Abstract
Brain tumors are one of the leading causes of death in adults with cancer; however, molecular classification of these tumors with in vivo magnetic resonance spectroscopy (MRS) is limited because of the small number of metabolites detected. In vitro MRS provides highly informative biomarker profiles at higher fields, but also consumes the sample so that it is unavailable for subsequent analysis. In contrast, ex vivo high-resolution magic angle spinning (HRMAS) MRS conserves the sample but requires large samples and can pose technical challenges for producing accurate data, depending on the sample testing temperature. We developed a novel approach that combines a two-dimensional (2D), solid-state, HRMAS proton (1H) NMR method, TOBSY (total through-bond spectroscopy), which maximizes the advantages of HRMAS and a robust classification strategy. We used 2 mg of tissue at -8°C from each of 55 brain biopsies, and reliably detected 16 different molecules. We compared two classification strategies, the support vector machine (SVM) classifier and a feed-forward neural network using the Levenberg-Marquardt back-propagation algorithm. We used the minimum redundancy/maximum relevance (MRMR) method as a powerful feature-selection scheme along with the SVM classifier. We also used the minimum redundancy/maximum relevance (MRMR) method as a powerful feature-selection scheme along with the SVM classifier.
Keywords: brain/CNS cancers, tumor biomarkers, ex vivo high-resolution magic angle spinning magnetic resonance spectroscopy, support vector machines, neural networks
Introduction
Brain tumors are the leading cause of cancer death in children, the second most common cause of cancer in young adults, and account for a high proportion of deaths in older adults (1). According to the US Central Brain Tumor Registry, between 2000 and 2004 35,347 males and 28,477 females died from primary malignant brain and other central nervous system (CNS) tumors (www.cbtrus.org). With current advances in imaging, surgery, radiotherapy, and chemotherapy, and in combination with new approaches in anti-angiogenesis, the outcome of individuals with CNS tumors has been improving (2-4). Even some high-grade tumors, such as ependymomas or medulloblastomas, can now be cured in children with the appropriate therapy (5,6).
The most widely used histological system of brain tumor typing is the classification scheme of the World Health Organization (WHO), in which tumors are classified according to histological features characteristic of the assumed cell of origin (7). However, diagnoses are frequently controversial, since tumors often do not follow classic histology, and pathological diagnosis can therefore be subjective (8). Thus, a non-subjective diagnostic approach that relies on highly informative biomarkers is needed to improve tumor typing accuracy and the appropriateness of the treatment course chosen. Such an approach would markedly improve the long-term prognosis, quality of life, and survival of patients with brain tumors.
Magnetic resonance spectroscopy (MRS) can provide statistically differentiable molecular biomarkers for tumor grade differentiation, treatment and patient survival prediction (9). Ex vivo high-resolution magic angle spinning (HRMAS) proton (1H) MRS of unprocessed tissue samples (10) can help to interpret in vivo 1H MRS results. This technique can not only improve micro-heterogeneity analysis of high-grade tumors (11), but can also elucidate the relationships between clinically-relevant cell processes and specific metabolites, such as choline-containing compounds involved in phospho-lipid metabolism and lipids involved in apoptosis leading to necrosis (12). Two-dimensional (2D) HRMAS 1H MRS enables more detailed and unequivocal assignments of biologically important metabolites in intact tissue samples (13-16). Furthermore, 2D MRS sequences have been implemented in vivo with relative success (17-20).
The use of MRS classification and statistical analyses for both in vivo and in vitro data has been widely reported (21-33). One of the principal difficulties in such analyses is the large number of metabolites that may contribute to the spectra, each with relative intensities that can vary greatly even in samples of the same type (22). Nonetheless, even early studies have reported that the spectra of body fluids obtained with MRS are systematically different between tumor patients and healthy individuals. In many cases, successful differentiation using both linear and nonlinear methods can be made, based on single resonance peaks or ratios of resonance ranges (23). Recent work on brain tumors has shown that classification according to histological type and grade is possible using similar approaches, such as linear discriminant analysis after feature extraction with independent components analysis in a Bayesian framework (27), correlation analysis and stepwise linear discriminant analysis (28), or belief networks (32). Support vector machines (SVMs) and probabilistic neural networks have also been employed in image-analysis systems to assist in brain tumor diagnosis (34-36).
Here, we present a brain tumor biopsy study that should enable the development of new clinical tools to better assess operable cancers via tissue molecular characterization and fingerprinting. This work should also facilitate the distinction of tumor types that cannot be readily distinguished histopathologically (37) or with routine neuroimaging (38). Such progress will enable neurooncologists, neuropathologists, neurosurgeons, and neurologists to make informed decisions related to tumor type, grade, and treatment options. This work will also facilitate the clinical application of lower resolution in vivo MRS to inoperable cancers using metabolic biomarkers to monitor anticancer therapies, in order to improve patient survival and quality of life.
Materials and methods
Tissue biopsies
Fifty-five samples of control biopsies from epileptic surgeries (n=9) and tumor biopsies (n=46) were analyzed. The tumor biopsies belonged to three categories: high-grade [20 cases: 12 glioblastoma multiforme (GBM); 8 anaplastic astrocytoma (AA)], low-grade (17 cases: 7 meningioma; 7 schwanoma; 3 pylocitic astrocytoma), and brain metastases (9 cases: 5 from adenocarcinoma; 4 from breast cancer). Subjects ranged in age from 17 to 54 years.
HRMAS 1H MRS using adiabatic TOBSY
We previously designed a 2D ex vivo HRMAS 1H MRS procedure for brain tumors, based on novel concepts rooted in solid-state NMR spectroscopy (39). The rationale for our approach is that as tumor biopsies are more solid than liquid in nature, conventional liquid-state NMR pulse sequences that rely on scalar-coupling-mediated magnetization transfer (i.e., total correlated spectroscopy or TOCSY) may not be the best choice for these purposes. Although magic angle spinning (MAS) averages anisotropic interactions such as chemical shielding anisotropy (CSA) or dipolar couplings (D), these can be reintroduced unintentionally by pulse sequences not designed to eliminate them; hence, the scalar-coupling transfer is compromised, and the efficiency of these pulse sequences can be dramatically altered under MAS conditions.
Briefly, we designed a pulse sequence, using symmetry principles (40) to maximize scalar-coupling transfer under MAS conditions, for samples that are predominantly solid in character. This was done by removing, based on the 1st order Average Hamiltonian Theory, the anisotropic interactions (CSA and D) and the offset frequency. We modified an existing pulse sequence, C9, which is based on a concept known as TOBSY in solid-state NMR (41). The C9 pulse sequence has been used successfully for 13C NMR spectroscopy to study the structure and dynamics of membrane proteins in proteoliposome samples that closely mimic tissue properties (42). For the purposes of our project, several modifications were made to: C9115, and b) construct C elements from adiabatic pulses [WURST (43)] that enhance signal-to-noise ratios and robustness with respect to radio-frequency field calibration (39).
All HRMAS 1H MRS using TOBSY experiments were performed on a Bruker Bio-Spin Avance NMR spectrometer (600.13 MHz) using a 4-mm triple resonance (1H, 13C, 2H) HRMAS probe (Bruker). Specimens were pre-weighed and transferred to a ZrO2 rotor tube (4 mm diameter, 50 μl), containing an external standard [trimethylsilyl propionic-2,2,3,3-d4 acid (TSP), Mw=172, d=0.00 ppm] that functioned as a reference both for both resonance chemical shift and quantification. The HRMAS 1H MRS was performed at -8°C with 3 kHz MAS speed to minimize tissue degradation. Typical acquisition parameters were: 2 k points direct dimension (13 ppm spectral width), 200 points indirect dimension (7.5 ppm spectral width), 8 scans with 2 dummy scans, 1 sec water pre-saturation, 2 sec total repetition time, 45 msec mixing time and total acquisition time 45 min.
Analysis of 2D TOBSY MR spectra
The spectra of intact specimens were analyzed using the XWINNMR 3.5 software package (Bruker Biospin Corp, Billerica, MA). Before Fourier transformation and phasing, the 2D free induction decays were subjected to QSINE = 3 window apodization. Baseline correction was then performed using a low order spline function. After Lorentzian and/or Gaussian fitting, the area under the curves or the volumes of the 15 most intense spectra resonances were calculated. Relative quantification using the TSP standard was performed as described below. These resonances were identified and assigned to the corresponding metabolites.
Quantification of brain metabolites from the 2D TOBSY MR spectra
To quantify the brain metabolites, we used the ratio of the cross peak volumes of the metabolites [CPV(M)] to the TSP diagonal peak volume [DPV(TSP)]. This ratio was further divided by the biopsy weight (w) to yield the normalized metabolite intensity: Ic (M) = 1/w × CPV (M)/DPV (TSP).
Brain tumor biopsy classification strategy
We applied two classifiers: a linear support vector machine (SVM) (44) and a feed-forward neural network using the Levenberg-Marquardt back-propagation (45) algorithm for training (NN MLP). The SVM methods classify data by first mapping the data in a hyper-dimensional space and subsequently separating the data into distinct clusters by defining a hyper-plane that ‘cuts’ the hyper-dimensional space into disjointed regions. The data located in one such region belong to a single cluster. The dimensionality of the hyper-dimensional space must be higher than that of the data space. The choice of how to map the data into the hyper-dimensional space may be informed by the problem, or the data may be embedded using some choice of basis functions (such as polynomial or Gaussian). Linear SVM methods search for the optimal hyper-plane to cut the hyper-dimensional space by solving a quadratic programming problem (44). Least-squares SVM (SVM-LS) search for the optimal separating hyper-plane by minimizing a least-squares cost function (46). The reason for trying both versions is that the computational cost of linear SVM methods is a rapidly increasing function of the SVM training set, whereas SVM-LS methods are far more tractable computationally.
Artificial neural networks (ANNs) have been established as useful tools for classification and function approximation problems (47). These networks are comprised of layers of nodes (‘neurons’). Each node receives N inputs from N other sources (the environment or other nodes), weighs each input by a weight (N weights total), and produces a nonlinear response - for example, a binary response (1 when a threshold is exceeded, otherwise 0) or a sigmoidal function for continuous output. An ANN can be built as a hierarchy of neuronal layers. The ANN computations use both forward propagation (‘feedforward’), from the input-layer to the output-layer nodes, and reverse propagation (‘back-propagation’), from the output-layer to the input-layer nodes, in order to satisfy criteria of numerical convergence to a solution. Typically, weights are initially chosen randomly, and the output is compared to the correct (known) output for the training set. A computational search is then performed to solve for the optimal set of N weights that minimizes the distortion of the computed output relative to the true one. An important characteristic of ANNs is that it is not necessary to have a complete knowledge of the relationship among the variables in the problem.
Both methods, SVM and ANN, face practical limitations from their unavoidable use of training sets. For example, the SVM hyper-plane separating disjointed regions may be erroneously drawn so that a new datum is placed in the wrong region (cluster); this is particularly likely when data points belonging to different clusters are close to each other in hyper-dimensional space. Similarly, for ANNs, the optimal weights for the training set may not accurately reflect the optimal weights for the problem. Therefore, for both methods it is important to have databases where new data are added and the algorithms are re-trained on the increasing total number of cases.
Fig. 1 represents the general architecture of the classification system we used. This classification system consists of a preprocessing step, where a subset of features is selected, and a main step, where the subset is used as input to the main body of the classifier. The primary reason for selecting the feature subset (or ‘reduced feature space’) was to speed up the numerical computation process. Two MRS feature spaces were selected: the full feature space comprised of all 16 MRS features, and a reduced feature space comprised of 4 specific features [choline (Cho), lactate (Lac), lipids (Lip), and n-acetyl aspartate (NAA)]. In the case of the SVM classifier, a feature selection scheme was also used, thus producing a third MRS feature space. Specifically, we selected the minimum redundancy/maximum relevance (MRMR) method (48), a powerful framework for feature selection that captures class characteristics in a broader spectrum by reducing mutual redundancy within the feature set.
Figure 1.
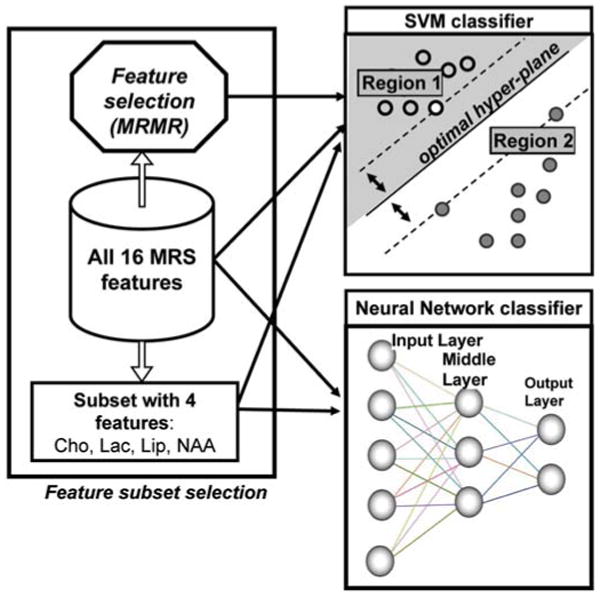
Building blocks of the classification system. Support vector machine (SVM) and neural network Levenberg-Marquardt back-propagation (NN-MLP) classifiers were used. First, the feature space is selected. All three selected feature spaces (full feature space with all 16 MRS features, reduced feature space using the MRMR feature selection, 4 MRS features) were input into the linear and least-squares SVM classifiers; and two feature spaces (all 16 MRS features, 4 MRS features) were input into the NN-MLP classifier. The SVM classifiers separate the data into clusters by drawing an optimal hyperplane through the hyper-dimensional space in which the original feature space is embedded such that it separates said hyper-dimensional space into respective disjointed regions. In the schematic, the nearest data points from the two regions (two clusters) are equidistant to the optimal hyperplane at a distance Δ. The NN classifiers use an iterative technique that combines the input/output of artificial neurons to achieve a similar result.
The classifiers were tested by comparing their discrimination behavior to a binary classification problem; namely, each individual class was compared against all other classes. We must note here that during all experiments with both classifiers we adopted the standard leave-one-out training/testing scheme; all but one element of the data was used as a training set, and the left-out case was used for testing the predictive performance of the resulting classifier. The performance of each classifier was evaluated using the classifications of ‘accuracy’ (the percentage of correctly classified cases), ‘sensitivity’ (the ratio of true positives to the sum of true positives and false negatives), and ‘specificity’ (the ratio of true negatives to the sum of false positives and true negatives).
During all SVM classification experiments we used the LIBSVM environment (www.csie.ntu.edu.tw/∼cjlin/libsvm), where the SVM soft-margin constant C was set to 100. For the feed-forward neural network, we used the Matlab trainlm routine, where the number of nodes in the hidden layer was set to 10 in all examined problems, and all nodes had the typical sigmoid function.
Results
Table I summarizes the results obtained for the three classification methods: the linear SVM classifier, the SVM-LS classifier, and the NN-MLP classifier. The results were quantified by three metrics, namely, sensitivity, specificity, and accuracy. Additionally, results for the linear SVM are presented for each of the three choices for reduced feature space, 4 features, all 16 features, and MRMR feature selection. While we might expect the full 16-feature space classifier to outperform the two reduced-feature spaces (4 feature and MRMR), it is possible that the other two methods could slightly outperform the full-feature space in subtyping (Table I and Fig. 2). For instance, the typing of controls was achieved with similar accuracy, sensitivity, and specificity whether the SVM used only 4 or all 16 features. On the other hand, in typing high-grade biopsies, although the specificity was higher based on the subset of 4 features, the accuracy and sensitivity were much higher when all 16 features were used by the SVM classifier. In typing low-grade biopsies, all metrics except sensitivity were higher when all 16 features were used. For biopsies of metastatic tumors, although the sensitivity was higher with 16 features, the specificity was very high with only 4 features. Furthermore, for metastatic tumors, the most important metric, accuracy, was the same regardless of whether 4 or 16 features were used.
Table I.
The accuracy, sensitivity and specificity of the three classifiers, estimated using reduced- and full-feature spaces.
| SVM linear | SVM-LS RBF kernel | MLP | ||||||
|---|---|---|---|---|---|---|---|---|
| Binary classification problem | ||||||||
| Metric | Class examined | using 4 features Cho, Lac, Lip, NAA | using 16 features | Feature selection first | using 4 features Cho, Lac, Lip, NAA | using 16 features | ||
| Selected features | ||||||||
| Accuracy | Normal | 98.2 | 98.2 | 3 features: | 100.0 | 96.4 | 96.4 | 98.2 |
| Sensitivity | 9 samples | 100 | 100 | ‘NAA’, ‘Asp’, ‘PC’ | 100.0 | 77.8 | 100.0 | 100.0 |
| Specificity | 97.8 | 97.8 | 100.0 | 100.0 | 95.7 | 97.8 | ||
| Accuracy | High-grade | 52.2 | 80.4 | 6 features: | 80.4 | 76.1 | 69.6 | 78.3 |
| Sensitivity | group | 5.0 | 85.0 | ‘Etn’, ‘PC’, ‘NAA’, ‘Gln’, | 85.0 | 70.0 | 60.0 | 70.0 |
| Specificity | 20 samples | 88.5 | 76.9 | ‘PUFA” ‘GABA’, ‘Ala’ | 76.9 | 80.8 | 76.9 | 84.6 |
| Accuracy | Low-grade | 69.6 | 76.1 | 8 features: | 78.3 | 73.9 | 69.6 | 80.4 |
| Sensitivity | group | 88.2 | 64.7 | ‘Lip’, ‘Etn’ ‘NAA’ ‘Glu” | 70.6 | 35.3 | 52.9 | 82.4 |
| Specificity | 17 samples | 58.6 | 82.8 | “Asp”, “Gln”, ‘Tau’ | 82.8 | 96.6 | 79.3 | 79.3 |
| Accuracy | Metastasis | 78.3 | 78.3 | 4 features | 91.3 | 84.8 | 78.3 | 84.8 |
| Sensitivity | group | 11.1 | 44.4 | ‘Asp’ ‘PUFA ‘Etn’ | 55.6 | 22.2 | 33.3 | 55.6 |
| Specificity | 9 samples | 94.6 | 86.5 | ‘Glu’ | 100.0 | 100.0 | 89.2 | 91.9 |
| Accuracy | Glioblastoma | 82.6 | 84.8 | 11 features: | 87.0 | 80.4 | 87.0 | 76.1 |
| Sensitivity | multiforme | 58.3 | 83.3 | ‘Etn’ ‘PC’ ‘NAA’ ‘Cho’ | 83.3 | 25.0 | 83.3 | 75.0 |
| Specificity | (subgroup) 12 samples | 91.2 | 85.3 | ‘GABA’ ‘PUFA’ ‘Gln’ ‘Ala’ ‘Asp’ ‘GPC’ ‘Lac’ | 88.2 | 100.0 | 88.2 | 76.5 |
| Accuracy | Anaplastic | 82.6 | 89.1 | 11 features: | 89.1 | 82.6 | 67.4 | 89.1 |
| Sensitivity | astrocytoma | 12.5 | 62.5 | NAA, Et, Lip, Gln, | 75.0 | 0.0 | 12.5 | 62.5 |
| Specificity | (subgroup) 8 samples | 97.4 | 94.7 | Tau, Ala, GABA, GPC, PUFA, PC, Glu | 92.1 | 100.0 | 78.9 | 94.7 |
| Accuracy | Menignioma | 84.8 | 89.1 | 4 features: | 93.5 | 87.0 | 73.9 | 82.6 |
| Sensitivity | (subgroup) | 0.0 | 57.1 | Etn, Li, Tau, Gln | 57.1 | 14.3 | 0.0 | 57.1 |
| Specificity | 7 samples | 100 | 94.9 | 100.0 | 100.0 | 87.2 | 87.2 | |
| Accuracy | Schwannoma | 87.0 | 89.1 | 4 features: | 91.3 | 84.8 | 80.4 | 89.1 |
| Sensitivity | (subgroup) | 71.4 | 71.4 | Gln, Cho, Etn, Lip | 42.9 | 0.0 | 42.9 | 57.1 |
| Specificity | 7 samples | 89.7 | 92.3 | 100.0 | 100.0 | 87.2 | 94.9 | |
| Accuracy | Piloytic | 93.5 | 89.1 | 1 feature: | 93.5 | 93.5 | 89.1 | 93.5 |
| Sensitivity | astrocytoma | 0 | 33.3 | Gln | 0.0 | 0.0 | 33.3 | 66.7 |
| Specificity | (subgroup) 3 samples | 100 | 93.0 | 100.0 | 100.0 | 93.0 | 95.3 | |
| Accuracy | Metastasis from | 87.0 | 89.1 | 6 features: | 91.3 | 89.1 | 87.0 | 89.1 |
| Sensitivity | adenocarcinoma | 0 | 80.0 | Gln, PUFA, GABA, | 40.0 | 0.0 | 20.0 | 60.0 |
| Specificity | (subgroup) 5 samples | 97.6 | 90.2 | Etn, Cho, Glu | 97.6 | 100.0 | 95.1 | 92.7 |
| Accuracy | Metastasis from | 91.3 | 82.6 | 1 feature: | 95.7 | 93.5 | 80.4 | 84.8 |
| Sensitivity | breast cancer | 0 | 25.0 | Asp | 50.0 | 25.0 | 25.0 | 0.0 |
| Specificity | (subgroup) 4 samples | 100 | 88.1 | 100.0 | 100.0 | 85.7 | 92.9 | |
Boldface denotes the maximum value for the respective metric across all classification methods for a particular group (control, tumor type, tumor subtype).
Figure 2.
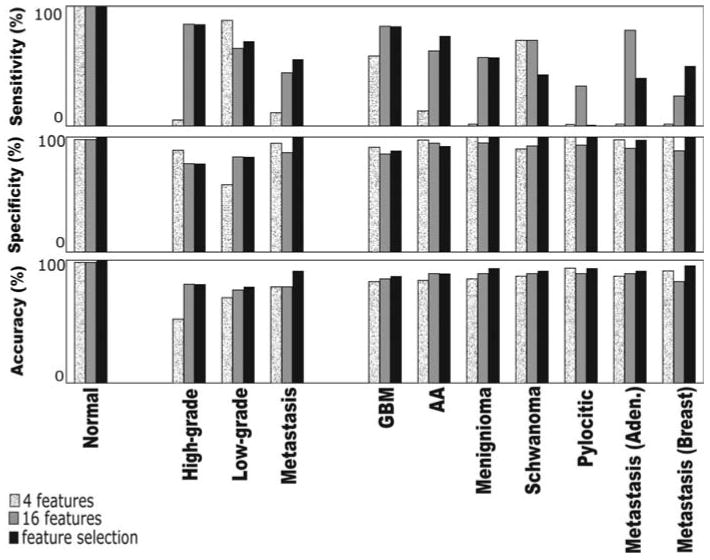
Performance of the linear SVM classifier. The performance of the linear SVM classifier was evaluated by three metrics (sensitivity, specificity, and accuracy), for each of the three feature selection schemes (4 features, feature selection, all features), and for tumor type (normal, high-grade, low-grade) and subtype [GBM, AA, meningioma, schwanoma, pylocytic, metastasis (aden.) and metastasis (breast cancer)]. Sensitivity was primarily increased with the combined SVM+MRMR approach (black bars) in high-grade biopsies, specificity was increased in low-grade biopsies, and both sensitivity and specificity were affected in metastases, resulting in higher accuracy.
In sub-typing the biopsies, the SVM showed a higher specificity for GBMs with only 4 features, a higher sensitivity with all 16 features, and a slightly increased accuracy when all 16 features were used. Similar results were observed for AAs and meningiomas. In schwanomas, the metrics did not differ whether 4 or 16 features were used, and in pilocytics, only the sensitivity seemed to increase when more features were used. Similarly, for sub-typing metastases, more features increased the sensitivity but did not necessarily improve the other metrics.
When feature selection was applied first using the MRMR method, most metrics increased, with some more than others (Table I). In particular, certain metrics reached 100%. For instance, the accuracy, specificity, and sensitivity in typing controls, and the specificity in typing metastases and in subtyping meningiomas, schwanomas, pilocytics, and breast cancer metastases reached 100%.
When an SVM-LS with a radial basis function kernel (RBF Kernel) was used, the SVM-LS slightly over-performed the linear SVM in some cases, but not consistently (Table I). For example, the accuracy in the high-grade group was higher using the LS-SVM than using the linear SVM with 4 features, but was less accurate than the linear SVM with 16 features or with MRMR feature selection. The sensitivity, as a rule, was considerably worse (reduced by 50% or more).
When we used NN MLP instead of the linear SVM (Table I), we received better results in a few cases. For instance, the accuracy in typing low-grade brain tumor biopsies was increased, as were the sensitivity and specificity of subtyping pilocytic astriocytomas (although the same accuracy was obtained). Fig. 2 summarizes the performance of the linear SVM classifier evaluated by the three metrics (sensitivity, specificity, and accuracy), for each of the three feature selection schemes (4 features, feature selection, all features), and for tumor types (normal, high-grade, low-grade) and subtypes [GBM, AA, meningioma, schwanoma, pylocytic, metastasis (aden.) and metastasis (breast cancer)].
All 16 metabolites were assigned by comparison to literature data (49-51) and Tugnoli et al (14) using 2D TOBSY spectra. Representative TOBSY MR spectra are shown in Fig. 3. Both saturated Lip and polyunsaturated fatty acids (PUFAs), proposed to be due to apoptosis, are prominent in high-grade tumors (i.e, GBM) and metastases. Also note that Asp is detected in metastases from breast cancer, while GPC has not been detected in metastases. Finally, PC levels seem to be highest in metastases. The rest of the metabolites are detected at various levels in all tumors.
Figure 3.
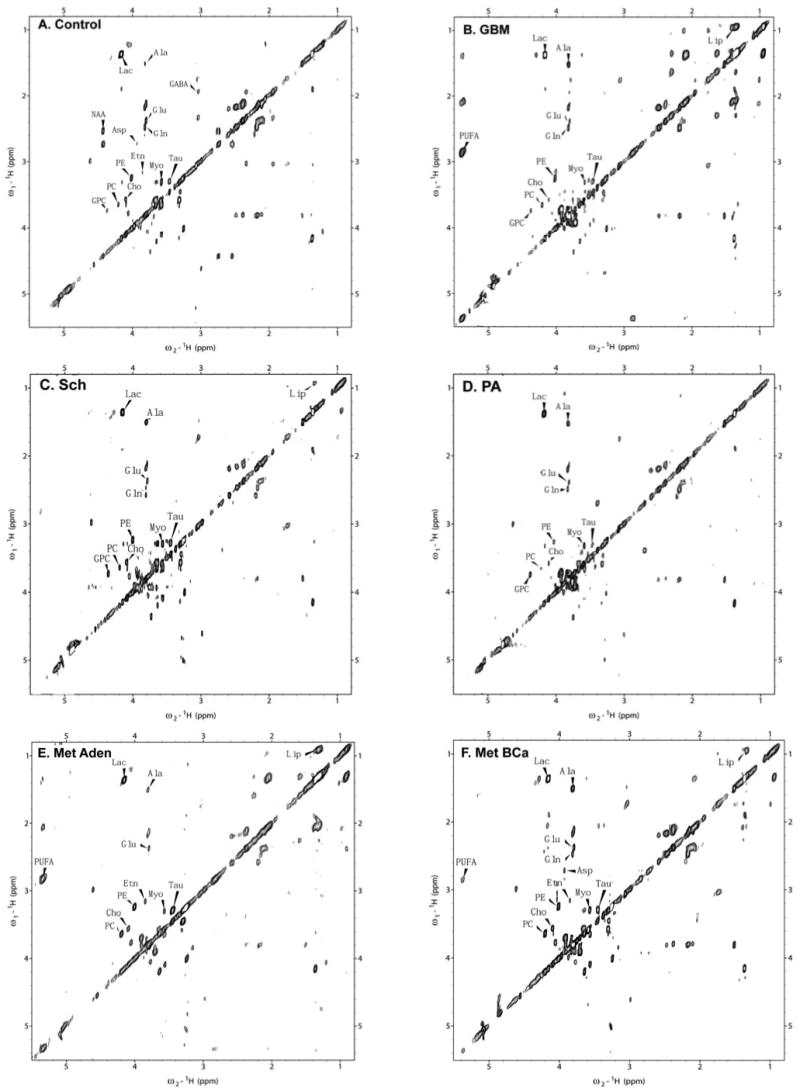
Total through-bond spectroscopy (TOBSY) using ex vivo HRMAS MRS on control and tumor tissue biopsies. HRMAS 1H MR spectra using TOBSY, with 45 msec mixing time, 3 kHz MAS speed, and -8°C at 600 MHz. A, Control; B, Glioblastoma multiforme (GBM); C, Schwannoma (Sch); D, Pilocytic astrocytoma (PA); E, Metastasis from adenocarcinoma (Met Aden); F, Metastasis from breast cancer (Met BCa). (Ala, alanine; Cho, choline; GABA, γ-amino-butyric acid; Gln, glutamine; Glu, glutamate; GPC, glycerophosphocholine; Lip, lipids; Myo, myoinositol; PC, phosphocholine; PE, phosphoethanolamine; PUFA, polyunsaturated fatty acids; Tau, taurine). Note that Cho, PC, GPC, PE, Etn, are clearly separable here due to the use of the 2D TOBSY method.
Discussion
Herein, we demonstrated for the first time that the novel 2D HRMAS 1H MRS method, which employs the adiabatic, solid-state NMR technique TOBSY, can be used in small intact samples to reliably detect at least 16 metabolites or biologically-relevant molecular species. We further showed that this can be achieved on a relatively short time scale of 45 min, which is shorter than has previously been discussed by others (52). This method was performed in combination with a robust artificial intelligence system to characterize brain tumor biopsies with high sensitivity, specificity, and accuracy. One principal finding of our study is that of the two classifiers used, a linear SVM and a feed-forward neural network, the linear SVM performs better. Also, feature selection using the MRMR method results in more robust selection of the reduced feature space, and provides a data-driven criterion for generalizing the feature selection as the training set changes (e.g., increases in size as more data are added). This leads to markedly improved classification performance, which is comparable to that obtained when using the full-feature space (Fig. 2).
Because we used a 2D MRS method, we were able to produce excellent quality data with little overlap (Fig. 3). This might be one reason why our accuracy is better than that reported by Poullet et al (33) who used a 1D HRMAS technique and found that SVM-LS performed better than linear SVM. This implies that the use of SNM-LS may further improve our accuracy, although we did find that the SVM-LS with the RBF Kernel performed worse than our linear SVM (Table I). Central nervous system tumor biopsies are often highly size restricted, which can make their analysis technically challenging, in particular for producing accurate MRS data. Our more sensitive TOBSY NMR approach (39) is optimal for identifying biomarkers that can facilitate the typing of tumor biopsies of only 2 mg in size. While we focused on brain tumors, the MRS of small biospecimens is equally relevant to other medically important tissues that are size limited, such as other clinical biopsies or stem cell populations.
The findings here enhance our knowledge from prior studies (9,11,12,24,26-33,53), since they suggest that the inclusion of more metabolites increases the performance of a given classification system, especially its sensitivity, resulting in increased accuracy. The use of in vivo MRS at higher field strengths, with 2D methods that allow the detection at least the same 16 metabolites that we reliably quantified here, should enable clinicians to characterize and diagnose even inoperable brain tumors with high accuracy. This should allow appropriate therapy selection and the non-invasive monitoring of such therapies in vivo, thereby avoiding serial biopsies. Indeed, using molecular information to guide brain tumor therapy has been previously suggested (54).
This study also demonstrates the feasibility of a novel and sensitive method for the molecular or metabolic assessment of brain tumor biopsies. Since this assessment is performed at a low temperature (-8°C), the tissue integrity is maintained, allowing clinicians to run the subsequent histopathological, genomic, and/or proteomic analyses needed to construct molecular cancer signatures (55-62). The biomarkers revealed by this method can be combined with those found by the aforementioned technologies and by clinical and demographic data (i.e., tumor location, MRI degree of contrast enhancement, MR perfusion and diffusion, age of the patient, treatment response, and survival), enabling the unique fingerprinting of brain tumors (54). Indeed, the impact of tumor location on the brain tumor classification by MRS has been recently reported (32).
In addition to demonstrating the capability of HRMAS 1H MRS to classify brain tumors, we also demonstrate here that certain molecules, or metabolites, may be putative biomarkers of importance in certain tumors. For instance, upon visual inspection of the TOBSY spectra, polyunsaturated fatty acids, PUFAs, a biomarker of apoptosis (63), Lip, proposed to predict clinical grade of brain tumors (53), accumulate in malignant tumors (i.e., GBMs and metastases) (Fig. 3). Despite being found in schwannomas, Lip were not detected in pilocytic astrocytomas, both of which are low-grade tumors (Table I and Fig. 3). Gln and Asp were the most significant features of pilocytic astrocytomas (an often difficult to diagnose tumor) and metastases, respectively. Also, the significant features for GBM and anaplastic astrocytomas were different - a distinction that is often difficult to make clinically (Table I). Others have claimed that glycine (not shown here) may be an important biomarker for glioma characterization (64,65), and taurine has been suggested as a putative biomarker for medulloblastomas and PNET (12,14,66-68). Although we cannot address this here, we did note that taurine was present in both high-grade and low-grade tumors, such as meningiomas; this is a finding that is in agreement with Tugnoli et al who analyzed in detail different types of meningiomas (15). As anticipated, the PC/GPC ratio, which corresponds to oncogenic transformation, was found to be highest in metastasis (9). In order to evaluate the biological significance of metabolites, we must first correlate metabolites with gene expression in the tumors (16). Once done, certain genes may be implicated and particular networks identified that will further assist the clinical guiding of brain tumor therapy and will further reveal molecular targets for novel therapies (54,69).
Finally, the current study has certain limitations. Specifically, the number of metabolites can be increased by further analysis of the 2D TOBSY spectra and we can train our classifiers with a greater number of samples. This will enhance the sensitivity and specificity, thereby allowing more accurate classification. On the other hand, we must again stress that our classification scheme was almost acceptable, even with only 4 metabolites being used. Based on this, we propose that the value of current clinical in vivo MRS, although revealing information of only a few metabolites, may have additional value when combined with conventional, perfusion, and diffusion MRIs, clinical and demographic data, and a powerful classification strategy. Although we are not aware of such on-going studies, certain prior reports also recommend this approach (32,70-73).
We conclude that the use of a sensitive multidimensional MRS technique, at higher magnetic fields and with a robust classification approach, should improve the characterization, typing, and prognostication of brain tumors in vivo. This approach should assist in stratifying patients for appropriate therapeutic protocols and in the monitoring of new therapies.
Acknowledgments
The study was supported in part by discretional funds to A. Aria Tzika. The authors thank the Departments of Surgery and Radiology at Massachusetts General Hospital, as well as Neurosurgery and Pathology at Brigham and Women's Hospital Boston for supporting this study. We also thank Drs Ann Power Smith and Kathryn Edmondson of Write Science Right for editorial assistance.
References
- 1.Legler JM, Gloeckler Ries LA, Smith MA, et al. RESPONSE: re: brain and other central nervous system cancers: recent trends in incidence and mortality. J Natl Cancer Inst. 2000;92:77–78. doi: 10.1093/jnci/92.1.77a. [DOI] [PubMed] [Google Scholar]
- 2.Eichler AF, Loeffler JS. Multidisciplinary management of brain metastases. Oncologist. 2007;12:884–898. doi: 10.1634/theoncologist.12-7-884. [DOI] [PubMed] [Google Scholar]
- 3.Jain RK, Di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 4.Jain RK. Taming vessels to treat cancer. Sci Am. 2008;298:56–63. doi: 10.1038/scientificamerican0108-56. [DOI] [PubMed] [Google Scholar]
- 5.Packer RJ, Gajjar A, Vezina G, et al. Phase III study of cranio-spinal radiation therapy followed by adjuvant chemotherapy for newly diagnosed average-risk medulloblastoma. J Clin Oncol. 2006;24:4202–4208. doi: 10.1200/JCO.2006.06.4980. [DOI] [PubMed] [Google Scholar]
- 6.Merchant TE, Zhu Y, Thompson SJ, Sontag MR, Heideman RL, Kun LE. Preliminary results from a Phase II trail of conformal radiation therapy for pediatric patients with localised low-grade astrocytoma and ependymoma. Int J Radiat Oncol Biol Phys. 2002;52:325–332. doi: 10.1016/s0360-3016(01)01807-7. [DOI] [PubMed] [Google Scholar]
- 7.Kleihues P, WK C. World Health Organization. IARC Press; Lyon: 2000. Classification of tumors. Pathology and genetics; p. 314. [Google Scholar]
- 8.Louis DN, Holland EC, Cairncross JG. Glioma classification: a molecular reappraisal. Am J Pathol. 2001;159:779–786. doi: 10.1016/S0002-9440(10)61750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzika AA. Proton magnetic resonance spectroscopic imaging as a cancer biomarker for pediatric brain tumors. Int J Oncol. 2008;32:517–526. doi: 10.3892/ijo_32_3_517. Review. [DOI] [PubMed] [Google Scholar]
- 10.Cheng LL, Ma MJ, Becerra L, et al. Quantitative neuropathology by high resolution magic angle spinning proton magnetic resonance spectroscopy. Proc Natl Acad Sci USA. 1997;94:6408–6413. doi: 10.1073/pnas.94.12.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng LL, Anthony DC, Comite AR, Black PM, Tzika AA, Gonzalez RG. Quantification of microheterogeneity in glioblastoma multiforme with ex vivo high-resolution magic-angle spinning (HRMAS) proton magetic resonance spectroscopy. Neuro-oncology. 2000;2:87–95. doi: 10.1093/neuonc/2.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzika AA, Cheng LL, Goumnerova L, et al. Biochemical characterization of pediatric brain tumors by using in vivo and ex vivo magnetic resonance spectroscopy. J Neurosurg. 2002;96:1023–1031. doi: 10.3171/jns.2002.96.6.1023. [DOI] [PubMed] [Google Scholar]
- 13.Morvan D, Demidem A, Papon J, De Latour M, Madelmont JC. Melanoma tumors acquire a new phospholipid metabolism phenotype under cystemustine as revealed by high-resolution magic angle spinning proton nuclear magnetic resonance spectroscopy of intact tumor samples. Cancer Res. 2002;62:1890–1897. [PubMed] [Google Scholar]
- 14.Tugnoli V, Schenetti L, Mucci A, et al. A comparison between in vivo and ex vivo HR-MAS 1H MR spectra of a pediatric posterior fossa lesion. Int J Mol Med. 2005;16:301–307. doi: 10.3892/ijmm.16.2.301. [DOI] [PubMed] [Google Scholar]
- 15.Tugnoli V, Schenetti L, Mucci A, et al. Ex vivo HR-MAS MRS of human meningiomas: a comparison with in vivo1H MR spectra. Int J Mol Med. 2006;18:859–869. [PubMed] [Google Scholar]
- 16.Tzika AA, Astrakas L, Cao HH, et al. Combination of high-resolution magic angle spinning proton magnetic resonance spectroscopy and microscale genomics to type brain tumor biopsies. Int J Mol Med. 2007;20:199–208. [PubMed] [Google Scholar]
- 17.Ryner LN, Sorenson JA, Thomas MA. 3D localized 2D NMR spectroscopy on an MRI scanner. J Magn Reson B. 1995;107:126–137. doi: 10.1006/jmrb.1995.1068. [DOI] [PubMed] [Google Scholar]
- 18.Thomas MA, Ryner LN, Mehta MP, Turski PA, Sorenson JA. Localized 2D J-resolved 1H MR spectroscopy of human brain tumors in vivo. J Magn Reson Imaging. 1996;6:453–459. doi: 10.1002/jmri.1880060307. [DOI] [PubMed] [Google Scholar]
- 19.Binesh N, Yue K, Fairbanks L, Thomas MA. Reproducibility of localized 2D correlated MR spectroscopy. Magn Reson Med. 2002;48:942–948. doi: 10.1002/mrm.10307. [DOI] [PubMed] [Google Scholar]
- 20.Thomas MA, Wyckoff N, Yue K, et al. Two-dimensional MR spectroscopic characterization of breast cancer in vivo. Technol Cancer Res Treat. 2005;4:99–106. doi: 10.1177/153303460500400113. [DOI] [PubMed] [Google Scholar]
- 21.Hagberg G, Burlina AP, Mader I, Roser W, Radue EW, Seelig J. In vivo proton MR spectroscopy of human gliomas: definition of metabolic coordinates for multi-dimensional classification. Magn Reson Med. 1995;34:242–252. doi: 10.1002/mrm.1910340216. [DOI] [PubMed] [Google Scholar]
- 22.Lisboa PJG, Kirby SPJ, Vellido A, Lee YYB, El-Deredy W. Assessment of statistical and neural networks methods in NMR spectral classification and metabolite selection. NMR Biomed. 1998;11:225–234. doi: 10.1002/(sici)1099-1492(199806/08)11:4/5<225::aid-nbm509>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 23.Hagberg G. From magnetic resonance spectroscopy to classification of tumors: a review of pattern recognition methods. NMR Biomed. 1998;11:148–156. doi: 10.1002/(sici)1099-1492(199806/08)11:4/5<148::aid-nbm511>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 24.Poptani H, Kaartinen J, Gupta RK, Niemitz M, Hiltunen Y, Kauppinen RA. Diagnostic assessment of brain tumours and non-neoplastic brain disorders in vivo using proton nuclear magnetic resonance spectroscopy and artificial neural networks. J Cancer Res Clin Oncol. 1999;125:343–349. doi: 10.1007/s004320050284. [DOI] [PubMed] [Google Scholar]
- 25.Tate AR, Foxall PJ, Holmes E, et al. Distinction between normal and renal cell carcinoma kidney cortical biopsy samples using pattern recognition of (1)H magic angle spinning (MAS) NMR spectra. NMR Biomed. 2000;13:64–71. doi: 10.1002/(sici)1099-1492(200004)13:2<64::aid-nbm612>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 26.Majos C, Alonso J, Aguilera C, et al. Adult primitive neuroectodermal tumor: proton MR spectroscopic findings with possible application for differential diagnosis. Radiology. 2002;225:556–566. doi: 10.1148/radiol.2252011592. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Lisboa PJG, El-Deredy W. Tumour grading from magnetic resonance spectroscopy: a comparison of feature extraction with variable selection. Stat Med. 2003;22:147–164. doi: 10.1002/sim.1321. [DOI] [PubMed] [Google Scholar]
- 28.Tate AR, Majos C, Moreno A, Howe FA, Griffiths JR, Arus C. Automated classification of short echo time in in vivo 1H brain tumor spectra: a multicenter study. Magn Reson Med. 2003;49:29–36. doi: 10.1002/mrm.10315. [DOI] [PubMed] [Google Scholar]
- 29.Devos A, Lukas L, Suykens JA, et al. Classification of brain tumours using short echo time 1H MR spectra. J Magn Reson. 2004;170:164–175. doi: 10.1016/j.jmr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Tate AR, Underwood J, Acosta DM, et al. Development of a decision support system for diagnosis and grading of brain tumours using in vivo magnetic resonance single voxel spectra. NMR Biomed. 2006;19:411–434. doi: 10.1002/nbm.1016. [DOI] [PubMed] [Google Scholar]
- 31.Opstad KS, Ladroue C, Bell BA, Griffiths JR, Howe FA. Linear discriminant analysis of brain tumour (1)H MR spectra: a comparison of classification using whole spectra versus metabolite quantification. NMR Biomed. 2007;20:763–770. doi: 10.1002/nbm.1147. [DOI] [PubMed] [Google Scholar]
- 32.Reynolds GM, Peet AC, Arvanitis TN. Generating prior probabilities for classifiers of brain tumours using belief networks. BMC Med Inform Decis Making. 2007;7:27. doi: 10.1186/1472-6947-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poullet JB, Martinez-Bisbal MC, Valverde D, et al. Quantification and classification of high-resolution magic angle spinning data for brain tumor diagnosis. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:5407–5410. doi: 10.1109/IEMBS.2007.4353565. [DOI] [PubMed] [Google Scholar]
- 34.Glotsos D, Spyridonos P, Cavouras D, Ravazoula P, Dadioti PA, Nikiforidis G. An image-analysis system based on support vector machines for automatic grade diagnosis of brain-tumour astrocytomas in clinical routine. Med Inform Internet Med. 2005;30:179–193. doi: 10.1080/14639230500077444. [DOI] [PubMed] [Google Scholar]
- 35.Glotsos D, Tohka J, Ravazoula P, Cavouras D, Nikiforidis G. Automated diagnosis of brain tumours astrocytomas using probabilistic neural network clustering and support vector machines. Int J Neural Syst. 2005;15:1–11. doi: 10.1142/S0129065705000013. [DOI] [PubMed] [Google Scholar]
- 36.Georgiadis P, Cavouras D, Kalatzis I, et al. Improving brain tumor characterization on MRI by probabilistic neural networks and non-linear transformation of textural features. Comput Methods Programs Biomed. 2008;89:24–32. doi: 10.1016/j.cmpb.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Lakhani SR, Ashworth A. Microarray and histopathological analysis of tumours: the future and the past? Nat Rev Cancer. 2001;1:151–157. doi: 10.1038/35101087. [DOI] [PubMed] [Google Scholar]
- 38.Julia-Sape M, Acosta D, Majos C, et al. Comparison between neuroimaging classifications and histopathological diagnoses using an international multicenter brain tumor magnetic resonance imaging database. J Neurosurg. 2006;105:6–14. doi: 10.3171/jns.2006.105.1.6. [DOI] [PubMed] [Google Scholar]
- 39.Andronesi OC, Mintzopoulos D, Struppe J, Black PM, Tzika AA. Solid-state NMR adiabatic TOBSY sequences provide enhanced sensitivity for multidimensional high-resolution magic-angle-spinning (1)H MR spectroscopy. J Magn Reson. doi: 10.1016/j.jmr.2008.05.017. In press. [DOI] [PubMed] [Google Scholar]
- 40.Levitt MH. Advances in NMR. In: Harris RK, G DM, editors. Encyclopedia of Nuclear Magnetic Resonance. John Wiley & Sons, Ltd.; Chichester: 2002. pp. 165–196. [Google Scholar]
- 41.Baldus M, Iuliucci RJ, Meier BH. Probing through-bond connectivities and through-space distances in solids by magic-angle-spinning nuclear magnetic resonance. J Am Chem Soc. 1997;119:1121–1124. [Google Scholar]
- 42.Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 43.Kupce E, Freeman R. Adiabatic pulses for wide-band inversion and broad-band decoupling. J Magn Reson A. 1995;115:273–276. [Google Scholar]
- 44.Vapnik V. Statistical Learning Theory: Wiley-Interscience. 1998. [Google Scholar]
- 45.Hagan MT, Menhaj MB. Training feedforward networks with the Marquardt algorithm. IEEE Trans Neural Netw. 1994;5:989–993. doi: 10.1109/72.329697. [DOI] [PubMed] [Google Scholar]
- 46.Suykens J, van Gestel T, De Brabanter J, De Moor B, Vandewalle J. Least squares support vector machines. KU Leuven; Belgium: 2002. [DOI] [PubMed] [Google Scholar]
- 47.Krogh A. What are artificial neural networks? Nat Biotechnol. 2008;26:195–197. doi: 10.1038/nbt1386. [DOI] [PubMed] [Google Scholar]
- 48.Ding C, Peng H. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol. 2005;3:185–205. doi: 10.1142/s0219720005001004. [DOI] [PubMed] [Google Scholar]
- 49.Remy C, Arus C, Ziegler A, et al. In vivo, ex vivo, and in vivo one- and two-dimensional nuclear magnetic resonance spectroscopy of an intracerebral glioma in rat brain: assignment of resonance. J Neurochem. 1994;62:166–179. doi: 10.1046/j.1471-4159.1994.62010166.x. [DOI] [PubMed] [Google Scholar]
- 50.Florian C, Preece N, Bhakoo K, Williams S. Characteristic metabolic profiles revealed by 1H NMR spectroscopy for three types of human brain and nervous system tumours. NMR Biomed. 1996;8:253–264. doi: 10.1002/nbm.1940080605. [DOI] [PubMed] [Google Scholar]
- 51.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 52.Cheng LL, Chang IW, Louis DN, Gonzalez RG. Correlation of high-resolution magic angle spinning proton magnetic resonance spectroscopy with histopathology of intact human brain tumor specimens. Cancer Res. 1998;58:1825–1832. [PubMed] [Google Scholar]
- 53.Astrakas LG, Zurakowski D, Tzika AA, et al. Noninvasive magnetic resonance spectroscopic imaging biomarkers to predict the clinical grade of pediatric brain tumors. Clin Cancer Res. 2004;10:8220–8228. doi: 10.1158/1078-0432.CCR-04-0603. [DOI] [PubMed] [Google Scholar]
- 54.Mischel PS, Cloughesy T. Using molecular information to guide brain tumor therapy. Nat Clin Pract Neurol. 2006;2:232–233. doi: 10.1038/ncpneuro0145. [DOI] [PubMed] [Google Scholar]
- 55.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 56.Ramaswamy S, Tamayo P, Rifkin R, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeang C, Ramaswamy S, Tamayo P, et al. Molecular classification of multiple tumor types. Bioinformatics. 2001;17:S316–S322. doi: 10.1093/bioinformatics/17.suppl_1.s316. [DOI] [PubMed] [Google Scholar]
- 58.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 59.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551–561. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 60.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz SA, Weil RJ, Johnson MD, Toms SA, Caprioli RM. Protein profiling in brain tumors using mass spectrometry: feasibility of a new technique for the analysis of protein expression. Clin Cancer Res. 2004;10:981–987. doi: 10.1158/1078-0432.ccr-0927-3. [DOI] [PubMed] [Google Scholar]
- 62.Kostka D, Spang R. Microarray based diagnosis profits from better documentation of gene expression signatures. PLoS Comput Biol. 2008;4:e22. doi: 10.1371/journal.pcbi.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hakumaki JM, Poptani H, Sandmair AM, Yla-Herttuala S, Kauppinen RA. 1H MRS detects polyunsaturated fatty acid accumulation during gene therapy of glioma: implications for the in vivo detection of apoptosis. Nat Med. 1999;5:1323–1327. doi: 10.1038/15279. [DOI] [PubMed] [Google Scholar]
- 64.Kinoshita Y, Kajiwara H, Yokota A, Koga Y. Proton magnetic resonance spectroscopy of astrocytic tumors: an in vitro study. Neurol Med Chir (Tokyo) 1993;33:350–359. doi: 10.2176/nmc.33.350. [DOI] [PubMed] [Google Scholar]
- 65.Kinoshita Y, Kajiwara H, Yokota A, Roga Y. Proton magnetic resonance spectroscopy of brain tumors: an in vitro study. Neurosurgery. 1994;35:606–613. doi: 10.1227/00006123-199410000-00005. [DOI] [PubMed] [Google Scholar]
- 66.Moreno-Torres A, Martinez-Perez I, Baquero M, et al. Taurine detection by proton magnetic resonance spectroscopy in medulloblastoma: contribution to noninvasive differential diagnosis with cerebellar astrocytoma. Neurosurgery. 2004;55:824–829. doi: 10.1227/01.neu.0000137655.53303.c8. [DOI] [PubMed] [Google Scholar]
- 67.Kinoshita Y, Yokota A. Absolute concentrations of metabolites in human brain tumors using in vitro proton magnetic resonance spectroscopy. NMR Biomed. 1997;10:2–12. doi: 10.1002/(sici)1099-1492(199701)10:1<2::aid-nbm442>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 68.Kovanlikaya A, Panigrahy A, Krieger MD, et al. Untreated pediatric primitive neuroectodermal tumor in vivo: quantitation of taurine with MR spectroscopy. Radiology. 2005;236:1020–1025. doi: 10.1148/radiol.2363040856. [DOI] [PubMed] [Google Scholar]
- 69.Chakravarti A, Tyndall E, Palanichamy K, Mehta M, Aldape K, Loeffler J. Impact of molecular profiling on clinical trial design for glioblastoma. Curr Oncol Rep. 2007;9:71–79. doi: 10.1007/BF02951429. [DOI] [PubMed] [Google Scholar]
- 70.Tzika AA, Zarifi MK, Goumnerova L, et al. Neuroimaging in pediatric brain tumors: Gd-DTPA-enhanced, hemodynamic, and diffusion MR imaging compared with MR spectroscopic imaging. AJNR Am J Neuroradiol. 2002;23:322–333. [PMC free article] [PubMed] [Google Scholar]
- 71.Tzika AA, Astrakas LG, Zarifi MK, et al. Multiparametric MR assessment of pediatric brain tumors. Neuroradiology. 2003;45:1–10. doi: 10.1007/s00234-002-0865-0. [DOI] [PubMed] [Google Scholar]
- 72.Tzika AA, Astrakas LG, Zarifi MK, et al. Spectroscopic and perfusion magnetic resonance imaging predictors of progression in pediatric brain tumors. Cancer. 2004;100:1246–1256. doi: 10.1002/cncr.20096. [DOI] [PubMed] [Google Scholar]
- 73.Lee MC, Nelson SJ. Supervised pattern recognition for the prediction of contrast-enhancement appearance in brain tumors from multivariate magnetic resonance imaging and spectroscopy. Artif Intell Med. 2008;43:61–74. doi: 10.1016/j.artmed.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


