense mutation and hexanucleotide duplication in the PAX2 gene in two unrelated families with renal-coloboma syndrome (MIM 120330). Hum Genet 103: 149–153 [DOI] [PubMed] [Google Scholar]
- DiTullio RA, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, Halazonetis TD (2002) 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol 4: 998–1002 [DOI] [PubMed] [Google Scholar]
- Gohler T, Munoz IM, Rouse J, Blow JJ (2008) PTIP/Swift is required for efficient PCNA ubiquitination in response to DNA damage. DNA Repair 7: 775–787 [DOI] [PubMed] [Google Scholar]
- Gong Z, Cho Y-W, Kim J-E, Chen J (2009) Accumulation of Pax2 transactivation-domain interaction protein (PTIP) to sites of DNA breaks via an RNF8-dependent pathway is required for cell survival following DNA damage. J Biol Chem [doi:10.1074/jbc.M809158200] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanway D, Chin JK, Xia G, Oshiro G, Winzeler EA, Romesberg FE (2002) Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc Natl Acad Sci USA 99: 10605–10610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister A et al. (2002) The HMG-I/Y-related protein p8 binds to p300 and Pax2 trans-activation domain-interacting protein to regulate the trans-activation activity of the Pax2A and Pax2B transcription factors on the glucagon gene promoter. J Biol Chem 277: 22314–22319 [DOI] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC (2008) Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem 283: 19478–19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MSY, Grant R, Manke I, Minn K, Yu XC, Yaffe MB, Chen JJ (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, DiTullio RA, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411 [DOI] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E (2007) Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27: 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS (2007) The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jowsey PA, Doherty AJ, Rouse J (2004) Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J Biol Chem 279: 55562–55569 [DOI] [PubMed] [Google Scholar]
- Kim D, Wang M, Cai Q, Brooks H, Dressler GR (2007) Pax transactivation-domain interacting protein is required for urine concentration and osmotolerance in collecting duct epithelia. J Am Soc Nephrol 18: 1458–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK et al. (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lechner MS, Levitan I, Dressler GR (2000) PTIP, a novel BRCT domain-containing protein interacts with Pax2 and is associated with active chromatin. Nucleic Acids Res 28: 2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB (2003) BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Massague J, Gomis RR (2006) The logic of TGF beta signaling. FEBS Lett 580: 2811–2820 [DOI] [PubMed] [Google Scholar]
- Motegi A et al. (2008) Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci USA 105: 12411–12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Jowsey PA, Toth R, Rouse J (2007) Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res 35: 5312–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR (2007) The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell 13: 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena PV et al. (2008) Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor. J Mol Biol 380: 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E et al. (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 16: 715–724 [DOI] [PubMed] [Google Scholar]
- Roberts TM, Kobor MS, Bastin-Shanower SA, Ii M, Horte SA, Gin JW, Emili A, Rine J, Brill SJ, Brown GW (2006) Slx4 regulates DNA damage phosphorylation of the BRCT checkpoint-dependent domain protein Rtt107/Esc4. Mol Biol Cell 17: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J (2004) Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J 23: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J (2007) Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Ann Rev Biochem 73: 39–85 [DOI] [PubMed] [Google Scholar]
- Sanyanusin P, Schimmenti LA, Mcnoe LA, Ward TA, Pierpont MEM, Sullivan MJ, Dobyns WB, Eccles MR (1995) Mutation of the Pax2 gene in a family with optic-nerve colobomas, renal anomalies and vesicoureteral reflux. Nat Genet 9: 358–364 [DOI] [PubMed] [Google Scholar]
- Shi XB et al. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y (2006) The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci 31: 402–410 [DOI] [PubMed] [Google Scholar]
- Shimizu K et al. (2001) Swift is a novel BRCT domain coactivator of Smad2 in transforming growth factor beta signaling. Mol Cell Biol 21: 3901–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S (2006) Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair 5: 1021–1029 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E (2007) PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 179: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M, GomezPardo E, Dressler GR, Gruss P (1995) Pax-2 controls multiple steps of urogenital development. Development 121: 4057–4065 [DOI] [PubMed] [Google Scholar]
- Ulrich HD (2005) The RAD6 pathway: control of DNA damage bypass and mutagenesis by ubiquitin and SUMO. Chembiochem 6: 1735–1743 [DOI] [PubMed] [Google Scholar]
- Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L (2008) Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci USA 105: 3768–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben J, Shivji MKK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, Wood RD (2000) Disruption of the developmentally regulated Rev31 gene causes embryonic lethality. Curr Biol 10: 1217–1220 [DOI] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J (2003) The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Maharaj ASR, Connelly JJ, Jockusch RA, Sternglanz R (2006) Rtt107/Esc4 binds silent chromatin and DNA repair proteins using different BRCT motifs. BMC Mol Biol 7: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgheib O, Huyen Y, DiTullio RA, Snyder A, Venere M, Stavridi ES, Halazonetis TD (2005) ATM signaling and 53BP1. Radiother Oncol 76: 119–122 [DOI] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, Elledge SJ (2006) A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 126: 529–542 [DOI] [PubMed] [Google Scholar]