Abstract
Background
Aldehydes are highly reactive molecules. While several non-P450 enzyme systems participate in their metabolism, one of the most important is the aldehyde dehydrogenase (ALDH) superfamily, composed of NAD(P)+-dependent enzymes that catalyze aldehyde oxidation.
Objective
This article presents a review of what is currently known about each member of the human ALDH superfamily including the pathophysiological significance of these enzymes.
Methods
Relevant literature involving all members of the human ALDH family was extensively reviewed, with the primary focus on recent and novel findings.
Conclusion
To date, 19 ALDH genes have been identified in the human genome and mutations in these genes and subsequent inborn errors in aldehyde metabolism are the molecular basis of several diseases, including Sjögren-Larsson syndrome, type II hyperprolinemia, γ-hydroxybutyric aciduria and pyridoxine-dependent seizures. ALDH enzymes also play important roles in embryogenesis and development, neurotransmission, oxidative stress and cancer. Finally, ALDH enzymes display multiple catalytic and non-catalytic functions including ester hydrolysis, antioxidant properties, xenobiotic bioactivation and UV light absorption.
Keywords: aldehyde dehydrogenase, aldehyde metabolism, ALDH
1. Introduction
Aldehydes are generated from a wide variety of endogenous and exogenous precursors during numerous physiological processes, including the biotransformation of endogenous compounds such as amino acids, neurotransmitters, carbohydrates, and lipids [1–3]. More than 200 aldehyde species arise from the oxidative degradation of cellular membrane lipids, also known as lipid peroxidation (LPO), including 4-hydroxy-2-nonenal (4-HNE) and malondialdehyde (MDA) [4]. Amino acid catabolism generates several aldehyde intermediates, including glutamate γ-semialdehyde, while neurotransmitters, such as gamma-aminobutyric acid (GABA), serotonin, noradrenaline, adrenaline, and dopamine, also give rise to aldehyde metabolites [2,5]. Xenobiotics and drugs – including ethanol, which generates acetaldehyde, and the anticancer drugs cyclophosphamide (CP) and ifosfamide, which generate acrolein – are important aldehyde precursors. Various aldehydes, including formaldehyde, acetaldehyde and acrolein, are also ubiquitous in the environment and are present in smog, cigarette smoke and motor vehicle exhaust. Aldehydes are also used or generated in a wide variety of industrial applications including in the production of resins, polyurethane and polyester plastics. In addition, numerous dietary aldehydes, including citral and benzaldehyde, naturally exist or are approved additives in various foods where they impart flavor and odor.
While some aldehydes play vital roles in normal physiological processes, including vision, embryonic development, and neurotransmission, many are cytotoxic and carcinogenic [6]. Aldehydes are strong electrophilic compounds with terminal carbonyl groups, making them highly reactive, and α,β-unsaturated aldehydes, such as 4-HNE and acrolein, also contain a second electrophile at the β-carbon. Unlike free radicals, aldehydes are relatively long-lived and not only react with cellular components in the vicinity of their formation but, through diffusion or transportation, also affect targets some distance away [4]. Aldehydes form adducts, believed to be the primary mechanism underlying their toxicity, with various cellular targets including glutathione (GSH), nucleic acids, and protein amino acids leading to impaired cellular homeostasis, enzyme inactivation, DNA damage, and cell death [7,8].
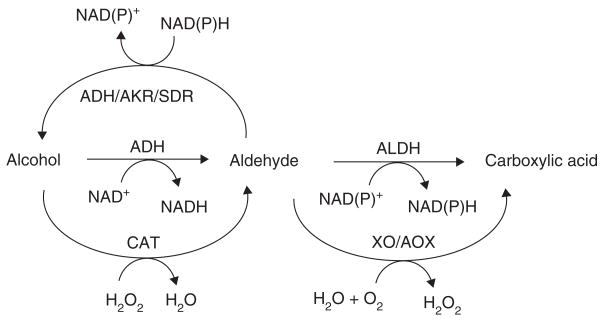
Aldehydes are detoxified primarily through reductive and oxidative Phase I enzyme-catalyzed reactions, including the non-P450 aldehyde reduction enzyme systems alcohol dehydrogenase (ADH), aldo-keto reductase (AKR) and short-chain dehydrogenase/reductase (SDR), and aldehyde oxidation enzyme systems xanthine oxidase (XO), aldehyde oxidase (AOX) and aldehyde dehydrogenase (ALDH) (Figure 1). The ALDH superfamily catalyzes the oxidation of numerous aldehyde substrates and, while other enzymes metabolize aldehydes, these enzymes play a particularly critical role in the cellular protection against these toxic species, as evidenced by the fact that mutations and polymorphisms in ALDH genes (leading to perturbations in aldehyde metabolism) are the molecular basis of several disease states and metabolic anomalies [2]. The present paper comprehensively reviews the 19 human ALDH proteins.
Figure 1. Non-P450 enzymatic metabolism of aldehydes.
ADH: Alcohol dehydrogenase; AKR: Aldo-keto reductase; ALDH: Aldehyde dehydrogenase; AO: Aldehyde oxidase; CAT: Catalase; SDR: Short chain dehydrogenase/reductase; XO: Xanthine oxidase.
2. The ALDH superfamily
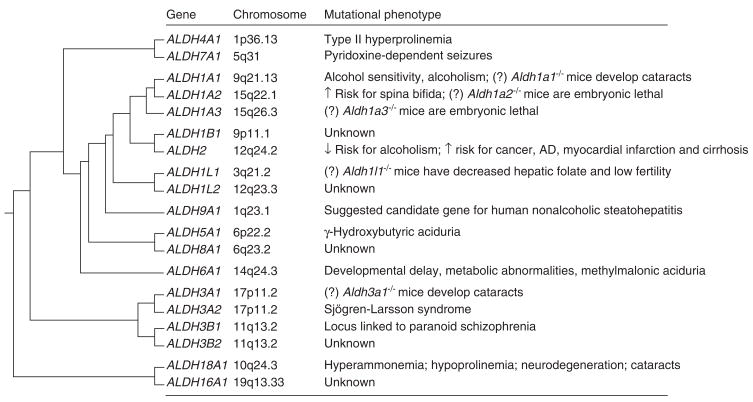
The human ALDH superfamily consists of 19 putatively functional genes with distinct chromosomal locations (Figure 2)[9]. A standardized gene nomenclature system based on divergent evolution and amino acid identity was established for the ALDH superfamily in 1998 [10]. The ALDH enzymes catalyze the NAD(P)+-dependent irreversible oxidation of a wide spectrum of endogenous and exogenous aldehydes (Table 1). ALDH proteins are found in all subcellular regions including cytosol, mitochondria, endoplasmic reticulum and nucleus, with several found in more than one compartment. ALDH isozymes found in organelles other than cytosol possess leader or signal sequences that allow their translocation to specific subcellular regions [11]. After translocation or import, mitochondrial sequences may be removed (resulting in shorter mature proteins), while microsomal and nuclear signals remain intact [12,13]. Most of the ALDHs have a wide tissue distribution and display distinct substrate specificity [2,14]. Generally regarded as detoxification enzymes, ALDHs serve to protect cells from the effects of aldehydes by oxidizing them to their respective carboxylic acids (Figure 1). This is evident from several studies in which an ALDH has been shown to protect against aldehyde-induced cytotoxicity [13]. However, the most compelling evidence relies on the observation that mutations and polymorphisms in ALDH genes (leading to loss of function) are associated with distinct phenotypes in humans and rodents [2,15], including Sjögren-Larsson syndrome (SLS) [16], type II hyperprolinemia [17], γ-hydroxybutyric aciduria [18], pyridoxine-dependent seizures [19], hyperammonemia [20], alcohol-related diseases [21], cancer [6] and late-onset alzheimer’s disease (AD) [22] (Figure 2). In addition to clinical phenotypes associated with mutations in ALDH genes, transgenic knockout mice have suggested a pivotal role of ALDHs in physiological functions and processes, such as embryogenesis and development [23,24].
Figure 2. Evolutionary relationship and mutational phenotypes of the nineteen human ALDH genes.
Clustering dendogram illustrates the evolutionary relationship of the nineteen human ALDH genes from a common ancestral gene ~ 3 billion years ago. Chromosomal location is also described. (?) indicates phenotype demonstrated in animals but not yet described in humans.
Table 1.
Human ALDH proteins.
| ALDH | Subcellular location | Preferred aldehyde substrate | Additional functions and characteristics |
|---|---|---|---|
| ALDH1A1 | Cytosol | Retinal | Ester hydrolysis; binds androgen, cholesterol, thyroid, daunorubicin, and flavopiridol; corneal and lens crystallin; oxidizes DOPAL, acetaldehyde |
| ALDH1A2 | Cytosol | Retinal | High affinity for LPO-derived aldehydes |
| ALDH1A3 | Cytosol | Retinal | High affinity for LPO-derived aldehydes |
| ALDH1B1 | Mitochondria | Acetaldehyde | May protect the cornea from UV-light |
| ALDH1L1 | Cytosol | 10-Formyltetrahydrofolate | Binds acetaminophen |
| ALDH1L2 | Unknown | Unknown | Induced by the anti-inflammatory agent indomethacin |
| ALDH2 | Mitochondria | Acetaldehyde | Ester hydrolysis; nitroglycerin bioactivation, oxidizes LPO-derived aldehydes; binds acetaminophen; oxidizes DOPAL and DOPEGAL |
| ALDH3A1 | Cytosol, nucleus* | Aromatic, aliphatic aldehydes | Ester hydrolysis; scavenges ROS; UV-filter; corneal crystallin; oxidizes LPO-derived aldehydes; regulation of cell-cycle; inducted by PAHs |
| ALDH3A2 | Microsomes, peroxisomes ‡ | Fatty aldehydes | Insulin regulates gene expression |
| ALDH3B1 | Cytosol § | Unknown | Oxidizes LPO-derived aldehydes |
| ALDH3B2 | Unknown | Unknown | Unknown |
| ALDH4A1 | Mitochondria | Glutamate γ-semialdehyde | Ester hydrolysis; may mitigate oxidative stress |
| ALDH5A1 | Mitochondria | Succinate semialdehyde | May be involved in neurotransmission efficiency |
| ALDH6A1 | Mitochondria | Malonate semialdehyde | Esterase activity; only known human CoA-dependent ALDH |
| ALDH7A1 | Cytosol, nucleus, mitochondria¶ | α-Aminoadipic semialdehyde | Closely related to plant osmoregulatory protein; may regulate cell cycle |
| ALDH8A1 | Cytosol | Retinal | Oxidizes LPO-derived aldehydes and acetaldehyde |
| ALDH9A1 | Cytosol | γ-Aminobutyraldehyde | Oxidizes betaine, acetaldehyde and DOPAL; involved in carnitine biosysnthesis; esterase activity |
| ALDH16A1 | Unknown | Unknown | Unknown |
| ALDH18A1 | Mitochondria | Glutamic γ-semialdehyde | Unknown |
While predominantly cytosolic, ALDH3A1 is also found in the nucleus.
A variant of ALDH3A2, FALDHv, is believed to be localized to the peroxisomes.
Marchitti SA, Vasiliou V. unpublished data.
Brocker C, Cantore M, Pappa A, et al. unpublished data.
Aside from their role in aldehyde detoxification, many ALDH enzymes possess multiple additional catalytic and non-catalytic functions (Table 1). Indeed, several ALDHs are known to catalyze ester hydrolysis [25] and act as binding proteins for various endogenous (e.g., androgen, cholesterol and thyroid hormone) and exogenous (e.g., acetaminophen) compounds [2]. Additionally, ALDH enzymes may have important antioxidant roles including the production of NAD(P)H [26,27], the absorption of UV light [28,29] and the scavenging of hydroxyl radicals via cysteine and methionine sulfhydryl groups [30].
ALDH enzymes share a number of highly conserved residues necessary for catalysis and cofactor binding [31–36]. The invariant catalytic cysteine Cys-302 (numbering based on the mature human ALDH2 protein), Glu-268, Gly-299, and Asn-169 are all essential for catalysis. Gly-245 and Gly-250 are essential residues of the ALDH Rossmann fold (GxxxxG) necessary for cofactor binding. In addition, Lys-192, Glu-399, and Phe-401 are believed to be integral for cofactor binding and may facilitate catalysis. Crystal structures of mammalian ALDH enzymes have revealed that each subunit contains three domains, namely an NAD(P)+ cofactor-binding domain, a catalytic domain, and a bridging domain [31,32]. At the interface of these domains lies a funnel passage leading to the catalytic pocket. The upper portion of the funnel, composed of residues from all three domains, is believed to confer the required ALDH specificity toward particular aldehyde substrates. The lower portion of the funnel, made up of highly conserved residues from both the cofactor and catalytic domains, appears to be the catalytic site where hydride transfer from substrate to cofactor takes place.
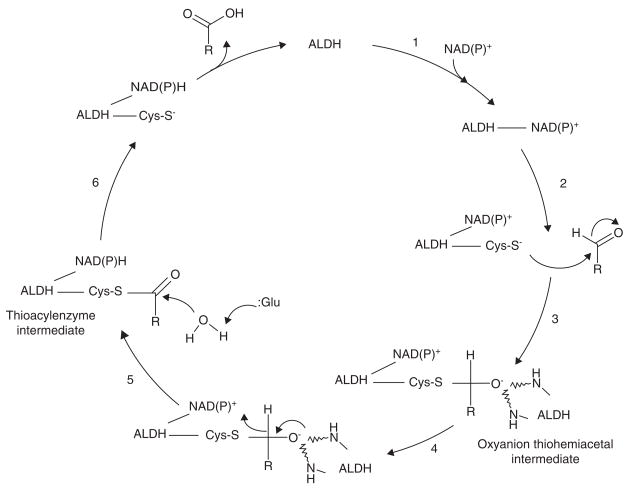
Based on crystallographic structures of ALDH enzymes, a catalytic mechanism has been proposed involving acylation, followed by deacylation (Figure 3)[31,32,37–39]. Briefly, cofactor binding results in a conformational change and activation of the catalytic Cys-302 nucleophile, which is positioned by Gly-299 [32]. Cys-302 then attacks the aldehydic function of the substrate and forms an oxyanion thiohemiacetal intermediate, stabilized in part by Asn-169 [31]. The negatively-charged oxygen of the oxyanion intermediate then facilitates hydride transfer to the cofactor, resulting in the formation of a thioacylenzyme intermediate. Hydrolysis of the thioaceylenzyme and release of carboxylic acid product takes place via Glu-268, which acts as a general base by activating the hydrolytic water after hydride transfer. For most ALDHs, the reduced cofactor is believed to dissociate from the enzyme last. However, one human ALDH, namely ALDH6A1, is CoA-dependent and has a slightly different catalysis mechanism in which the reduced cofactor is released prior to the deacylation step and yields a CoA ester product instead of a free acid [40].
Figure 3. Proposed non-CoA dependent ALDH catalysis mechanism.
1, Cofactor binding results in a conformation change of the enzyme and activation of the catalytic thiol (Cys-S−); 2, Nucleophilic attack of the aldehyde substrate; 3, Oxyanion intermediate stabilized by two NH groups of the ALDH peptide chain; 4, Hydride transfer to cofactor. 5; Glutamate residue acts as a base catalyst in the hydrolysis of the thioacylenzyme intermediate; 6, Release of carboxylic acid product followed by cofactor.
3. ALDH1A1
ALDH1A1 encodes a homotetramer ubiquitously distributed in the adult epithelium of various organs including testis, brain, eye lens, liver, kidney, lung and retina [41,42]. ALDH1A1 is one of three highly conserved cytosolic isozymes (see ALDH1A2 and ALDH1A3) that catalyze the oxidation of the retinol metabolite, retinal (retinaldehyde), to retinoic acid (RA) [43,44]. ALDH1A1 has high affinity for the oxidation of both all-trans-(Km < 0.1 μM) and 9-cis-retinal[45].
RA regulates gene expression by serving as a ligand for nuclear RA receptors (RAR) and retinoid X receptors (RXR). Its synthesis is critical for normal growth, differentiation, development and maintenance of adult epithelia in vertebrate animals [46]. In retinoid-dependent tissues (including the retina), retinal-oxidizing ALDHs have been shown to exhibit differential expression patterns during rodent organogenesis [47–49], indicating that RA signaling is necessary for embryogenesis [50,51]. The in vivo role of ALDH1A1 in RA synthesis is evidenced by the fact that, while Aldh1a1−/− mice are viable and have normal morphology of the retina, the livers of Aldh1a1−/− mice display reduced RA synthesis and increased serum retinal levels after retinol treatment [52,53]. Interestingly, Aldh1a1−/− mice are protected against both diet-induced obesity and insulin resistance, suggesting that retinal may transcriptionally regulate the metabolic response to high-fat diets and that ALDH1A1 may be a candidate gene for therapeutic targeting [54]. In cultured hepatocytes, supression of ALDH1A1 reduces both the omega oxidation of free fatty acids and the production of reactive oxygen species (ROS) [55]. RXRα−/− mice display decreased liver ALDH1A1 levels, suggesting that RA binding is an activating factor in ALDH1A1 gene expression [56]. The androgen receptor may also be involved in regulating levels of ALDH1A1 [57], which is known to be an androgen binding protein [58]. RA is required for testicular development and ALDH1A1 is absent in genital tissues of humans with androgen receptor-negative testicular feminization [57,59].
In the human brain, ALDH1A1 is highly expressed in dopaminergic neurons [60], which are known to require RA for their differentiation and development [61]. In these neurons, ALDH1A1 is under the control of Pitx3, a homeodomain transcription factor [61] that may regulate the specification and maintenance of distinct populations of dopaminergic neurons through ALDH1A1 upregulation [62]. Decreased levels of ALDH1A1 occur in dopaminergic neurons of the substantia nigra in Parkinson’s disease (PD) patients [63] and in those of the ventral tegmental area in schizophrenic patients [60]. In the central nervous system (CNS), monoamine oxidase (MAO) metabolizes dopamine to its aldehyde metabolite, 3,4-dihydroxyphenylacetaldehyde (DOPAL). Increasing evidence suggests that DOPAL may be neurotoxic, and its accumulation may lead to cell death associated with neurological pathologies [5]. ALDH1A1 may play a critical role in maintaining low intraneuronal levels of DOPAL by catalyzing its metabolism to 3,4-dihydroxyphenylacetic acid (DOPAC) [5,60].
ALDH1A1 is one of 139 genes that are differentially expressed in primary human hematopoietic stem cells (HSCs) and, through the production of RA, ALDH1A1 has been shown to promote their differentiation [64,65]. These data suggest that ALDH1A1 inhibition could potentially be used for the therapeutic amplification of HSCs.
ALDH1A1 is one of the major enzymes involved in the metabolism of the ethanol metabolite, acetaldehyde (Km 50 – 180 μM), to which many of the deleterious effects of ethanol are attributed [66]. Indeed, low ALDH1A1 activity may account for alcohol sensitivity in some Caucasian populations [67,68]. Decreased levels of ALDH1A1 are reported in RXRα−/− mice, which are more susceptible to alcoholic liver injury [56], while increased ALDH1A1 expression occurs in brains of DBA/2 mice, a mouse strain exhibiting alcohol avoidance [69]. These data in rodents have led to the suggestion that acetaldehyde accumulation in peripheral organs is aversive, while acetaldehyde produced in the brain may be reinforcing.
ALDH1A1 also plays a key role in the cellular defense against oxidative stress. Human ALDH1A1 efficiently oxidizes LPO-derived aldehydes, including 4-HNE (Km 17.9 μM), hexanal (Km 13.4 μM), and MDA (Km 114.4 μM) (Figure 4)[41,70]. Using various Aldh1a1−/−mouse models, ALDH1A1 has been demonstrated to play a key role in protecting the mouse eye lens and cornea by detoxifying LPO-derived aldehydes and preventing cataract formation induced by oxidative stresses, including ageing and UV radiation [29].
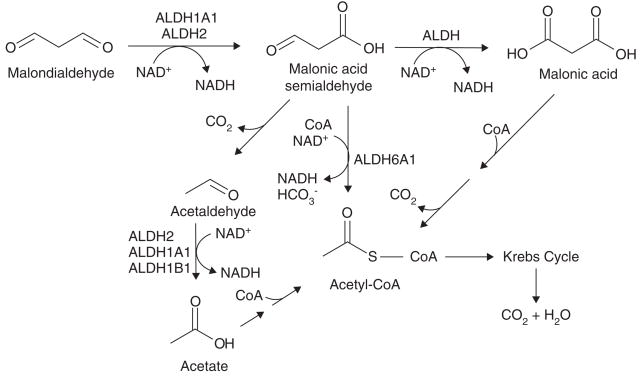
Figure 4. The role of ALDH isozymes in the metabolism of malondialdehyde (MDA).
MDA is the major aldehyde product of lipid peroxidation. The proposed pathway of MDA metabolism involves ALDH1A1, ALDH2 and ALDH6A1.
Similar to other ALDHs, ALDH1A1 may also play an important role in cancer therapeutics (Table 2). ALDH1A1 activity has been reported to decrease the effectiveness of some oxazaphosphorine anticancer drugs, such as CP and ifosfamide, by detoxifying their major active aldehyde metabolites [71]. Indeed, inhibition of ALDH1A1 activity leads to increased toxicity of the major metabolite of CP, 4-hydroperoxycyclophosphamide [72]. Accordingly, patients with low breast tumor ALDH1A1 levels have been reported to respond to CP-based treatment significantly more often than those with high levels, indicating that ALDH1A1 may be a predictor of the drug’s therapeutic effectiveness [73]. Various noncancerous cells, such as hematopoietic progenitor cells, express relatively high ALDH1A1 levels and thus are relatively resistant to oxazaphosphorine-induced toxicity [74]. ALDH1A1 has also been shown to bind to certain anticancer drugs, including daunorubicin [75] and flavopiridol [76], and is downregulated in certain carcinomas [77].
Table 2.
ALDH isozymes putatively involved in cancer.
| ALDH | Cancer phenotype | Ref. |
|---|---|---|
| ALDH1A1 | Increases the resistance of tumor cells to oxazaphosphorine anticancer drugs | [71,72] |
| Increases the resistance of normal cells to the oxazaphosphorine anticancer drugs | [74] | |
| Downregulated in human adrenocortical carcinomas | [77] | |
| Downregulated in mouse liver tumors | [75] | |
| Binds to anticancer drugs (e.g., daunorubicin, flavopiridol) | [75,76] | |
| ALDH1A2 | Candidate tumor suppressor in prostate cancer | [101] |
| ALDH1A3 | Downregulated in human breast cancer cell lines (e.g., MCF-7) | [110] |
| Downregulated in mammary tumor-susceptible BALB/cJ mice and upregulated in mammary tumor-resistant mice | [112] | |
| Upregulated by induction of wild-type p53 in cultured human colon cancer cells | [111] | |
| Methylation-silenced in gastric cancer | [113] | |
| Upregulated by the anticancer cytotoxic agent interleukin-13 in human glioma cells | [114] | |
| ALDH1L1 | Downregulated in human liver, lung, prostate, pancreas and ovarian cancers | [122] |
| Over-expression in cancer cell lines suppresses cellular proliferation and induces apoptosis | [127] | |
| Anticancer drug methotrexate induces ALDH1L1 activity | [132] | |
| ALDH2 | ALDH2 dysfunction is associated with increased risk for multiple cancers including upper digestive track cancers, stomach, colon, lung, head and neck cancers | [6] |
| ALDH3A1 | Increases the resistance of tumor cells to oxazaphosporine anticancer drugs | [177] |
| Increases chemotherapy resistance of human PBPCs to4-hydroperoxycyclophosphamide | [179] | |
| Detoxifies LPO-derived aldehydes in human hepatoma and lung tumor cells | [174] | |
| Upregulated in human cancer tissues and cell lines | [180] | |
| May regulate cell cycle and facilitate DNA repair | [13] | |
| ALDH4A1 | Upregulated as a result of DNA damage; may be a direct target of tumor suppressor gene p53 | [212] |
| ALDH7A1 | May regulate cell cycle | [231] |
Recently, ALDH1A1 was found to be downregulated in cell cultures and whole-skin tissue samples from patients with atopic dermatitis, suggesting its use as a potential dermal biomarker of this disease [78].
Aside from aldehyde metabolism, ALDH1A1 possesses esterase activity [79] and has been proposed to be the major enzyme catalyzing the oxidation of 3-deoxyglucosome, a potent glycating agent [80]. In addition, ALDH1A1 binds thyroid hormone [81] and is induced by estrogens [82], suggesting it may be regulated by or involved in hormone signaling.
4. ALDH1A2
ALDH1A2 is a cytosolic homotetramer expressed in various embryonic and adult tissues including intestine, testis, lung, kidney, liver, brain and retina [48,83]. Like ALDH1A1, ALDH1A2 catalyzes the oxidation of both all-trans-retinal and 9-cis-retinal to RA [23]. Compared with other ALDH isozymes [84], ALDH1A2 appears to exhibit the highest specificity (Vmax/Km = 49 nmol·min−1·mg−1·μM−1) for all-trans-retinal[43,44]. This property may be due to a unique disordered loop in its active site that binds all-trans-retinal in a distinct manner [85].
ALDH1A2 is involved in several developmental processes and may be a key regulator of RA synthesis in developing tissues [86]. Aldh1α2−/− mice die in early embryonic stages due to defects in early heart morphogenesis [23,87]. They display a lack of axial rotation, incomplete neural tube closure, reduction of the trunk region [23], and many of the features of human DiGeorge/velocardiofacial syndrome, a disorder characterized by cleft palate, heart abnormalities and learning disabilities [88]. Abnormalities in endothelial cell cycle progression during early vascular development have also been identified in Aldh1α2−/− embryos [89]. Various animal models have identified Aldh1a2 as a key regulator in the development of numerous tissues including kidney [90], retina [91], lung [92], forebrain [93], pancreas [94], and spinal cord [23].
A significant association between spina bifida in humans and three distinct ALDH1A2 single nucleotide polymorphisms (SNPs), including one silent (A151A; c.453A > G) and two intronic (rs3784259 and rs3784260), has been found; however, their functional significance remains unclear [95]. ALDH1A2 may also play a role in congenital diaphragmatic hernia (CDH), which is associated with chromosomal 15q defects within a region that includes both ALDH1A2 and ALDH1A3[96]. In addition, compounds known to induce CDH have been shown to inhibit ALDH1A2 [97].
ALDH1A2 may play a role in the defense against ethanol toxicity through either acetaldehyde detoxification or the synthesis of RA [98]. While rat ALDH1A2 oxidizes acetaldehyde inefficiently in vitro (Km 0.65 mM) [44], ALDH1A2 induction by lens epithelium-derived growth factor protects cells from ethanol-induced toxicity [99]. In addition, ALDH1A2 is decreased in RXRα−/− mice, which display increased susceptibility to alcohol-induced liver injury [56].
Similar to other ALDHs, ALDH1A2 metabolizes LPO-derived aldehydes and may protect against oxidative stress. Rat ALDH1A2 oxidizes medium-chain saturated LPO-derived aliphatic aldehydes, including hexanal (Km 28 μM), octanal (Km 5 μM) and decanal (Km 3 μM), with high affinity [44].
RA synthesis by ALDH1A2 promotes differentiation, cell growth arrest and apoptosis and may have an anticancer effect (Table 2)[100,101]. Indeed, ALDH1A2 has been suggested as a candidate tumor suppressor gene in prostate cancer [101]. ALDH1A2 expression is induced by DNA demethylation in human prostate cancer cell lines, downregulated in prostate tumors, and low expression of ALDH1A2 is associated with shorter recurrence-free survival. In addition, overexpression of wild-type ALDH1A2 in prostate cancer cells inhibits cell growth [101].
5. ALDH1A3
ALDH1A3 is a cytosolic homodimer that participates in the synthesis of RA and plays an important role in embryonic development [102]. ALDH1A3 oxidizes both all-trans-retinal and 9-cis-retinal (Km 0.2 μM for all-trans-retinal) to RA. ALDH1A3 is expressed in various late-stage embryonic and adult rodent tissues including tooth buds, intestine, kidney, brain, retina, prostate, skeletal muscle, lung, liver and pancreas [48]. In humans, ALDH1A3 expression has been observed in salivary gland, stomach, breast, kidney and fetal nasal mucosa [103,104]. Aldh1a3−/− mouse embryos die from defects in nasal development [24].
ALDH1A3 has been shown to participate in the development of the eye [105], nucleus accumbens and olfactory bulbs [106], hair follicles [107], the forebrain [108] and the cerebral cortex [109].
A number of studies have demonstrated that ALDH1A3 deficiency may play a critical role in cancer (Table 2). For example, ALDH1A3 expression is downregulated in human breast cancer MCF-7 cells [110] and ALDH1A3 is one of two genes that are upregulated by induction of wild type p53 in cultured human colon cancer cells [111]. In mammary tumor-susceptible BALB/cJ mice heterozygous for p53, Aldh1a3 is one of five candidate genes located within a region identified for its linkage to mammary tumorigenesis [112]. In mice resistant to induced mammary tumors (C57BL/6J), Aldh1a3 is one of two upregulated genes [112]. ALDH1A3 is methylation-silenced in gastric cancer cells [113] and induced by the antitumor agent IL-13 cytotoxin in glioblastoma cells [114].
Similar to ALDH1A2, ALDH1A3 may also play a role in CDH. Nitrofen-induced CDH and associated pulmonary hypoplasia of mouse fetuses is associated with ALDH1A3 upregulation, postulated to be the result of lung retinol deficiency [115]. As mentioned above, CDH is associated with human chromosome defects within a region including ALDH1A3[96].
ALDH1A3 also may have a role in mitigating oxidative stress by detoxifying LPO-derived aldehydes. Indeed, mouse ALDH1A3 has been shown to have very high affinity for octanal (Km 0.7 μM), decanal (Km 6.5 μM) and hexanal (Km 22.1 μM) [102].
6. ALDH1B1
ALDH1B1 is a mitochondrial homotetramer expressed in various adult and fetal human tissues including liver, testis, kidney, skeletal muscle, heart, placenta, brain and lung [116,117]. To date, little is known about ALDH1B1; however, it shares 75% sequence homology with ALDH2, the primary enzyme involved in the metabolism of the ethanol metabolite, acetaldehyde. ALDH1B1 displays relatively high affinity for acetaldehyde (Km 30 μM) and is believed to play a major role in acetaldehyde oxidation in vivo[117]. In vitro, ALDH1B1 is upregulated in response to UV light [118] and may play a role in protecting the cornea from this environmental insult [119].
7. ALDH1L1
ALDH1L1 is a multi-domain homotetramer consisting of two distinct catalytic domains, namely an amino-terminal formyl transferase domain and a carboxy-terminal ALDH domain [120]. ALDH1L1, through a unique mode of cofactor binding, appears to prefer NADP+ over NAD+[121]. ALDH1L1 is expressed at high levels in human liver, kidney and pancreas and at moderate levels in lung, prostate, brain, skeletal muscle, heart, ovary, thymus and testis [122]. ALDH1L1 appears to be present in multiple subcellular regions including both cytosol and mitochondria [123].
ALDH1L1, also known as 10-formyltetrahydrofolate (10-FTHF) dehydrogenase (10-FTHFD), catalyzes the formation of tetrahydrofolate (THF) from 10-FTHF [124]. THF is a major metabolite of dietary folate and an important substrate in one-carbon metabolism while 10-FTHF participates in purine biosynthesis and may influence DNA replication and repair [122]. NEUT2 mice, which are ALDH1L1 deficient, have reduced reproductive efficiency and display a substantial decrease in hepatic 10-FTHF and THF, indicating ALDH1L1 may regulate levels of 10-FTHF and THF [125].
ALDH1L1 may have a role in cancer by regulating cellular proliferation (Table 2). In vitro, ALDH1L1 overexpression in various cancer cell lines results in suppressed cellular proliferation and increased cytotoxicity, believed to be due to a catalytic function of ALDH1L1 [122]. In A549 cells, overexpression of ALDH1L1 induces the phosphorylation and translocation of p53 into the nucleus [126], resulting in a G1 cell cycle arrest and caspase-dependent apoptosis [127]. ALDH1L1 is significantly downregulated in human liver, lung, prostate, pancreas and ovarian cancers, which may enhance tumor proliferation [122]. During murine embryogenesis, ALDH1L1 expression correlates to regions lacking proliferating cells and is restricted to the midline of the developing CNS, suggesting it may also play a role in human neural tube defects [128]. Two intronic SNPs in ALDH1L1 are associated with an increased (intron 4) and a decreased (intron 13) risk for postmenopausal breast cancer [129].
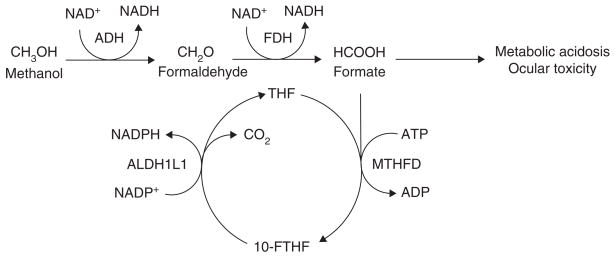
ALDH1L1 has an important role in methanol toxicity (Figure 5). Methanol is metabolized in the liver in two steps to the toxic metabolite, formate, which is then oxidized to carbon dioxide by a process dependent on THF, ATP, methyl-enetetrahydrofolate dehydrogenase (MTHFD) and ALDH1L1. Compared with other species, humans are particularly susceptible to methanol toxicity as a result of low formate oxidation rates due, in part, to lower liver ALDH1L1 activity [130]. In addition, methanol is highly toxic to the ocular system [123] and retinal Müller cells appear to represent a specific target [131]. Müller cells strongly and preferentially express ALDH1L1, suggesting that it may have a protective role [123]. However, low retinal THF levels may affect the ability of ALDH1L1 to participate in formate oxidation and, along with its specific localization in this target cell, suggest it may have an additional role in methanol toxicity.
Figure 5. The role of ALDH1L1 in methanol metabolism and toxicity.
Methanol is metabolized to the end product, carbon dioxide, through a pathway in which it is first converted to formaldehyde by ADH, which in turn is rapidly metabolized to formate by FDH. A build-up of formate is thought to cause the majority of the deleterious effects associated with methanol poisoning. Formate can enter the folate pathway through its conjugation to THF by MTHFD to form 10-FTHF, which is converted back into THF by ALDH1L1, accompanied by the release of carbon dioxide.
10-FTHF: 10-formyltetrahydrofolate; ADH: Alcohol dehydrogenase; ALDH1L1: Aldehyde dehydrogenase 1 family, member L1; FDH: Formaldehyde dehydrogenase; MTHFD: Methylenetetrahydrofolate dehydrogenase; THF: Tetrahydrofolate.
ALDH1L1 may be regulated by xenobiotics. For example, rats treated with ethanol or the chemotherapeutic drug methotrexate, both known to deplete cellular folate levels, display lower hepatic ALDH1L1 activity [132,133]. In addition, acetaminophen covalently adducts to ALDH1L1 and reduces its activity [134].
8. ALDH1L2
Human ALDH1L2, one of the most recently discovered ALDH genes, is located on chromosome 12q23.3 and is composed of 23 exons that encode a protein of 923 amino acids (102 kDa). ALDH1L2 shares 72% sequence identity with ALDH1L1 and has three domains which closely correspond to those of ALDH1L1, including a formyl-trans-N-formyl transferase at the amino terminal (residues 23 – 202), a formyltransferase domain in the middle (residues 226 – 327), and an ALDH domain at the carboxyl terminal (residues 451 – 910) [9]. To date, gene expression profiling has identified high expression of ALDH1L2 mRNA in the spleen and the corpus callosum [135]. In vitro, the treatment of human breast cancer cells with the anti-inflammatory agent, indometacin, upregulates ALDH1L2 gene expression [136]. To date, no further information exists regarding the properties or physiological significance of ALDH1L2.
9. ALDH2
ALDH2 encodes a mitochondrial matrix protein that is constituitively expressed in a variety of tissues including liver, kidney, heart, lung and brain [137]. ALDH2 is the primary enzyme involved in the oxidation of acetaldehyde (Km < 1 μM) during ethanol metabolism [138]. To date, several mutant ALDH2 alleles have been described, including the widely studied ALDH2*2 allele (single base pair mutation G/C → A/T), which results in an E487K substitution and consequent catalytic inactivation of ALDH2 due to a conformational change that leads to decreased nucleophilicity of the active site cysteine residue and decreased NAD+ affinity [31,139,140]. Glu-487, located in the bridging domain, appears to maintain a stable structural scaffold and facilitate catalysis by linking together the cofactor-binding and catalytic domains through its interactions with Arg-264 and Arg-475 [31,141]. ALDH2 functions as a homotetramer; however, the mutant ALDH2*2 allele is dominant, and heterotetrameric ALDH2 proteins containing even one ALDH2*2 subunit are enzymatically inactive [142]. The ALDH2*2 allele is found in approximately 40 – 50% of individuals of Asian descent [143] and, in those who drink alcohol, alcohol-induced toxicity occurs [144] due primarily to acetaldehyde accumulation and its effects [145]. This results in a lower alcoholism rate in Asian populations [146], but a number of studies have demonstrated the association of ALDH2*2 with an increased risk for various cancers, including oropharyngolaryngeal, esophageal, stomach, colon, lung, head and neck cancers [6,147]. Alcoholic ALDH2*2 individuals also display increased levels of acetaldehyde-derived DNA adducts, indicating a potential mechanism of DNA damage and cancer development [148]. ALDH2*2 has also been associated with alcoholic liver disease and cirrhosis in Asian individuals, even with moderate alcohol intake [21]. The ALDH2*2 allele may also be a risk factor for increased DNA damage in workers exposed to polyvinyl chloride, a carcinogen that is metabolized to the ALDH2 substrate chloroacetaldehyde [149], which produces DNA crosslinks and strand breaks [150].
ALDH2, acting as a nitrate reductase, is the principal enzyme necessary for the activation of nitroglycerin, used to treat angina and heart failure, and ALDH2*2 is associated with a lack of nitroglycerin efficacy in Chinese patients [151]. ALDH2*2 is also associated with myocardial infarction in Korean patients [152] and hypertension in Japanese patients [153]. Aldh2−/− mice display increased alcohol toxicity correlating with increased brain and blood acetaldehyde levels [154,155] and increased urinary 8-hydroxydeoxyguanosine and DNA-acetaldehyde adducts after both inhalation exposure to acetaldehyde [156] or oral ethanol gavage [157], as compared to Aldh2+/+ wild-type mice, indicating that both oral ingestion and inhalation of ethanol may pose a significant risk to ALDH2*2 individuals.
Recently, an additional polymorphic locus in the promoter (G/A) region of ALDH2, believed to affect ALDH2 activity by transcriptional mechanisms, has been found to be associated with variations in alcohol consumption habits among an American Jewish population [158].
Hepatotoxicity in alcoholics may occur, in part, to competition of LPO-derived aldehydes with acetaldehyde for ALDH2-mediated metabolism. ALDH2 is believed to have a major role in the metabolism of LPO-derived aldehydes, including 4-HNE and MDA (Figure 4)[2] and, specifically, appears to be responsible for 4-HNE elimination in hepatic Ito [159] and Kupffer cells [160]. In the brain, ALDH2 enzymatic activity is elevated in the cerebral cortex of AD patients, which may be a protective mechanism against high 4-HNE levels [161]. Indeed, in vitro, ALDH2-deficient cells are highly vulnerable to 4-HNE-induced apoptosis [162] and, in humans, ALDH2*2 is associated with an elevated risk for late-onset AD in Japanese [22] and Chinese [163] individuals. ALDH2 is also believed to play a major role in the metabolism of the neurotoxic aldehyde metabolite of dopamine, DOPAL (Km 4.2 μM), and ALDH2 deficiencies may contribute to the etiology of PD [5,164]. Significant impairment of DOPAL metabolism is reported in the presence of acetaldehyde, indicating competition between these substrates for metabolism by ALDH2 [165].
10. ALDH3A1
ALDH3A1 is a homodimer constitutively expressed in various tissues, including cornea, stomach, esophagus and lung, and is believed to have a crucial role in the cellular defense against oxidative stress [166]. ALDH3A1 catalyzes the oxidation of various LPO-derived aldehydes including α,β-hydroxyalkenals such as 4-HNE (Km 45 μM) [167].
ALDH3A1, considered a corneal crystallin, is one of the most abundantly expressed proteins in mammalian corneal epithelium, accounting for upwards of 50% of the total water-soluble protein fraction [166,168]. ALDH3A1 is undetectable in eye lens; however, corneal ALDH3A1 protects both the cornea and underlying lens against UV-induced oxidative stress [29]. Aldh3a1−/− mice display clear corneas [169]; however, upon exposure to UV light, accelerated lens opacification and cataract formation occur [29]. The lens of Aldh3a1−/− mice have increased levels of 4-HNE- and MDA-protein adducts and decreased proteasome activity associated with increased protein oxidation [29]. In humans, diseased corneas exhibit reduced ALDH3A1 expression [170] and, in vitro, human cornea epithelial (HCE) cells overexpressing ALDH3A1 are less sensitive to UV light and associated cytotoxicity [26]. In addition, ALDH3A1 may scavenge ROS and prevent proteins from hydroxyl radical-induced modifications through a conserved free cysteinyl residue in its active site [30]. ALDH3A1 also generates NADPH, critical for GSH maintenance, which may also absorb UV light [28] and act as a direct antioxidant [171]. Given that the water-soluble protein fraction of the cornea consists of only 17% of total protein but accounts for upwards of 50% of the total UV light absorption capacity, ALDH3A1 itself may directly absorb UV light [172]. In vitro, ALDH3A1 prevents UV-induced protein inactivation and, in vivo, UV light inactivates ALDH3A1 while other metabolic enzymes are unaffected, suggesting that ALDH3A1 may absorb UV light through a ‘suicide’ response [28,173].
ALDH3A1 may also regulate cellular proliferation and the cell cycle. Cell lines expressing high ALDH3A1 levels are more resistant to the antiproliferative effects of LPO-derived aldehydes [174] and ALDH3A1 inhibition or deficiency reduces cellular growth rates, believed to be due to aldehyde accumulation [175]. In addition to its cytosolic location, ALDH3A1 is present in the nucleus, where it may exert cell cycle regulation by reducing DNA synthesis and proliferation rates through the downregulation of cyclins A, B and E, the transcription factor E2F1, and the cell-regulatory protein p21, and through the regulation of kinase activities [13]. In vitro, ALDH3A1 has been shown to prevent DNA damage and reduce apoptosis from various toxins including hydrogen peroxide, mitomycin C and etoposide, indicating that ALDH3A1-mediated cell cycle delay and subsequent decreased cell growth is associated with resistance to DNA damage and may serve to facilitate DNA repair [27].
ALDH3A1, through the oxidation of oxazaphosphorines such as CP, contributes to drug resistance in various tumor types (Table 2)[176,177], and interindividual variations in ALDH3A1 activity may account for varying clinical responses to CP in certain cancers [73,178]. ALDH3A1 knockdown increases cellular sensitivity to CP and its metabolite, 4-hydroperoxycyclophosphamide, in cancer cell lines [72], while ALDH3A1 transfection in normal human peripheral blood hematopoietic progenitor cells (PBPCs) results in increased chemotherapy resistance to CP [179]. ALDH3A1 upregulation in non-cancerous cells could protect them during cancer treatment and have clinical applications [71]. Unexpectedly, however, a study using human breast tumor samples demonstrated that ALDH3A1 levels were not a predictor of the therapeutic efficacy of CP-based chemotherapy [73]. On the other hand, ALDH3A1 has been identified as a potential diagnostic marker for non-small-cell lung cancer [180] and ALDH3A1 may be a candidate gene in the pathogenesis of esophageal squamous cell carcinoma [181]. Interestingly, while ALDH3A1 is poorly expressed in normal liver, its expression in hepatoma cells increases in direct correlation with the degree of tumor growth [182]. ALDH3A1 is induced in other neoplastic tissues and cell lines [183] and its expression is differentially affected by hormones such as progesterone (downregulates) and cortisone (upregulates), suggesting a potential role in hormone-dependent tumors [184]. ALDH3A1 expression is also induced by various xenobiotics, including polycyclic hydrocarbons (PAHs) and 3-methylcholanthrene, through multiple ALDH3A1 xenobiotic response elements (XREs) [185,186].
11. ALDH3A2
ALDH3A2 is a microsomal homodimer expressed in various human tissues including liver, kidney, intestine, stomach, skeletal muscles, skin, lung, pancreas, placenta, heart and brain [187,188]. Apart from the major ALDH3A2 mRNA transcript, a splice variant transcript has been identified, namely the minor fatty ALDH variant (FALDHv)[188], which appears to be exclusively localized in peroxisomal membranes, where it is involved in phytanic acid metabolism [189].
ALDH3A2, also known as fatty aldehyde dehydrogenase (FALDH), forms, along with fatty ADH, the fatty alcohol:NAD oxidoreductase (FAO) enzyme complex that catalyzes the oxidation of fatty alcohol to fatty acid [190]. ALDH3A2 has high affinity for straight- and branched-chain aliphatic aldehydes of medium- and long-chain length, including both saturated and unsaturated aldehydes [191]. The most relevant substrates of ALDH3A2 are fatty aldehydes derived from the metabolism of fatty alcohol [192], phytanic acid [193], leukotriene B4[194], and ether glycerolipids [195]. Dysfunction of fatty aldehyde metabolism due to ALDH3A2 deficiency results in SLS, a rare autosomal recessive neurocutaneous disorder characterized by congenital icthyosis, mental retardation and spastic tetraplegia [16] specifically diagnosed by measuring ALDH3A2 activity in cultured human fibroblasts [192]. More than 72 ALDH3A2 mutations have been discovered in SLS patients including amino acid substitutions, deletions, insertions, and splicing errors [196]. SLS pathogenesis is attributed to the abnormal accumulation of lipids in the membranes of skin and brain, aldehyde Schiff-base adducts with amine-containing lipids and proteins, and defective eicosanoid metabolism [196]. In a recent study, human ALDH3A2 was delivered to keratinocytes of SLS patients using recombinant adeno-associated virus-2 vectors, which resulted in both augmented ALDH3A2 activity comparable to phenotypically normal heterozygous carriers and decreased toxicity of long-chain aldehydes to near the level of unaffected keratinocytes, indicating that ALDH3A2 gene therapy could be an effective treatment for SLS [197].
ALDH3A2 may have a role in diabetes and associated oxidative stress-induced complications. Insulin increases ALDH3A2 expression through the phosphatidylinositol 3-kinase (PI3K)-dependent pathway in the liver and white adipose tissues of normal rats, while no increase is seen in insulin-resistant mouse models [198]. In addition, fatty aldehydes accumulate in insulin-resistant diabetic rat livers due to failure of ALDH3A2 induction [199]. In vitro, ALDH3A2 protects against cytotoxicity induced by the LPO-derived aldehyde, dodecanal [189]; and 4-HNE treatment in mouse adipocytes overexpressing ALDH3A2 results in decreased ROS production [198].
12. ALDH3B1
ALDH3B1 encodes a protein highly expressed in the kidney and liver and moderately expressed in lung and various regions of the brain including the cortex, striatum, hippocampus, brainstem and cerebellum [200]. While relatively little is known about ALDH3B1, it is a catalytically active enzyme that has distinct substrate specificity towards medium- and long-chain (six carbons and longer) saturated and unsaturated aliphatic aldehydes including the LPO-derived aldehydes hexanal, 4-HNE, nonanal, octanal, trans-2-hexenal, trans-2-nonenal, and trans-2-octenal, and the aromatic aldehyde benzaldehyde [200]. By contrast, short-chain aldehydes, such as MDA and acetaldehyde, appear to be poor substrates. While ALDH3B1 is capable of utilizing either NAD+ or NADP+ as cofactor, cofactor preference may be substrate-specific [200].
A SNP in intron 2 (rs581105; T/G) of ALDH3B1 has recently been linked to the development of paranoid schizophrenia and proposed to be involved in an alteration of dopamine metabolism [201,202]. While ALDH3B1 is present in the brain, initial studies indicate that the dopamine-derived aldehyde DOPAL is a poor substrate [200]. However, ALDH3B1 may be involved in protecting the brain through the detoxification of other aldehydes, such as those produced during oxidative stress. In support of this, ALDH3B1 overexpression protects cells in vitro from toxicity induced by the LPO-derived aldehyde octanal [200]. An alternative splice variant of human ALDH3B1 exists that lacks both exons 3 and 4 (corresponding to amino acids 55 – 91) and encodes a shorter ALDH3B1 protein isoform b, which may prove to have altered enzymatic activity and pathophysiological implications [200].
13. ALDH3B2
ALDH3B2, located upstream from ALDH3B1 on chromosome 11q13.2, encodes a protein that shares 83% sequence identity with ALDH3B1. ALDH3B2 contains an in-frame stop codon at codon 17, suggesting it may be a pseudogene; however, ALDH3B2 transcripts have been found in human salivary gland tissue, indicating that ALDH3B2 retains promoter activity [203]. Microarray data indicate that ALDH3B2 is expressed in various organs including prostate, lung, kidney, liver and brain [204].
14. ALDH4A1
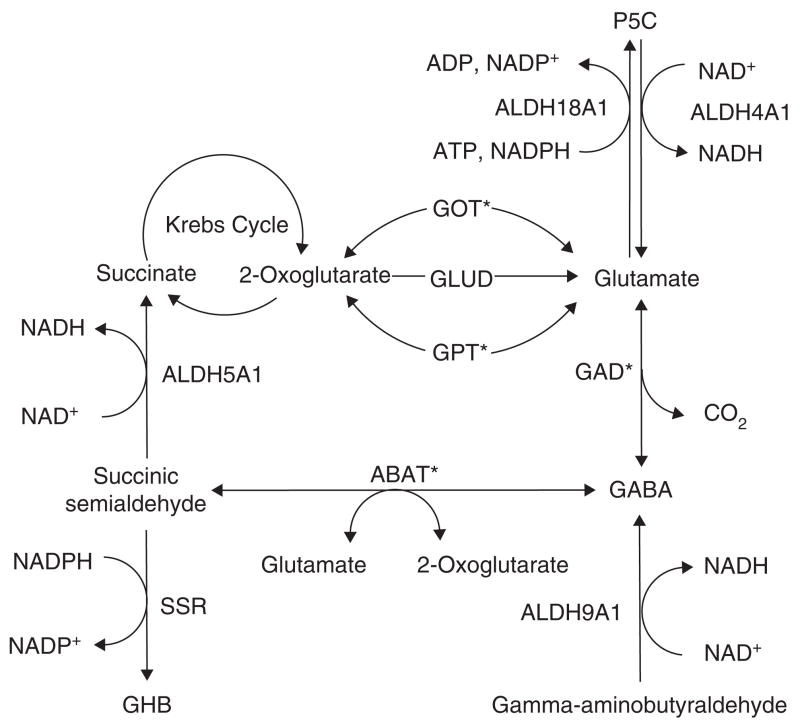
ALDH4A1, also known as pyrroline-5-carboxylate (P5C) dehydrogenase, is a mitochondrial matrix homodimer highly expressed in liver, skeletal muscle and kidney [205]. ALDH4A1 is involved in proline degradation and catalyzes the NAD+-dependent conversion of P5C to the neurotransmitter, glutamate (Figure 6), which prevents the accumulation of P5C [206,207]. ALDH4A1 mutations cause type II hyperprolinemia, an autosomal recessive disease characterized by seizures, mental retardation and high levels of P5C in physiological fluids [17,208]. Patients with type II hyperprolinemia have a frame shift (G521fs(+1)) or missense (S352L) mutation in ALDH4A1 that results in ablated enzyme activity [208]. The phenotype associated with type II hyperprolinemia is the result of P5C-mediated deactivation of the vitamin B6 derivative pyridoxal phosphate (PLP) through a Knoevenagel-type condensation reaction (see ALDH7A1; Figure 7)[209]. PLP is a required cofactor in a wide range of reactions, including the biosynthesis of neurotransmitters, and its inactivation is known to cause deleterious effects [210].
Figure 6. The role of ALDH isozymes in glutamate and GABA pathways.
ALDH4A1, ALDH5A1, ALDH9A1 and ALDH18A1 play many important roles in the synthesis and catabolism of the neurotransmitters glutamate and γ-aminobutyric acid, and mutational defects in these ALDH genes result in a variety of neurological disorders.
*Indicates enzymes that require pyridoxal phosphate (PLP) as cofactor.
ABAT: 4-Aminobutyrate aminotransferase GABA, γ-aminobutyric acid; GAD: Glutamate decarboxylase; GHB: γ-hydroxybutyric acid; GLUD: Glutamate dehydrogenase; GOT: Glutamic-oxaloacetic transaminase; GPT: Glutamic-pyruvate transaminase; SSR: Succinic semialdehyde reductase.
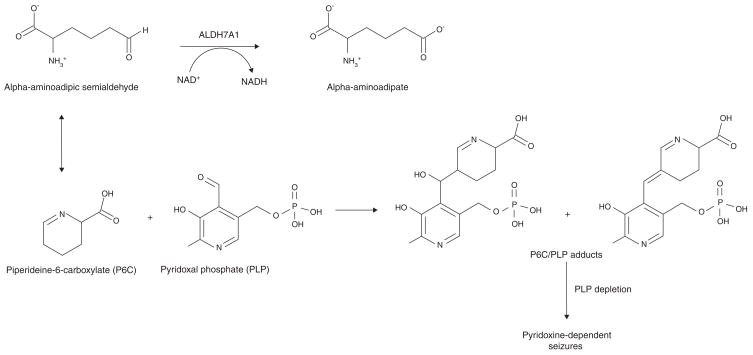
Figure 7. The role of ALDH7A1 as an alpha-aminoadipic semialdehyde (AASA) dehydrogenase.
AASA is in equilibrium with its cyclic Schiff base, piperideine-6-carboxylate (P6C). Pyridoxal phosphate (PLP) is the active form of vitamin B6 and an important cofactor in many types of reactions. Mutations in ALDH7A1 result in the accumulation of P6C and Knoevenagel adduct products between P6C and PLP, leading to the inactivation and depletion of PLP and pyridoxine-dependent seizures.
In addition to its role in proline metabolism, ALDH4A1 may be involved in protection against oxidative stress. ALDH4A1 appears to be a major enzyme responsible for the oxidation of short- and medium-chain aliphatic LPO-derived aldehydes [211] and, in vitro, cells overexpressing ALDH4A1 produce lower levels of intracellular ROS after hydrogen peroxide and UV treatment [212]. ALDH4A1 is upregulated in response to DNA damage; this process appears to be p53-dependent, indicating that ALDH4A1 may have a role in DNA repair and cell survival [212]. Indeed, inhibition of ALDH4A1 expression in human glioblastoma cells results in increased susceptibility to p53-mediated apoptosis [212].
15. ALDH5A1
ALDH5A1 is a mitochondrial homotetramer found in liver, kidney, skeletal muscle, and brain [213]. ALDH5A1, also known as succinic semialdehyde (SSA) dehydrogenase (SSADH), catalyzes the NAD+-dependent conversion of SSA (Km 6.3 μM) to succinate (Figure 6) in the last step of GABA catabolism [213]. While SSA is primarily oxidized by ALDH5A1 to succinate, a small fraction is reduced by cytosolic SSA reductase to γ-hydroxybutyric acid [214], a compound with neurotransmitter- and neuromodulator-like properties normally only found in small quantities in the CNS [215]. ALDH5A1 mutations, of which more than 50 have been identified, are responsible for γ-hydroxybutyric aciduria, a rare autosomal recessive disorder characterized by neurological and cognitive defects due to the accumulation of GABA, γ-hydroxybutyric acid, and SSA in tissues and physiological fluids [18].
Aldh5A1−/− mice, used as a model to study γ-hydroxybutyric aciduria, display absence seizure onset at 2 weeks that progresses to generalized tonic-clonic seizures and early death [216]. These mice also display altered brain phospholipid composition and significant downregulation of genes associated with myelin sheath thickness and compaction, indicating that ALDH5A1 may have a role in neurotransmission efficiency [217,218]. While the effects of γ-hydroxybutyric acid are not completely understood, the compound increases thiobarbituric acid-reactive substance (TBARS) levels and decreases the total antioxidant potential in rat cerebral cortex homogenates, suggesting elevated LPO and an association with oxidative stress processes [219]. Interestingly, ALDH5A1 is inhibited by the LPO-derived aldehydes acrolein and 4-HNE (IC50 15 μM and 110 μM, respectively) [220].
16. ALDH6A1
ALDH6A1 is a mitochondrial tetramer expressed at high levels in the liver, kidney and heart and at lower levels in muscle and brain [221]. Also known as methylmalonate semialdehyde (MMS) dehydrogenase (MMSDH), ALDH6A1, the only known CoA-dependent human ALDH, is involved in valine and pyrimidine catabolism and catalyzes the oxidative decarboxylation of malonate semialdehyde (Km 4.5 μM) and MMS (Km 5.3 μM) to acetyl-CoA and propionyl-CoA, respectively [222].
ALDH6A1 mutations lead to a disorder characterized by a variety of metabolic abnormalities, including increased levels of 3-amino and 3-hydroxyisobutyric acids, β-alanine and 3-hydroxypropionic acid, usually accompanied by some degree of psychomotor delay [223]. To date, the ALDH6A1 mutations involved are not well characterized; however, one patient was found to have a transversion (1336G > A) in ALDH6A1 leading to the replacement of a highly conserved glycine with an arginine (G446R) [223].
ALDH6A1 is upregulated by valine carbon utilization for lipogenesis during the differentiation of 3T3-L1 fibroblasts into mature adipocytes [224]. ALDH6A1 has also been identified as a cardiac protein that undergoes oxidative modification via tyrosine nitration in aged rats, possibly contributing to age-dependent heart degeneration [225]. ALDH6A1, through the metabolism of malonate semi-aldehyde, is also involved in the metabolism of the LPO-derived aldehyde, MDA, to acetyl-CoA (Figure 4).
17. ALDH7A1
ALDH7A1 is a homotetramer expressed in a wide range of tissues. High levels of ALDH7A1 are observed in rat heart, liver and kidney [226], while ALDH7A1 in black seabream fish (sbALDH7A1) is highly expressed in the liver and kidney but not the heart [227]. In human fetal tissues, ALDH7A1 has been detected at high levels in the cochlea, eye, ovary, heart and kidney, while moderate levels are observed in the liver, spleen, muscle, lung and brain [228].
Human ALDH7A1 has a primary role in the pipecolic acid pathway of lysine catabolism, catalyzing the oxidation of alpha-aminoadipic semialdehyde (AASA) (Km 180 μM) to alpha-aminoadipate (Figure 7)[229]. ALDH7A1 mutations are the molecular basis for pyridoxine-dependent epilepsy (PDE), an autosomal recessive disorder characterized by the onset of intractable seizures during infancy and early childhood, preventable by daily, high dose supplementation with pyridoxine (Vitamin B6)[19]. Due to ALDH7A1 deficiency, increased concentrations of AASA and its cyclic Schiff base, piperidiene-6-carbxylate (P6C), occur in patients, which results in the inactivation and depletion of the coenzyme PLP through a Knoevenagel adduction reaction (Figure 7)[19]. PLP is a required coenzyme that is especially important in amino acid and neurotransmitter pathways, including those involving GABA, serotonin and noradrenaline [210]. The underlying cause of seizures in PDE has been attributed to PLP inactivation and subsequent disruption in neurotransmitter metabolism [19].
Aside from its role in AASA metabolism, little kinetic information exists regarding mammalian ALDH7A1 activity towards other substrates; however, sbALDH7A1 shares 83% identity to the human protein and is active, albeit with low affinity, with a variety of common aldehydes including acetaldehyde (Km 3.6 mM), propionaldehyde (Km 1.2 mM), and benzaldehyde (Km 0.45 mM). It has similar affinity for NAD+ as mammalian ALDH7A1 [227]. sbALDH7A1 is not active towards 4-HNE, MDA, SSA, betaine aldehyde or all-trans retinaldehyde.
ALDH7A1 shares 60% amino acid sequence identity with the osmotic stress-induced 26 g pea turgor plant protein, refered to as plant ALDH7B1, thought to be involved in the regulation of osmotic pressure within plant cells [226]. Plant ALDH7B1 expression increases in response to cellular stresses such as dehydration, temperature flunctuations and high salinity [230]. Sequence conservation between evolutionarily distant species, such as human ALDH7A1 and plant ALDH7B1, often indicates a functional similarity; however, ALDH7A1 levels in vitro do not appear to be affected by exposure to a variety of stress-inducing treatments such as heat shock, dehydration, ionizing irradiation, glucocorticoids, iron or t-butylhydroperoxide [226]. In addition, screening of the sbALDH7A1 promoter region has identified no regions with homology to known osmotic response elements [231]. Interestingly, ALDH7A1 expression in the cochlea of the ear, dependent on the proper maintenance of internal hydrostatic pressure, indicates that mammalian ALDH7A1 may be involved in osmotic regulation with a potential role in hearing disorders. However, to date, no connection has been found, including in patients with Menière disease, an inner-ear disorder affecting hearing and balance [232].
ALDH7A1 is highly and differentially expressed during porcine oocyte maturation, including during the first and second meiotic stages [233]. Screening of the sbALDH7A1 promoter region has revealed cis-elements related to cell cycle regulation [231].
18. ALDH8A1
While little information exists regarding ALDH8A1, it is a cytosolic enzyme with high expression in liver and kidney and moderate expression in brain, spinal cord, mammary gland, thymus, adrenal, testis, prostate and gastrointestinal tract [234]. Along with the ALDH1 family, ALDH8A1 is also believed to participate in the biosynthesis of RA through the oxidation of retinal and displays a distinct preference for 9-cis-retinal, as opposed to all-trans-retinal[234], which is unique among the retinal-oxidizing ALDHs [235]. ALDH8A1 also metabolizes aliphatic aldehydes, including acetaldehyde (Km 10.2 mM), decanal, octanal, hexanal and propanal, with activity that increases with chain length [234]. ALDH8A1 is also active towards several important metabolic aldehydes including SSA and glutaraldehyde [234].
19. ALDH9A1
ALDH9A1 encodes a cytosolic tetramer that is highly expressed in the liver, skeletal muscle, kidney and brain [236]. ALDH9A1, involved in an alternate biosynthesis pathway of GABA, catalyzes the oxidation of γ-aminobutyraldehyde (Km 8 – 14 μM), the metabolite of the biogenic amine putrescine, to GABA (Figure 6)[237,238]. ALDH9A1 may also play a role in the metabolism of catecholamine-derived aldehydes such as the toxic dopamine metabolite, DOPAL (Km 2.6 μM) [5,238]. ALDH9A1 also oxidizes betaine aldehyde [239], γ-trimethylaminobutyraldehyde, which is involved in carnitine biosynthesis [240], and acetaldehyde (Km 40 – 50 μM) [237]. High affinity of ALDH9A1 for γ-aminobutyraldehyde, DOPAL, and acetaldehyde indicates that these substrates may compete for metabolism by ALDH9A1, which may affect GABA and dopamine pathways and brain development. In a Japanese fish model, ALDH9A1 is developmentally regulated and ethanol treatment downregulates ALDH9A1 expression during embryogenesis, suggesting ALDH9A1 may play a role in the teratogenic effects of ethanol [241]. ALDH9A1 is also a candidate gene in inflammation-mediated aggravation of human non-alcoholic steatohepatitis [242].
20. ALDH16A1
ALDH16A1, one of the most recently identified ALDHs, was sequenced from a human uterine cDNA library by the Mammalian Gene Collection (MGC) consortium [243]. The 2627 bp transcript has been traced to human chromosome 19q13.33 and is composed of 17 exons, which encode an 802 amino acid protein containing a putative NAD+-dependent ALDH domain, with a theoretical molecular weight of 85 kDa and an isoelectric point of 6.77. ALDH16A1 orthologues have since been identified in a variety of species including chimpanzee (98% identity), mouse (81% identity), rat, dog and zebra fish. Microarray data indicate that ALDH16A1 is widely expressed in a variety of tissues including bone marrow, heart, kidney and lung [204]. To date, the physiological significance and function of ALDH16A1 remains to be established.
21. ALDH18A1
ALDH18A1, also known as Δ-pyrroline-5-carboxylate synthase (P5CS), is a bifunctional inner mitochondrial membrane enzyme containing an N-terminal γ-glutamyl kinase domain and a C-terminal γ-glutamyl phosphate reductase domain that is expressed at high levels in human pancreas, ovary, testis and kidney, and moderate levels in colon, small intestine, placenta, heart and skeletal muscle [244]. ALDH18A1 catalyzes the ATP- and NADPH-dependent reduction of glutamate to P5C, which is further converted to ornithine (Figure 6)[244]. This pathway is extremely important in the de novo synthesis of the amino acids proline and arginine; defects in ALDH18A1 lead to a variety of metabolic and neurologic abnormalities, including hypoprolinemia, hypoornithinemia, hypocitrullinemia, hypoargininemia and hyperammonemia with cataract formation, neurodegeneration and connective tissue anomalies [245,246]. Various ALDH18A1 mutations have been described, including a missense mutation resulting in the replacement of a highly conserved leucine with a serine (L396S) [245], and an arginine to glutamine substitution (R84Q) at a conserved residue within the γ-glutamyl kinase domain [20]. While alternative splicing of ALDH18A1 gives rise to two distinct isoforms, namely ALDH18A1_i1 and ALDH18A1_i2, differing by a two amino acid insert very close to the γ-glutamyl kinase active site, mutations in either lead to deficient ALDH18A1 activity and metabolic abnormalities [20]. However, sensitivity to ornithine inhibition (Ki 0.25 mM), thought to play a regulatory role, is observed only in the shorter ALDH18A1_i2, which is expressed primarily in the gut, where it contributes to arginine biosynthesis [20]. ALDH18A1_i1 is expressed in multiple tissues and is predominantly involved in proline biosynthesis. ALDH18A1 is downregulated in the auditory midbrain of mice displaying age-dependent hearing loss, potentially due to accumulation of the ALDH18A1 substrate, glutamate, indicating that ALDH18A1 may play a role in preventing auditory toxicity by modulating glutamate levels [247].
22. Expert opinion and conclusion
Aldehydes are ubiquitous in nature and the environment, and are produced during numerous physiological processes and biotransformation events. It is known that several are essential to certain physiological processes (e.g., retinal for vision), and many aldehydes previously believed to be merely cytotoxic intermediates appear to have multiple functions including roles in signal transduction, gene regulation and cellular proliferation. Nonetheless, the toxicity of aldehydes is well characterized, most notably their ability to adduct macromolecules such as proteins and DNA. In this regard, it is not surprising that several enzyme systems (e.g., ADH, AKR, AO, XO, SDR, and ALDH) catalyze the metabolism of aldehyde species, many with broad overlapping substrate specificity. Of these, the ALDH superfamily clearly plays a very important role in the oxidative pathway of aldehyde detoxification.
Although the physiological role of several of the 19 human ALDH isozymes is unknown or not fully characterized, many have been shown to be critical in the detoxification of specific aldehyde substrates. The clinical importance of the ALDH superfamily is evidenced from human phenotypes directly linked to mutations in ALDH genes leading to the absence, deficiency or inactivation of ALDH proteins. Multiple disease states are associated with ALDH dysfunction including many cancers, metabolic diseases and neurological abnormalities. Aside from those that have been causally linked to the ALDH family, numerous other pathologies have been proposed to be influenced by these isozymes. ALDH genotype has also been shown to affect the efficacy of drug treatment for various diseases and disorders, including cancer. In this regard, it has been demonstrated for several drug therapies (e.g., nitroglycerin, cyclophosphamide) that patient ALDH genotype should be taken into consideration in order to design the most efficacious treatment strategy. Investigations focusing on ALDH genes and isozymes as therapeutic targets in certain disease states have shown promise and will no doubt be a part of future research efforts.
Aside from their demonstrated importance in aldehyde metabolism, the ALDH superfamily also exhibits multiple catalytic and non-catalytic functions. These enzymes may have important, yet to be clearly defined, roles in ester metabolism, drug bioactivation, ROS scavenging and the absorption of UV light. The significance of the capacity of several ALDHs to bind various endogenous (e.g., thyroid hormone) and exogenous (e.g., acetaminophen, daunorubicin) compounds remains to be elucidated. The recent discovery of ALDH isozymes present in multiple subcellular compartments – including the nucleus, where they may exert effects on gene expression and cellular proliferation – is exciting, and points to unique physiological roles of these enzymes as governed by subcellular localization. It is our belief that as research of the ALDH superfamily expands, these isozymes will be found to be critical in a number of diseases and developmental processes, making human ALDH genotype an important factor in clinical treatment strategies.
Acknowledgments
We thank our colleagues, especially Dr David Thompson, for valuable discussions and critical reading of this manuscript. This work was supported by NIH/NEI grants EY11490 and EY17963. S Marchitti was supported by NIH/NIAAA predoctoral Fellowship AA016875.
Footnotes
Declaration of interest
The authors state no conflict of interest and have received no payment in preparation of this manuscript.
Bibliography
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Vasiliou V, Pappa A, Petersen DR. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem Biol Interact. 2000;129:1–19. doi: 10.1016/s0009-2797(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 2.Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 2004;36:279–99. doi: 10.1081/dmr-120034001. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien PJ, Siraki AG, Shangari N. Aldehyde sources, metabolism, molecular toxicity mechanisms, and possible effects on human health. Crit Rev Toxicol. 2005;35:609–62. doi: 10.1080/10408440591002183. [DOI] [PubMed] [Google Scholar]
- 4.••.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. A landmark paper concerning the chemical properties and reactivity of aldehydes. [DOI] [PubMed] [Google Scholar]
- 5.••.Marchitti SA, Deitrich RA, Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol Rev. 2007:59125–50. doi: 10.1124/pr.59.2.1. A comprehensive recent review paper. [DOI] [PubMed] [Google Scholar]
- 6.Yokoyama A, Muramatsu T, Omori T, et al. Alcohol and aldehyde dehydrogenase gene polymorphisms and oropharyngolaryngeal, esophageal and stomach cancers in Japanese alcoholics. Carcinogenesis. 2001;22:433–9. doi: 10.1093/carcin/22.3.433. [DOI] [PubMed] [Google Scholar]
- 7.Nadkarni DV, Sayre LM. Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem Res Toxicol. 1995;8:284–91. doi: 10.1021/tx00044a014. [DOI] [PubMed] [Google Scholar]
- 8.•.Brooks PJ, Theruvathu JA. DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol. 2005;35:187–93. doi: 10.1016/j.alcohol.2005.03.009. A very detailed paper concerning aldehyde-DNA adducts. [DOI] [PubMed] [Google Scholar]
- 9.Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2005;2:138–43. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasiliou V, Bairoch A, Tipton KF, Nebert DW. Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics. 1999;9:421–34. [PubMed] [Google Scholar]
- 11.Braun T, Bober E, Singh S, et al. Evidence for a signal peptide at the amino-terminal end of human mitochondrial aldehyde dehydrogenase. FEBS Lett. 1987;215:233–6. doi: 10.1016/0014-5793(87)80152-7. [DOI] [PubMed] [Google Scholar]
- 12.Vasiliou V, Kozak CA, Lindahl R, Nebert DW. Mouse microsomal Class 3 aldehyde dehydrogenase: AHD3 cDNA sequence, inducibility by dioxin and clofibrate, and genetic mapping. DNA Cell Biol. 1996;15:235–45. doi: 10.1089/dna.1996.15.235. [DOI] [PubMed] [Google Scholar]
- 13.Pappa A, Brown D, Koutalos Y, et al. Human aldehyde dehydrogenase 3A1 inhibits proliferation and promotes survival of human corneal epithelial cells. J Biol Chem. 2005;280:27998–8006. doi: 10.1074/jbc.M503698200. [DOI] [PubMed] [Google Scholar]
- 14.Sladek NE. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol. 2003;17:7–23. doi: 10.1002/jbt.10057. [DOI] [PubMed] [Google Scholar]
- 15.Vasiliou V, Pappa A. Polymorphisms of human aldehyde dehydrogenases. Consequences for drug metabolism and disease. Pharmacology. 2000;61:192–8. doi: 10.1159/000028400. [DOI] [PubMed] [Google Scholar]
- 16.Rizzo WB, Carney G. Sjogren-Larsson syndrome: diversity of mutations and polymorphisms in the fatty aldehyde dehydrogenase gene (ALDH3A2) Hum Mutat. 2005;26:1–10. doi: 10.1002/humu.20181. [DOI] [PubMed] [Google Scholar]
- 17.Onenli-Mungan N, Yuksel B, Elkay M, et al. Type II hyperprolinemia: a case report. Turk J Pediatr. 2004;46:167–9. [PubMed] [Google Scholar]
- 18.Akaboshi S, Hogema BM, Novelletto A, et al. Mutational spectrum of the succinate semialdehyde dehydrogenase (ALDH5A1) gene and functional analysis of 27 novel disease-causing mutations in patients with SSADH deficiency. Hum Mutat. 2003;22:442–50. doi: 10.1002/humu.10288. [DOI] [PubMed] [Google Scholar]
- 19.Mills PB, Struys E, Jakobs C, et al. Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat Med. 2006;12:307–9. doi: 10.1038/nm1366. [DOI] [PubMed] [Google Scholar]
- 20.Baumgartner MR, Hu CA, Almashanu S, et al. Hyperammonemia with reduced ornithine, citrulline, arginine and proline: a new inborn error caused by a mutation in the gene encoding delta(1)-pyrroline-5-carboxylate synthase. Hum Mol Genet. 2000;9:2853–8. doi: 10.1093/hmg/9.19.2853. [DOI] [PubMed] [Google Scholar]
- 21.Enomoto N, Takase S, Takada N, Takada A. Alcoholic liver disease in heterozygotes of mutant and normal aldehyde dehydrogenase-2 genes. Hepatology. 1991;13:1071–5. [PubMed] [Google Scholar]
- 22.Kamino K, Nagasaka K, Imagawa M, et al. Deficiency in mitochondrial aldehyde dehydrogenase increases the risk for late-onset alzheimer’s disease in the Japanese population. Biochem Biophys Res Commun. 2000;273:192–6. doi: 10.1006/bbrc.2000.2923. [DOI] [PubMed] [Google Scholar]
- 23.Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–8. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 24.Dupe V, Matt N, Garnier JM, et al. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci USA. 2003;100:14036–41. doi: 10.1073/pnas.2336223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sydow K, Daiber A, Oelze M, et al. Central role of mitochondrial aldehyde dehydrogenase and reactive oxygen species in nitroglycerin tolerance and cross-tolerance. J Clin Invest. 2004;113:482–9. doi: 10.1172/JCI19267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pappa A, Chen C, Koutalos Y, et al. Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic Biol Med. 2003;34:1178–89. doi: 10.1016/s0891-5849(03)00070-4. [DOI] [PubMed] [Google Scholar]
- 27.Lassen N, Pappa A, Black WJ, et al. Antioxidant function of corneal ALDH3A1 in cultured stromal fibroblasts. Free Radic Biol Med. 2006;41:1459–69. doi: 10.1016/j.freeradbiomed.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Estey T, Cantore M, Weston PA, et al. Mechanisms involved in the protection of UV-induced protein inactivation by the corneal crystallin ALDH3A1. J Biol Chem. 2007;282:4382–92. doi: 10.1074/jbc.M607546200. [DOI] [PubMed] [Google Scholar]
- 29.Lassen N, Bateman JB, Estey T, et al. Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem. 2007;282:25668–76. doi: 10.1074/jbc.M702076200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uma L, Hariharan J, Sharma Y, Balasubramanian D. Corneal aldehyde dehydrogenase displays antioxidant properties. Exp Eye Res. 1996;63:117–20. doi: 10.1006/exer.1996.0098. [DOI] [PubMed] [Google Scholar]
- 31.Steinmetz CG, Xie P, Weiner H, Hurley TD. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5:701–11. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- 32.Liu ZJ, Sun YJ, Rose J, et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4:317–26. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 33.Hempel J, Liu ZJ, Perozich J, et al. Conserved residues in the aldehyde dehydrogenase family. Locations in the class 3 tertiary structure. Adv Exp Med Biol. 1997;414:9–13. doi: 10.1007/978-1-4615-5871-2_2. [DOI] [PubMed] [Google Scholar]
- 34.Perozich J, Kuo I, Lindahl R, Hempel J. Coenzyme specificity in aldehyde dehydrogenase. Chem Biol Interact. 2001;130–132:115–24. doi: 10.1016/s0009-2797(00)00227-1. [DOI] [PubMed] [Google Scholar]
- 35.Hempel J, Kuo I, Perozich J, et al. Aldehyde dehydrogenase. Maintaining critical active site geometry at motif 8 in the class 3 enzyme. Eur J Biochem. 2001;268:722–6. doi: 10.1046/j.1432-1327.2001.01926.x. [DOI] [PubMed] [Google Scholar]
- 36.Abriola DP, Fields R, Stein S, et al. Active site of human liver aldehyde dehydrogenase. Biochemistry. 1987;26:5679–84. doi: 10.1021/bi00392a015. [DOI] [PubMed] [Google Scholar]
- 37.D’Ambrosio K, Pailot A, Talfournier F, et al. The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis. Biochemistry. 2006;45:2978–86. doi: 10.1021/bi0515117. [DOI] [PubMed] [Google Scholar]
- 38.Hammen PK, Lali-Hassani A, Hallenga K, et al. Multiple conformations of NAD and NADH when bound to human cytosolic and mitochondrial aldehyde dehydrogenase. Biochemistry. 2002;41:7156–68. doi: 10.1021/bi012197t. [DOI] [PubMed] [Google Scholar]
- 39.Hurley TD, Steinmetz CG, Weiner H. Three-dimensional structure of mitochondrial aldehyde dehydrogenase. Mechanistic implications. Adv Exp Med Biol. 1999;463:15–25. doi: 10.1007/978-1-4615-4735-8_3. [DOI] [PubMed] [Google Scholar]
- 40.Sohling B, Gottschalk G. Purification and characterization of a coenzyme-A-dependent succinate-semialdehyde dehydrogenase from Clostridium kluyveri. Eur J Biochem. 1993;212:121–7. doi: 10.1111/j.1432-1033.1993.tb17641.x. [DOI] [PubMed] [Google Scholar]
- 41.King G, Holmes R. Human corneal and lens aldehyde dehydrogenases. Purification and properties of human lens ALDH1 and differential expression as major soluble proteins in human lens (ALDH1) and cornea (ALDH3) Adv Exp Med Biol. 1997;414:19–27. [PubMed] [Google Scholar]
- 42.Zhai Y, Sperkova Z, Napoli JL. Cellular expression of retinal dehydrogenase types 1 and 2: effects of vitamin A status on testis mRNA. J Cell Physiol. 2001;186:220–32. doi: 10.1002/1097-4652(200102)186:2<220::AID-JCP1018>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 43.Zhao D, McCaffery P, Ivins KJ, et al. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur J Biochem. 1996;240:15–22. doi: 10.1111/j.1432-1033.1996.0015h.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Penzes P, Napoli JL. Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. Recognition of retinal as substrate. J Biol Chem. 1996;271:16288–93. doi: 10.1074/jbc.271.27.16288. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida A, Hsu LC, Dave V. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme. 1992;46:239–44. doi: 10.1159/000468794. [DOI] [PubMed] [Google Scholar]
- 46.Ross SA, McCaffery PJ, Drager UC, De Luca LM. Retinoids in embryonal development. Physiol Rev. 2000;80:1021–54. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- 47.Haselbeck RJ, Hoffmann I, Duester G. Distinct functions for Aldh1 and Raldh2 in the control of ligand production for embryonic retinoid signaling pathways. Dev Genet. 1999;25:353–64. doi: 10.1002/(SICI)1520-6408(1999)25:4<353::AID-DVG9>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niederreither K, Fraulob V, Garnier JM, et al. Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech Dev. 2002;110:165–71. doi: 10.1016/s0925-4773(01)00561-5. [DOI] [PubMed] [Google Scholar]
- 49.Marlier A, Gilbert T. Expression of retinoic acid-synthesizing and -metabolizing enzymes during nephrogenesis in the rat. Gene Expr Patterns. 2004;5:179–85. doi: 10.1016/j.modgep.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Dickman ED, Thaller C, Smith SM. Temporally-regulated retinoic acid depletion produces specific neural crest, ocular and nervous system defects. Development. 1997;124:3111–21. doi: 10.1242/dev.124.16.3111. [DOI] [PubMed] [Google Scholar]
- 51.Duester G. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur J Biochem. 2000;267:4315–24. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- 52.Fan X, Molotkov A, Manabe S, et al. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol. 2003;23:4637–48. doi: 10.1128/MCB.23.13.4637-4648.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molotkov A, Duester G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J Biol Chem. 2003;278:36085–90. doi: 10.1074/jbc.M303709200. [DOI] [PubMed] [Google Scholar]
- 54.Ziouzenkova O, Orasanu G, Sharlach M, et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13:695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z, Srivastava S, Yang X, et al. A hierarchical approach employing metabolic and gene expression profiles to identify the pathways that confer cytotoxicity in HepG2 cells. BMC Syst Biol. 2007;1:21. doi: 10.1186/1752-0509-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gyamfi MA, Kocsis MG, He L, et al. The role of retinoid X receptor alpha in regulating alcohol metabolism. J Pharmacol Exp Ther. 2006;319:360–8. doi: 10.1124/jpet.106.108175. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem. 1998;251:549–57. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida A, Hsu LC, Yanagawa Y. Biological role of human cytosolic aldehyde dehydrogenase 1: hormonal response, retinal oxidation and implication in testicular feminization. Adv Exp Med Biol. 1993;328:37–44. doi: 10.1007/978-1-4615-2904-0_5. [DOI] [PubMed] [Google Scholar]
- 59.Pereira F, Rosenmann E, Nylen E, et al. The 56 kDa androgen binding protein is an aldehyde dehydrogenase. Biochem Biophys Res Commun. 1991;175:831–8. doi: 10.1016/0006-291x(91)91640-x. [DOI] [PubMed] [Google Scholar]
- 60.Galter D, Buervenich S, Carmine A, et al. ALDH1 mRNA: presence in human dopamine neurons and decreases in substantia nigra in Parkinson’s disease and in the ventral tegmental area in schizophrenia. Neurobiol Dis. 2003;14:637–47. doi: 10.1016/j.nbd.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 61.Jacobs FM, Smits SM, Noorlander CW, et al. Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development. 2007;134:2673–84. doi: 10.1242/dev.02865. [DOI] [PubMed] [Google Scholar]
- 62.Chung S, Hedlund E, Hwang M, et al. The homeodomain transcription factor Pitx3 facilitates differentiation of mouse embryonic stem cells into AHD2-expressing dopaminergic neurons. Mol Cell Neurosci. 2005;28:241–52. doi: 10.1016/j.mcn.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Mandel S, Grunblatt E, Riederer P, et al. Gene expression profiling of sporadic Parkinson’s disease substantia nigra pars compacta reveals impairment of ubiquitin-proteasome subunits, SKP1A, aldehyde dehydrogenase, and chaperone HSC-70. Ann NY Acad Sci. 2005;1053:356–75. doi: 10.1196/annals.1344.031. [DOI] [PubMed] [Google Scholar]
- 64.Chute JP, Muramoto GG, Whitesides J, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;103:11707–12. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He X, Gonzalez V, Tsang A, et al. Differential gene expression profiling of CD34+ CD133+ umbilical cord blood hematopoietic stem progenitor cells. Stem Cells Dev. 2005;14:188–98. doi: 10.1089/scd.2005.14.188. [DOI] [PubMed] [Google Scholar]
- 66.Ueshima Y, Matsuda Y, Tsutsumi M, Takada A. Role of the aldehyde dehydrogenase-1 isozyme in the metabolism of acetaldehyde. Alcohol Alcohol. 1993;1B(Suppl):15–9. doi: 10.1093/alcalc/28.supplement_1b.15. [DOI] [PubMed] [Google Scholar]
- 67.Ward RJ, McPherson AJ, Chow C, et al. Identification and characterisation of alcohol-induced flushing in Caucasian subjects. Alcohol Alcohol. 1994;29:433–8. [PubMed] [Google Scholar]
- 68.Yoshida A, Dave V, Ward RJ, Peters TJ. Cytosolic aldehyde dehydrogenase (ALDH1) variants found in alcohol flushers. Ann Hum Genet. 1989;53:1–7. doi: 10.1111/j.1469-1809.1989.tb01116.x. [DOI] [PubMed] [Google Scholar]
- 69.Bhave SV, Hoffman PL, Lassen N, et al. Gene array profiles of alcohol and aldehyde metabolizing enzymes in brains of C57BL/6 and DBA/2 mice. Alcohol Clin Exp Res. 2006;30:1659–69. doi: 10.1111/j.1530-0277.2006.00201.x. [DOI] [PubMed] [Google Scholar]
- 70.Manzer R, Qamar L, Estey T, et al. Molecular cloning and baculovirus expression of the rabbit corneal aldehyde dehydrogenase (ALDH1A1) cDNA. DNA Cell Biol. 2003;22:329–38. doi: 10.1089/104454903322216671. [DOI] [PubMed] [Google Scholar]
- 71.Sladek NE. Aldehyde dehydrogenase-mediated cellular relative insensitivity to the oxazaphosphorines. Curr Pharm Des. 1999;5:607–25. [PubMed] [Google Scholar]
- 72.Moreb JS, Mohuczy D, Ostmark B, Zucali JR. RNAi-mediated knockdown of aldehyde dehydrogenase class-1A1 and class-3A1 is specific and reveals that each contributes equally to the resistance against 4-hydroperoxycyclophosphamide. Cancer Chemother Pharmacol. 2007;59:127–36. doi: 10.1007/s00280-006-0233-6. [DOI] [PubMed] [Google Scholar]
- 73.Sladek NE, Kollander R, Sreerama L, Kiang DT. Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Rational individualization of oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer Chemother Pharmacol. 2002;49:309–21. doi: 10.1007/s00280-001-0412-4. [DOI] [PubMed] [Google Scholar]
- 74.Sladek NE. Metabolism and pharmacokinetic behavior of cyclophosphamide and related oxazaphosphorines. Oxford; England: 1994. pp. 79–156. [Google Scholar]
- 75.Banfi P, Lanzi C, Falvella FS, et al. The daunorubicin-binding protein of Mr 54,000 is an aldehyde dehydrogenase and is down-regulated in mouse liver tumors and in tumor cell lines. Mol Pharmacol. 1994;46:896–900. [PubMed] [Google Scholar]
- 76.Schnier JB, Kaur G, Kaiser A, et al. Identification of cytosolic aldehyde dehydrogenase 1 from non-small cell lung carcinomas as a flavopiridol-binding protein. FEBS Lett. 1999;454:100–4. doi: 10.1016/s0014-5793(99)00773-5. [DOI] [PubMed] [Google Scholar]
- 77.Velazquez-Fernandez D, Laurell C, Geli J, et al. Expression profiling of adrenocortical neoplasms suggests a molecular signature of malignancy. Surgery. 2005;138:1087–94. doi: 10.1016/j.surg.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 78.Park YD, Lyou YJ, Yang JM. Detection of down-regulated acetaldehyde dehydrogenase 1 in atopic dermatitis patients by two-dimensional electrophoresis. Exp Dermatol. 2007;16:130–4. doi: 10.1111/j.1600-0625.2006.00524.x. [DOI] [PubMed] [Google Scholar]
- 79.Sidhu RS, Blair AH. Human liver aldehyde dehydrogenase. Esterase activity. J Biol Chem. 1975;250:7894–8. [PubMed] [Google Scholar]
- 80.Collard F, Vertommen D, Fortpied J, et al. Identification of 3-deoxyglucosone dehydrogenase as aldehyde dehydrogenase 1A1 (retinaldehyde dehydrogenase 1) Biochimie. 2007;89:369–73. doi: 10.1016/j.biochi.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou J, Weiner H. Binding of thyroxine analogs to human liver aldehyde dehydrogenases. Eur J Biochem. 1997;245:123–8. doi: 10.1111/j.1432-1033.1997.00123.x. [DOI] [PubMed] [Google Scholar]
- 82.Deng L, Shipley GL, Loose-Mitchell DS, et al. Coordinate regulation of the production and signaling of retinoic acid by estrogen in the human endometrium. J Clin Endocrinol Metab. 2003;88:2157–63. doi: 10.1210/jc.2002-021844. [DOI] [PubMed] [Google Scholar]
- 83.Hsu LC, Chang WC, Yoshida A. Mouse type-2 retinaldehyde dehydrogenase (RALDH2): genomic organization, tissue-dependent expression, chromosome assignment and comparison to other types. Biochim Biophys Acta. 2000;1492:289–93. doi: 10.1016/s0167-4781(00)00108-1. [DOI] [PubMed] [Google Scholar]
- 84.Gagnon I, Duester G, Bhat PV. Kinetic analysis of mouse retinal dehydrogenase type-2 (RALDH2) for retinal substrates. Biochim Biophys Acta. 2002;1596:156–62. doi: 10.1016/s0167-4838(02)00213-3. [DOI] [PubMed] [Google Scholar]
- 85.Lamb AL, Newcomer ME. The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity. Biochemistry. 1999;38:6003–11. doi: 10.1021/bi9900471. [DOI] [PubMed] [Google Scholar]
- 86.Maden M. Heads or tails? Retinoic acid will decide. Bioessays. 1999;21:809–12. doi: 10.1002/(SICI)1521-1878(199910)21:10<809::AID-BIES2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 87.Niederreither K, Vermot J, Le RI, et al. The regional pattern of retinoic acid synthesis by RALDH2 is essential for the development of posterior pharyngeal arches and the enteric nervous system. Development. 2003;130:2525–34. doi: 10.1242/dev.00463. [DOI] [PubMed] [Google Scholar]
- 88.Vermot J, Niederreither K, Garnier JM, et al. Decreased embryonic retinoic acid synthesis results in a DiGeorge syndrome phenotype in newborn mice. Proc Natl Acad Sci USA. 2003;100:1763–8. doi: 10.1073/pnas.0437920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lai L, Bohnsack BL, Niederreither K, Hirschi KK. Retinoic acid regulates endothelial cell proliferation during vasculogenesis. Development. 2003;130:6465–74. doi: 10.1242/dev.00887. [DOI] [PubMed] [Google Scholar]
- 90.Cartry J, Nichane M, Ribes V, et al. Retinoic acid signalling is required for specification of pronephric cell fate. Dev Biol. 2006;299:35–51. doi: 10.1016/j.ydbio.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 91.Mic FA, Molotkov A, Molotkova N, Duester G. Raldh2 expression in optic vesicle generates a retinoic acid signal needed for invagination of retina during optic cup formation. Dev Dyn. 2004;231:270–7. doi: 10.1002/dvdy.20128. [DOI] [PubMed] [Google Scholar]
- 92.Desai TJ, Chen F, Lu J, et al. Distinct roles for retinoic acid receptors alpha and beta in early lung morphogenesis. Dev Biol. 2006;291:12–24. doi: 10.1016/j.ydbio.2005.10.045. [DOI] [PubMed] [Google Scholar]
- 93.Ribes V, Wang Z, Dolle P, Niederreither K. Retinaldehyde dehydrogenase 2 (RALDH2)-mediated retinoic acid synthesis regulates early mouse embryonic forebrain development by controlling FGF and sonic hedgehog signaling. Development. 2006;133:351–61. doi: 10.1242/dev.02204. [DOI] [PubMed] [Google Scholar]
- 94.Martin M, Gallego-Llamas J, Ribes V, et al. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- 95.Deak KL, Dickerson ME, Linney E, et al. Analysis of ALDH1A2, CYP26A1, CYP26B1, CRABP1, and CRABP2 in human neural tube defects suggests a possible association with alleles in ALDH1A2. Birth Defects. Res A Clin Mol Teratol. 2005;73:868–75. doi: 10.1002/bdra.20183. [DOI] [PubMed] [Google Scholar]
- 96.Kristoffersson U, Heim S, Mandahl N, et al. Monosomy and trisomy of 15q24 – qter in a family with a translocation t(6;15)(p25;q24) Clin Genet. 1987;32:169–71. doi: 10.1111/j.1399-0004.1987.tb03348.x. [DOI] [PubMed] [Google Scholar]
- 97.Mey J, Babiuk RP, Clugston R, et al. Retinal dehydrogenase-2 is inhibited by compounds that induce congenital diaphragmatic hernias in rodents. Am J Pathol. 2003;162:673–9. doi: 10.1016/s0002-9440(10)63861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seitz HK. Alcohol and retinoid metabolism. Gut. 2000;47:748–50. doi: 10.1136/gut.47.6.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fatma N, Kubo E, Chylack LT, Jr, et al. LEDGF regulation of alcohol and aldehyde dehydrogenases in lens epithelial cells: stimulation of retinoic acid production and protection from ethanol toxicity. Am J Physiol Cell Physiol. 2004;287:C508–16. doi: 10.1152/ajpcell.00076.2004. [DOI] [PubMed] [Google Scholar]
- 100.De Luca LM. Retinoids and their receptors in differentiation, embryogenesis, and neoplasia. FASEB J. 1991;5:2924–33. [PubMed] [Google Scholar]
- 101.Kim H, Lapointe J, Kaygusuz G, et al. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 2005;65:8118–24. doi: 10.1158/0008-5472.CAN-04-4562. [DOI] [PubMed] [Google Scholar]
- 102.Graham CE, Brocklehurst K, Pickersgill RW, Warren MJ. Characterization of retinaldehyde dehydrogenase 3. Biochem J. 2006;394:67–75. doi: 10.1042/BJ20050918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hsu LC, Chang WC, Hiraoka L, Hsieh CL. Molecular cloning, genomic organization, and chromosomal localization of an additional human aldehyde dehydrogenase gene, ALDH6. Genomics. 1994;24:333–41. doi: 10.1006/geno.1994.1624. [DOI] [PubMed] [Google Scholar]
- 104.Zhang X, Zhang QY, Liu D, et al. Expression of cytochrome p450 and other biotransformation genes in fetal and adult human nasal mucosa. Drug Metab Dispos. 2005;33:1423–8. doi: 10.1124/dmd.105.005769. [DOI] [PubMed] [Google Scholar]
- 105.Molotkov A, Molotkova N, Duester G. Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development. 2006;133:1901–10. doi: 10.1242/dev.02328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wagner E, Luo T, Drager UC. Retinoic acid synthesis in the postnatal mouse brain marks distinct developmental stages and functional systems. Cereb Cortex. 2002;12:1244–53. doi: 10.1093/cercor/12.12.1244. [DOI] [PubMed] [Google Scholar]
- 107.Everts HB, Sundberg JP, King LE, Jr, Ong DE. Immunolocalization of enzymes, binding proteins, and receptors sufficient for retinoic acid synthesis and signaling during the hair cycle. J Invest Dermatol. 2007;127:1593–604. doi: 10.1038/sj.jid.5700753. [DOI] [PubMed] [Google Scholar]
- 108.Molotkova N, Molotkov A, Duester G. Role of retinoic acid during forebrain development begins late when Raldh3 generates retinoic acid in the ventral subventricular zone. Dev Biol. 2007;303:601–10. doi: 10.1016/j.ydbio.2006.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wagner E, Luo T, Sakai Y, et al. Retinoic acid delineates the topography of neuronal plasticity in postnatal cerebral cortex. Eur J Neurosci. 2006;24:329–40. doi: 10.1111/j.1460-9568.2006.04934.x. [DOI] [PubMed] [Google Scholar]
- 110.Rexer BN, Zheng WL, Ong DE. Retinoic acid biosynthesis by normal human breast epithelium is via aldehyde dehydrogenase 6, absent in MCF-7 cells. Cancer Res. 2001;61:7065–70. [PubMed] [Google Scholar]
- 111.Okamura S, Ng CC, Koyama K, et al. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncol Res. 1999;11:281–5. [PubMed] [Google Scholar]
- 112.Koch JG, Gu X, Han Y, et al. Mammary tumor modifiers in BALB/cJ mice heterozygous for p53. Mamm Genome. 2007;18:300–9. doi: 10.1007/s00335-007-9028-2. [DOI] [PubMed] [Google Scholar]
- 113.Yamashita S, Tsujino Y, Moriguchi K, et al. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2′-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97:64–71. doi: 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han J, Yang L, Puri RK. Analysis of target genes induced by IL-13 cytotoxin in human glioblastoma cells. J Neurooncol. 2005;72:35–46. doi: 10.1007/s11060-004-3119-7. [DOI] [PubMed] [Google Scholar]
- 115.Nakazawa N, Montedonico S, Takayasu H, et al. Disturbance of retinol transportation causes nitrofen-induced hypoplastic lung. J Pediatr Surg. 2007;42:345–9. doi: 10.1016/j.jpedsurg.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 116.Hsu LC, Chang WC. Cloning and characterization of a new functional human aldehyde dehydrogenase gene. J Biol Chem. 1991;266:12257–65. [PubMed] [Google Scholar]
- 117.Stewart MJ, Malek K, Xiao Q, et al. The novel aldehyde dehydrogenase gene, ALDH5, encodes an active aldehyde dehydrogenase enzyme. Biochem Biophys Res Commun. 1995;211:144–51. doi: 10.1006/bbrc.1995.1789. [DOI] [PubMed] [Google Scholar]
- 118.Crabb DW, Stewart MJ, Xiao Q. Hormonal and chemical influences on the expression of class 2 aldehyde dehydrogenases in rat H4IIEC3 and human HuH7 hepatoma cells. Alcohol Clin Exp Res. 1995;19:1414–9. doi: 10.1111/j.1530-0277.1995.tb01000.x. [DOI] [PubMed] [Google Scholar]
- 119.Algar EM, Cheung B, Hayes J, et al. Bovine corneal aldehyde dehydrogenases: evidence for multiple gene products (ALDH3 and ALDHX) Adv Exp Med Biol. 1993;328:153–7. doi: 10.1007/978-1-4615-2904-0_17. [DOI] [PubMed] [Google Scholar]
- 120.Krupenko SA, Horstman DA, Wagner C, Cook RJ. Baculovirus expression and purification of rat 10-formyltetrahydrofolate dehydrogenase. Protein Expr Purif. 1995;6:457–64. doi: 10.1006/prep.1995.1061. [DOI] [PubMed] [Google Scholar]
- 121.Tsybovsky Y, Donato H, Krupenko NI, et al. Crystal structures of the carboxyl terminal domain of rat 10-formyltetrahydrofolate dehydrogenase: implications for the catalytic mechanism of aldehyde dehydrogenases. Biochemistry. 2007;46:2917–29. doi: 10.1021/bi0619573. [DOI] [PubMed] [Google Scholar]
- 122.Krupenko SA, Oleinik NV. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth Differ. 2002;13:227–36. [PubMed] [Google Scholar]
- 123.Martinasevic MK, Green MD, Baron J, Tephly TR. Folate and 10-formyltetrahydrofolate dehydrogenase in human and rat retina: relation to methanol toxicity. Toxicol Appl Pharmacol. 1996;141:373–81. doi: 10.1006/taap.1996.0302. [DOI] [PubMed] [Google Scholar]
- 124.Kutzbach C, Stokstad EL. Mammalian methylenetetrahydrofolate reductase. Partial purification, properties, and inhibition by S-adenosylmethionine. Biochim Biophys Acta. 1971;250:459–77. doi: 10.1016/0005-2744(71)90247-6. [DOI] [PubMed] [Google Scholar]
- 125.Champion KM, Cook RJ, Tollaksen SL, Giometti CS. Identification of a heritable deficiency of the folate-dependent enzyme 10-formyltetrahydrofolate dehydrogenase in mice. Proc Natl Acad Sci USA. 1994;91:11338–42. doi: 10.1073/pnas.91.24.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Oleinik NV, Krupenko NI, Priest DG, Krupenko SA. Cancer cells activate p53 in response to 10-formyltetrahydrofolate dehydrogenase expression. Biochem J. 2005;391:503–11. doi: 10.1042/BJ20050533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Oleinik NV, Krupenko SA. Ectopic expression of 10-formyltetrahydrofolate dehydrogenase in A549 cells induces G1 cell cycle arrest and apoptosis. Mol Cancer Res. 2003;1:577–88. [PubMed] [Google Scholar]
- 128.Anthony TE, Heintz N. The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. J Comp Neurol. 2007;500:368–83. doi: 10.1002/cne.21179. [DOI] [PubMed] [Google Scholar]
- 129.Stevens VL, McCullough ML, Pavluck AL, et al. Association of polymorphisms in one-carbon metabolism genes and postmenopausal breast cancer incidence. Cancer Epidemiol Biomarkers Prev. 2007;16:1140–7. doi: 10.1158/1055-9965.EPI-06-1037. [DOI] [PubMed] [Google Scholar]
- 130.Johlin FC, Swain E, Smith C, Tephly TR. Studies on the mechanism of methanol poisoning: purification and comparison of rat and human liver 10-formyltetrahydrofolate dehydrogenase. Mol Pharmacol. 1989;35:745–50. [PubMed] [Google Scholar]
- 131.Garner CD, Lee EW, Louis-Ferdinand RT. Muller cell involvement in methanol-induced retinal toxicity. Toxicol Appl Pharmacol. 1995;130:101–7. doi: 10.1006/taap.1995.1014. [DOI] [PubMed] [Google Scholar]
- 132.Schalinske KL, Steele RD. Methotrexate alters carbon flow through the hepatic folate-dependent one-carbon pool in rats. Carcinogenesis. 1996;17:1695–700. doi: 10.1093/carcin/17.8.1695. [DOI] [PubMed] [Google Scholar]
- 133.Min H, Im ES, Seo JS, et al. Effects of chronic ethanol ingestion and folate deficiency on the activity of 10-formyltetrahydrofolate dehydrogenase in rat liver. Alcohol Clin Exp Res. 2005;29:2188–93. doi: 10.1097/01.alc.0000191756.02856.a8. [DOI] [PubMed] [Google Scholar]
- 134.Pumford NR, Halmes NC, Martin BM, et al. Covalent binding of acetaminophen to N-10-formyltetrahydrofolate dehydrogenase in mice. J Pharmacol Exp Ther. 1997;280:501–5. [PubMed] [Google Scholar]
- 135.Jongeneel CV, Delorenzi M, Iseli C, et al. An atlas of human gene expression from massively parallel signature sequencing (MPSS) Genome Res. 2005;15:1007–14. doi: 10.1101/gr.4041005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ackerstaff E, Gimi B, Artemov D, Bhujwalla ZM. Anti-inflammatory agent indomethacin reduces invasion and alters metabolism in a human breast cancer cell line. Neoplasia. 2007;9:222–35. doi: 10.1593/neo.06673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Goedde HW, Agarwal DP. Pharmacogenetics of aldehyde dehydrogenase (ALDH) Pharmacol Ther. 1990;45:345–71. doi: 10.1016/0163-7258(90)90071-9. [DOI] [PubMed] [Google Scholar]
- 138.Klyosov AA, Rashkovetsky LG, Tahir MK, Keung WM. Possible role of liver cytosolic and mitochondrial aldehyde dehydrogenases in acetaldehyde metabolism. Biochemistry. 1996;35:4445–56. doi: 10.1021/bi9521093. [DOI] [PubMed] [Google Scholar]
- 139.Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA. 1984;81:258–61. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Farres J, Wang X, Takahashi K, et al. Effects of changing glutamate 487 to lysine in rat and human liver mitochondrial aldehyde dehydrogenase. A model to study human (Oriental type) class 2 aldehyde dehydrogenase. J Biol Chem. 1994;269:13854–60. [PubMed] [Google Scholar]
- 141.Larson HN, Zhou J, Chen Z, et al. Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487. J Biol Chem. 2007;282:12940–50. doi: 10.1074/jbc.M607959200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chao YC, Liou SR, Tsai SF, Yin SJ. Dominance of the mutant ALDH2 2;allele in the expression of human stomach aldehyde dehydrogenase-2 activity. Proc Natl Sci Counc Repub China B. 1993;17:98–102. [PubMed] [Google Scholar]
- 143.Goedde HW, Agarwal DP, Fritze G, et al. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet. 1992;88:344–6. doi: 10.1007/BF00197271. [DOI] [PubMed] [Google Scholar]
- 144.Wall TL, Johnson ML, Horn SM, et al. Evaluation of the self-rating of the effects of alcohol form in Asian Americans with aldehyde dehydrogenase polymorphisms. J Stud Alcohol. 1999;60:784–9. doi: 10.15288/jsa.1999.60.784. [DOI] [PubMed] [Google Scholar]
- 145.Peng GS, Chen YC, Tsao TP, et al. Pharmacokinetic and pharmacodynamic basis for partial protection against alcoholism in Asians, heterozygous for the variant ALDH2*2 gene allele. Pharmacogenet Genomics. 2007;17:845–55. doi: 10.1097/FPC.0b013e3282609e67. [DOI] [PubMed] [Google Scholar]
- 146.Luczak SE, Elvine-Kreis B, Shea SH, et al. Genetic risk for alcoholism relates to level of response to alcohol in Asian-American men and women. J Stud Alcohol. 2002;63:74–82. [PubMed] [Google Scholar]
- 147.Muto M, Hitomi Y, Ohtsu A, et al. Association of aldehyde dehydrogenase 2 gene polymorphism with multiple oesophageal dysplasia in head and neck cancer patients. Gut. 2000;47:256–61. doi: 10.1136/gut.47.2.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Matsuda T, Yabushita H, Kanaly RA, et al. Increased DNA damage in ALDH2-deficient alcoholics. Chem Res Toxicol. 2006;19:1374–8. doi: 10.1021/tx060113h. [DOI] [PubMed] [Google Scholar]
- 149.Wong RH, Wang JD, Hsieh LL, et al. Effects on sister chromatid exchange frequency of aldehyde dehydrogenase 2 genotype and smoking in vinyl chloride workers. Mutat Res. 1998;420:99–107. [PubMed] [Google Scholar]
- 150.Spengler SJ, Singer B. Formation of interstrand cross-links in chloroacetaldehyde-treated DNA demonstrated by ethidium bromide fluorescence. Cancer Res. 1988;48:4804–6. [PubMed] [Google Scholar]
- 151.Li Y, Zhang D, Jin W, et al. Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J Clin Invest. 2006;116:506–11. doi: 10.1172/JCI26564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jo SA, Kim EK, Park MH, et al. A Glu487Lys polymorphism in the gene for mitochondrial aldehyde dehydrogenase 2 is associated with myocardial infarction in elderly Korean men. Clin Chim Acta. 2007;382:43–7. doi: 10.1016/j.cca.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 153.Hui P, Nakayama T, Morita A, et al. Common single nucleotide polymorphisms in Japanese patients with essential hypertension: aldehyde dehydrogenase 2 gene as a risk factor independent of alcohol consumption. Hypertens Res. 2007;30:585–92. doi: 10.1291/hypres.30.585. [DOI] [PubMed] [Google Scholar]
- 154.Isse T, Oyama T, Matsuno K, et al. Paired acute inhalation test reveals that acetaldehyde toxicity is higher in aldehyde dehydrogenase 2 knockout mice than in wild-type mice. J Toxicol Sci. 2005;30:329–37. doi: 10.2131/jts.30.329. [DOI] [PubMed] [Google Scholar]
- 155.Isse T, Oyama T, Matsuno K, et al. Aldehyde dehydrogenase 2 activity affects symptoms produced by an intraperitoneal acetaldehyde injection, but not acetaldehyde lethality. J Toxicol Sci. 2005;30:315–28. doi: 10.2131/jts.30.315. [DOI] [PubMed] [Google Scholar]
- 156.Ogawa M, Isse T, Oyama T, et al. Urinary 8-hydoxydeoxyguanosine (8-OHdG) and plasma malondialdehyde (MDA) levels in Aldh2 knock-out mice under acetaldehyde exposure. Ind Health. 2006;44:179–83. doi: 10.2486/indhealth.44.179. [DOI] [PubMed] [Google Scholar]
- 157.Ogawa M, Oyama T, Isse T, et al. A comparison of covalent binding of ethanol metabolites to DNA according to Aldh2 genotype. Toxicol Lett. 2007;168:148–54. doi: 10.1016/j.toxlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 158.Fischer M, Wetherill LF, Carr LG, et al. Association of the aldehyde dehydrogenase 2 promoter polymorphism with alcohol consumption and reactions in an American Jewish population. Alcohol Clin Exp Res. 2007;31:1654–9. doi: 10.1111/j.1530-0277.2007.00471.x. [DOI] [PubMed] [Google Scholar]
- 159.Reichard JF, Vasiliou V, Petersen DR. Characterization of 4-hydroxy-2-nonenal metabolism in stellate cell lines derived from normal and cirrhotic rat liver. Biochim Biophys Acta. 2000;1487:222–32. doi: 10.1016/s1388-1981(00)00095-0. [DOI] [PubMed] [Google Scholar]
- 160.Luckey SW, Petersen DR. Metabolism of 4-hydroxynonenal by rat Kupffer cells. Arch Biochem Biophys. 2001;389:77–83. doi: 10.1006/abbi.2001.2307. [DOI] [PubMed] [Google Scholar]
- 161.Picklo MJ, Olson SJ, Markesbery WR, Montine TJ. Expression and activities of aldo-keto oxidoreductases in Alzheimer disease. J Neuropathol Exp Neurol. 2001;60:686–95. doi: 10.1093/jnen/60.7.686. [DOI] [PubMed] [Google Scholar]
- 162.Ohsawa I, Nishimaki K, Yasuda C, et al. Deficiency in a mitochondrial aldehyde dehydrogenase increases vulnerability to oxidative stress in PC12 cells. J Neurochem. 2003;84:1110–7. doi: 10.1046/j.1471-4159.2003.01619.x. [DOI] [PubMed] [Google Scholar]
- 163.Wang B, Wang J, Zhou S, et al. The association of mitochondrial aldehyde dehydrogenase gene (ALDH2) polymorphism with susceptibility to late-onset alzheimer’s disease in Chinese. J neurol Sci. 2008;268(1–2):172–5. doi: 10.1016/j.jns.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 164.Maring JA, Deitrich RA, Little R. Partial purification and properties of human brain aldehyde dehydrogenases. J Neurochem. 1985;45:1903–10. doi: 10.1111/j.1471-4159.1985.tb10550.x. [DOI] [PubMed] [Google Scholar]
- 165.MacKerell AD, Jr, Blatter EE, Pietruszko R. Human aldehyde dehydrogenase: kinetic identification of the isozyme for which biogenic aldehydes and acetaldehyde compete. Alcohol Clin Exp Res. 1986;10:266–70. doi: 10.1111/j.1530-0277.1986.tb05087.x. [DOI] [PubMed] [Google Scholar]
- 166.•.Estey T, Piatigorsky J, Lassen N, Vasiliou V. ALDH3A1: a corneal crystallin with diverse functions. Exp Eye Res. 2007;84:3–12. doi: 10.1016/j.exer.2006.04.010. A very informative paper on the many roles of ALDH3A1. [DOI] [PubMed] [Google Scholar]
- 167.Pappa A, Estey T, Manzer R, et al. Human aldehyde dehydrogenase 3A1 (ALDH3A1): biochemical characterization and immunohistochemical localization in the cornea. Biochem J. 2003;376:615–23. doi: 10.1042/BJ20030810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pappa A, Sophos NA, Vasiliou V. Corneal and stomach expression of aldehyde dehydrogenases: from fish to mammals. Chem Biol Interact. 2001;130–2:181–91. doi: 10.1016/s0009-2797(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 169.Nees DW, Wawrousek EF, Robison WG, Jr, Piatigorsky J. Structurally normal corneas in aldehyde dehydrogenase 3a1-deficient mice. Mol Cell Biol. 2002;22:849–55. doi: 10.1128/MCB.22.3.849-855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pei Y, Reins RY, McDermott AM. Aldehyde dehydrogenase (ALDH) 3A1 expression by the human keratocyte and its repair phenotypes. Exp Eye Res. 2006;83:1063–73. doi: 10.1016/j.exer.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 171.Kirsch M, De GH. NAD(P)H, a directly operating antioxidant? FASEB J. 2001;15:1569–74. doi: 10.1096/fj.00-0823hyp. [DOI] [PubMed] [Google Scholar]
- 172.Abedinia M, Pain T, Algar EM, Holmes RS. Bovine corneal aldehyde dehydrogenase: the major soluble corneal protein with a possible dual protective role for the eye. Exp Eye Res. 1990;51:419–26. doi: 10.1016/0014-4835(90)90154-m. [DOI] [PubMed] [Google Scholar]
- 173.Downes JE, Swann PG, Holmes RS. Ultraviolet light-induced pathology in the eye: associated changes in ocular aldehyde dehydrogenase and alcohol dehydrogenase activities. Cornea. 1993;12:241–8. doi: 10.1097/00003226-199305000-00010. [DOI] [PubMed] [Google Scholar]
- 174.Canuto RA, Muzio G, Ferro M, et al. Inhibition of class-3 aldehyde dehydrogenase and cell growth by restored lipid peroxidation in hepatoma cell lines. Free Radic Biol Med. 1999;26:333–40. doi: 10.1016/s0891-5849(98)00206-8. [DOI] [PubMed] [Google Scholar]
- 175.Muzio G, Trombetta A, Martinasso G, et al. Antisense oligonucleotides against aldehyde dehydrogenase 3 inhibit hepatoma cell proliferation by affecting MAP kinases. Chem Biol Interact. 2003;143–4:37–43. doi: 10.1016/s0009-2797(02)00170-9. [DOI] [PubMed] [Google Scholar]
- 176.Manthey CL, Landkamer GJ, Sladek NE. Identification of the mouse aldehyde dehydrogenases important in aldophosphamide detoxification. Cancer Res. 1990;50:4991–5002. [PubMed] [Google Scholar]
- 177.Sreerama L, Sladek NE. Identification and characterization of a novel class 3 aldehyde dehydrogenase overexpressed in a human breast adenocarcinoma cell line exhibiting oxazaphosphorine-specific acquired resistance. Biochem Pharmacol. 1993;45:2487–505. doi: 10.1016/0006-2952(93)90231-k. [DOI] [PubMed] [Google Scholar]
- 178.Sreerama L, Sladek NE. Cellular levels of class 1 and class 3 aldehyde dehydrogenases and certain other drug-metabolizing enzymes in human breast malignancies. Clin Cancer Res. 1997;3:1901–14. [PubMed] [Google Scholar]
- 179.Wang JS, Fang Q, Sun DJ, et al. Genetic modification of hematopoietic progenitor cells for combined resistance to 4-hydroperoxycyclophosphamide, vincristine, and daunorubicin. Acta Pharmacol Sin. 2001;22:949–55. [PubMed] [Google Scholar]
- 180.Kim B, Lee HJ, Choi HY, et al. Clinical validity of the lung cancer biomarkers identified by bioinformatics analysis of public expression data. Cancer Res. 2007;67:7431–8. doi: 10.1158/0008-5472.CAN-07-0003. [DOI] [PubMed] [Google Scholar]
- 181.Huang J, Hu N, Goldstein AM, et al. High frequency allelic loss on chromosome 17p13.3-p11.1 in esophageal squamous cell carcinomas from a high incidence area in northern China. Carcinogenesis. 2000;21:2019–26. doi: 10.1093/carcin/21.11.2019. [DOI] [PubMed] [Google Scholar]
- 182.Canuto RA, Ferro M, Muzio G, et al. Role of aldehyde metabolizing enzymes in mediating effects of aldehyde products of lipid peroxidation in liver cells. Carcinogenesis. 1994;15:1359–64. doi: 10.1093/carcin/15.7.1359. [DOI] [PubMed] [Google Scholar]
- 183.Sreerama L, Sladek NE. Class 1 and class 3 aldehyde dehydrogenase levels in the human tumor cell lines currently used by the National Cancer Institute to screen for potentially useful antitumor agents. Adv Exp Med Biol. 1997;414:81–94. doi: 10.1007/978-1-4615-5871-2_11. [DOI] [PubMed] [Google Scholar]
- 184.Stephanou P, Pappas P, Vasiliou V, Marselos M. Prepubertal regulation of the rat dioxin-inducible aldehyde dehydrogenase (ALDH3) Adv Exp Med Biol. 1999;463:143–50. doi: 10.1007/978-1-4615-4735-8_17. [DOI] [PubMed] [Google Scholar]
- 185.Vasiliou V, Reuter SF, Kozak CA, Nebert DW. Mouse dioxin-inducible cytosolic aldehyde dehydrogenase-3: AHD4 cDNA sequence, genetic mapping, and differences in mRNA levels. Pharmacogenetics. 1993;3:281–90. doi: 10.1097/00008571-199312000-00002. [DOI] [PubMed] [Google Scholar]
- 186.Reisdorph R, Lindahl R. Constitutive and 3-methylcholanthrene-induced rat ALDH3A1 expression is mediated by multiple xenobiotic response elements. Drug Metab Dispos. 2007;35:386–93. doi: 10.1124/dmd.106.012393. [DOI] [PubMed] [Google Scholar]
- 187.Chang C, Yoshida A. Human fatty aldehyde dehydrogenase gene (ALDH10): organization and tissue-dependent expression. Genomics. 1997;40:80–5. doi: 10.1006/geno.1996.4547. [DOI] [PubMed] [Google Scholar]
- 188.Rogers GR, Markova NG, De LV, et al. Genomic organization and expression of the human fatty aldehyde dehydrogenase gene (FALDH) Genomics. 1997;39:127–35. doi: 10.1006/geno.1996.4501. [DOI] [PubMed] [Google Scholar]
- 189.Ashibe B, Hirai T, Higashi K, et al. Dual subcellular localization in the endoplasmic reticulum and peroxisomes and a vital role in protecting against oxidative stress of fatty aldehyde dehydrogenase are achieved by alternative splicing. J Biol Chem. 2007;282:20763–73. doi: 10.1074/jbc.M611853200. [DOI] [PubMed] [Google Scholar]
- 190.Ichihara K, Kusunose E, Noda Y, Kusunose M. Some properties of the fatty alcohol oxidation system and reconstitution of microsomal oxidation activity in intestinal mucosa. Biochim Biophys Acta. 1986;878:412–8. doi: 10.1016/0005-2760(86)90250-x. [DOI] [PubMed] [Google Scholar]
- 191.Kelson TL, Secor M, Jr, Rizzo WB. Human liver fatty aldehyde dehydrogenase: microsomal localization, purification, and biochemical characterization. Biochim Biophys Acta. 1997;1335:99–110. doi: 10.1016/s0304-4165(96)00126-2. [DOI] [PubMed] [Google Scholar]
- 192.Rizzo WB, Craft DA. Sjogren-Larsson syndrome. Deficient activity of the fatty aldehyde dehydrogenase component of fatty alcohol:NAD+ oxidoreductase in cultured fibroblasts. J Clin Invest. 1991;88:1643–8. doi: 10.1172/JCI115478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Verhoeven NM, Jakobs C, Carney G, et al. Involvement of microsomal fatty aldehyde dehydrogenase in the alpha-oxidation of phytanic acid. FEBS Lett. 1998;429:225–8. doi: 10.1016/s0014-5793(98)00574-2. [DOI] [PubMed] [Google Scholar]
- 194.Willemsen MA, de Jong JG, van Domburg PH, et al. Defective inactivation of leukotriene B4 in patients with Sjogren-Larsson syndrome. J Pediatr. 2000;136:258–60. doi: 10.1016/s0022-3476(00)70113-2. [DOI] [PubMed] [Google Scholar]
- 195.Rizzo WB, Heinz E, Simon M, Craft DA. Microsomal fatty aldehyde dehydrogenase catalyzes the oxidation of aliphatic aldehyde derived from ether glycerolipid catabolism: implications for Sjogren-Larsson syndrome. Biochim Biophys Acta. 2000;1535:1–9. doi: 10.1016/s0925-4439(00)00077-6. [DOI] [PubMed] [Google Scholar]
- 196.Rizzo WB. Sjogren-Larsson syndrome: molecular genetics and biochemical pathogenesis of fatty aldehyde dehydrogenase deficiency. Mol Genet Metab. 2007;90:1–9. doi: 10.1016/j.ymgme.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Haug S, Braun-Falco M. Restoration of fatty aldehyde dehydrogenase deficiency in Sjogren-Larsson syndrome. Gene Ther. 2006;13:1021–6. doi: 10.1038/sj.gt.3302743. [DOI] [PubMed] [Google Scholar]
- 198.Demozay D, Rocchi S, Mas JC, et al. Fatty aldehyde dehydrogenase: potential role in oxidative stress protection and regulation of its gene expression by insulin. J Biol Chem. 2004;279:6261–70. doi: 10.1074/jbc.M312062200. [DOI] [PubMed] [Google Scholar]
- 199.Traverso N, Menini S, Odetti P, et al. Diabetes impairs the enzymatic disposal of 4-hydroxynonenal in rat liver. Free Radic Biol Med. 2002;32:350–9. doi: 10.1016/s0891-5849(01)00811-5. [DOI] [PubMed] [Google Scholar]
- 200.Marchitti SA, Orlicky DJ, Vasiliou V. Expression and initial characterization of human ALDH3B1. Biochem Biophys Res Commun. 2007;356:792–8. doi: 10.1016/j.bbrc.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xu Q, Jia YB, Zhang BY, et al. Association study of an SNP combination pattern in the dopaminergic pathway in paranoid schizophrenia: a novel strategy for complex disorders. Mol Psychiatry. 2004;9:510–21. doi: 10.1038/sj.mp.4001472. [DOI] [PubMed] [Google Scholar]
- 202.Sun X, Jia Y, Zhang X, et al. Multi-locus association study of schizophrenia susceptibility genes with a posterior probability method. Sci China C Life Sci. 2005;48:263–9. doi: 10.1007/BF03183620. [DOI] [PubMed] [Google Scholar]
- 203.Hsu LC, Chang WC, Yoshida A. Human aldehyde dehydrogenase genes, ALDH7 and ALDH8: genomic organization and gene structure comparison. Gene. 1997;189:89–94. doi: 10.1016/s0378-1119(96)00839-6. [DOI] [PubMed] [Google Scholar]
- 204.Shmueli O, Horn-Saban S, Chalifa-Caspi V, et al. GeneNote: whole genome expression profiles in normal human tissues. C R Biol. 2003;326:1067–72. doi: 10.1016/j.crvi.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 205.Hu CA, Lin WW, Valle D. Cloning, characterization, and expression of cDNAs encoding human delta 1-pyrroline-5-carboxylate dehydrogenase. J Biol Chem. 1996;271:9795–800. doi: 10.1074/jbc.271.16.9795. [DOI] [PubMed] [Google Scholar]
- 206.Forte-McRobbie CM, Pietruszko R. Purification and characterization of human liver ‘high Km’ aldehyde dehydrogenase and its identification as glutamic gamma-semialdehyde dehydrogenase. J Biol Chem. 1986;261:2154–63. [PubMed] [Google Scholar]
- 207.Haslett MR, Pink D, Walters B, Brosnan ME. Assay and subcellular localization of pyrroline-5-carboxylate dehydrogenase in rat liver. Biochim Biophys Acta. 2004;1675:81–6. doi: 10.1016/j.bbagen.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 208.Geraghty MT, Vaughn D, Nicholson AJ, et al. Mutations in the Delta1-pyrroline 5-carboxylate dehydrogenase gene cause type II hyperprolinemia. Hum Mol Genet. 1998;7:1411–5. doi: 10.1093/hmg/7.9.1411. [DOI] [PubMed] [Google Scholar]
- 209.Farrant RD, Walker V, Mills GA, et al. Pyridoxal phosphate de-activation by pyrroline-5-carboxylic acid. Increased risk of vitamin B6 deficiency and seizures in hyperprolinemia type II. J Biol Chem. 2001;276:15107–16. doi: 10.1074/jbc.M010860200. [DOI] [PubMed] [Google Scholar]
- 210.Lheureux P, Penaloza A, Gris M. Pyridoxine in clinical toxicology: a review. Eur J Emerg Med. 2005;12:78–85. doi: 10.1097/00063110-200504000-00007. [DOI] [PubMed] [Google Scholar]
- 211.Farres J, Julia P, Pares X. Aldehyde oxidation in human placenta. Purification and properties of 1-pyrroline-5-carboxylate dehydrogenase. Biochem J. 1988;256:461–7. doi: 10.1042/bj2560461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yoon KA, Nakamura Y, Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49:134–40. doi: 10.1007/s10038-003-0122-3. [DOI] [PubMed] [Google Scholar]
- 213.Kang JH, Park YB, Huh TL, et al. High-level expression and characterization of the recombinant enzyme, and tissue distribution of human succinic semialdehyde dehydrogenase. Protein Expr Purif. 2005;44:16–22. doi: 10.1016/j.pep.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 214.Gold BI, Roth RH. Kinetics of in vivo conversion of gamma-[3H] aminobutyric acid to gamma-[3H] hydroxybutyric acid by rat brain. J Neurochem. 1977;28:1069–73. doi: 10.1111/j.1471-4159.1977.tb10670.x. [DOI] [PubMed] [Google Scholar]
- 215.Maitre M, Andriamampandry C, Kemmel V, et al. Gamma-hydroxybutyric acid as a signaling molecule in brain. Alcohol. 2000;20:277–83. doi: 10.1016/s0741-8329(99)00092-0. [DOI] [PubMed] [Google Scholar]
- 216.Sauer SW, Kolker S, Hoffmann GF, et al. Enzymatic and metabolic evidence for a region specific mitochondrial dysfunction in brains of murine succinic semialdehyde dehydrogenase deficiency (Aldh5a1−/− mice) Neurochem Int. 2007;50:653–9. doi: 10.1016/j.neuint.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 217.Donarum EA, Stephan DA, Larkin K, et al. Expression profiling reveals multiple myelin alterations in murine succinate semialdehyde dehydrogenase deficiency. J Inherit Metab Dis. 2006;29:143–56. doi: 10.1007/s10545-006-0247-6. [DOI] [PubMed] [Google Scholar]
- 218.Barcelo-Coblijn G, Murphy EJ, Mills K, et al. Lipid abnormalities in succinate semialdehyde dehydrogenase (Aldh5a1−/−) deficient mouse brain provide additional evidence for myelin alterations. Biochim Biophys Acta. 2007;1772:556–62. doi: 10.1016/j.bbadis.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Sgaravatti AM, Sgarbi MB, Testa CG, et al. Gamma-hydroxybutyric acid induces oxidative stress in cerebral cortex of young rats. Neurochem Int. 2007;50:564–70. doi: 10.1016/j.neuint.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 220.Nguyen E, Picklo MJ., Sr Inhibition of succinic semialdehyde dehydrogenase activity by alkenal products of lipid peroxidation. Biochim Biophys Acta. 2003;1637:107–12. doi: 10.1016/s0925-4439(02)00220-x. [DOI] [PubMed] [Google Scholar]
- 221.Kedishvili NY, Popov KM, Rougraff PM, et al. CoA-dependent methylmalonate-semialdehyde dehydrogenase, a unique member of the aldehyde dehydrogenase superfamily. cDNA cloning, evolutionary relationships, and tissue distribution. J Biol Chem. 1992;267:19724–9. [PubMed] [Google Scholar]
- 222.Goodwin GW, Rougraff PM, Davis EJ, Harris RA. Purification and characterization of methylmalonate-semialdehyde dehydrogenase from rat liver. Identity to malonate-semialdehyde dehydrogenase. J Biol Chem. 1989;264:14965–71. [PubMed] [Google Scholar]
- 223.Chambliss KL, Gray RG, Rylance G, et al. Molecular characterization of methylmalonate semialdehyde dehydrogenase deficiency. J Inherit Metab Dis. 2000;23:497–504. doi: 10.1023/a:1005616315087. [DOI] [PubMed] [Google Scholar]
- 224.Kedishvili NY, Popov KM, Jaskiewicz JA, Harris RA. Coordinated expression of valine catabolic enzymes during adipogenesis: analysis of activity, mRNA, protein levels, and metabolic consequences. Arch Biochem Biophys. 1994;315:317–22. doi: 10.1006/abbi.1994.1506. [DOI] [PubMed] [Google Scholar]
- 225.Kanski J, Behring A, Pelling J, Schoneich C. Proteomic identification of 3-nitrotyrosine-containing rat cardiac proteins: effects of biological aging. Am J Physiol Heart Circ Physiol. 2005;288:H371–81. doi: 10.1152/ajpheart.01030.2003. [DOI] [PubMed] [Google Scholar]
- 226.Lee P, Kuhl W, Gelbart T, et al. Homology between a human protein and a protein of the green garden pea. Genomics. 1994;21:371–8. doi: 10.1006/geno.1994.1279. [DOI] [PubMed] [Google Scholar]
- 227.Tang WK, Chan CB, Cheng CH, Fong WP. Seabream antiquitin: molecular cloning, tissue distribution, subcellular localization and functional expression. FEBS Lett. 2005;579:3759–64. doi: 10.1016/j.febslet.2005.05.070. [DOI] [PubMed] [Google Scholar]
- 228.Skvorak AB, Robertson NG, Yin Y, et al. An ancient conserved gene expressed in the human inner ear: identification, expression analysis, and chromosomal mapping of human and mouse antiquitin (ATQ1) Genomics. 1997;46:191–9. doi: 10.1006/geno.1997.5026. [DOI] [PubMed] [Google Scholar]
- 229.Chang YF, Ghosh P, Rao VV. L-pipecolic acid metabolism in human liver: L-alpha-aminoadipate delta-semialdehyde oxidoreductase. Biochim Biophys Acta. 1990;1038:300–5. doi: 10.1016/0167-4838(90)90241-7. [DOI] [PubMed] [Google Scholar]
- 230.Stroeher VL, Boothe JG, Good AG. Molecular cloning and expression of a turgor-responsive gene in Brassica napus. Plant Mol Biol. 1995;27:541–51. doi: 10.1007/BF00019320. [DOI] [PubMed] [Google Scholar]
- 231.Chan CB, Tang WK, Cheng CH, Fong WP. Cloning of the black seabream (Acanthopagrus schlegeli) antiquitin gene and functional characterization of its promoter region. Mol Cell Biochem. 2007;297:151–60. doi: 10.1007/s11010-006-9340-2. [DOI] [PubMed] [Google Scholar]
- 232.Lynch M, Cameron TL, Knight M, et al. Structural and mutational analysis of antiquitin as a candidate gene for Meniere disease. Am J Med Genet. 2002;110:397–99. doi: 10.1002/ajmg.10494. [DOI] [PubMed] [Google Scholar]
- 233.Ellederova Z, Halada P, Man P, et al. Protein patterns of pig oocytes during in vitro maturation. Biol Reprod. 2004;71:1533–9. doi: 10.1095/biolreprod.104.030304. [DOI] [PubMed] [Google Scholar]
- 234.Lin M, Napoli JL. cDNA cloning and expression of a human aldehyde dehydrogenase (ALDH) active with 9-cis-retinal and identification of a rat ortholog, ALDH12. J Biol Chem. 2000;275:40106–12. doi: 10.1074/jbc.M008027200. [DOI] [PubMed] [Google Scholar]
- 235.Gagnon I, Duester G, Bhat PV. Enzymatic characterization of recombinant mouse retinal dehydrogenase type 1. Biochem Pharmacol. 2003;65:1685–90. doi: 10.1016/s0006-2952(03)00150-3. [DOI] [PubMed] [Google Scholar]
- 236.Lin SW, Chen JC, Hsu LC, et al. Human gamma-aminobutyraldehyde dehydrogenase (ALDH9): cDNA sequence, genomic organization, polymorphism, chromosomal localization, and tissue expression. Genomics. 1996;34:376–80. doi: 10.1006/geno.1996.0300. [DOI] [PubMed] [Google Scholar]
- 237.Kurys G, Ambroziak W, Pietruszko R. Human aldehyde dehydrogenase. Purification and characterization of a third isozyme with low Km for gamma-aminobutyraldehyde. J Biol Chem. 1989;264:4715–21. [PubMed] [Google Scholar]
- 238.Ambroziak W, Pietruszko R. Human aldehyde dehydrogenase. Activity with aldehyde metabolites of monoamines, diamines, and polyamines. J Biol Chem. 1991;266:13011–8. [PubMed] [Google Scholar]
- 239.Chern MK, Pietruszko R. Human aldehyde dehydrogenase E3 isozyme is a betaine aldehyde dehydrogenase. Biochem Biophys Res Commun. 1995;213:561–8. doi: 10.1006/bbrc.1995.2168. [DOI] [PubMed] [Google Scholar]
- 240.Vaz FM, Fouchier SW, Ofman R, et al. Molecular and biochemical characterization of rat gamma-trimethylaminobutyraldehyde dehydrogenase and evidence for the involvement of human aldehyde dehydrogenase 9 in carnitine biosynthesis. J Biol Chem. 2000;275:7390–4. doi: 10.1074/jbc.275.10.7390. [DOI] [PubMed] [Google Scholar]
- 241.Wang X, Zhu S, Khan IA, Dasmahapatra AK. Ethanol attenuates Aldh9 mRNA expression in Japanese medaka (Oryzias latipes) embryogenesis. Comp Biochem Physiol B Biochem Mol Biol. 2007;146:357–63. doi: 10.1016/j.cbpb.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 242.Sato W, Horie Y, Kataoka E, et al. Hepatic gene expression in hepatocyte-specific Pten deficient mice showing steatohepatitis without ethanol challenge. Hepatol Res. 2006;34:256–65. doi: 10.1016/j.hepres.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 243.Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–03. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hu CA, Lin WW, Obie C, Valle D. Molecular enzymology of mammalian Delta1-pyrroline-5-carboxylate synthase. Alternative splice donor utilization generates isoforms with different sensitivity to ornithine inhibition. J Biol Chem. 1999;274:6754–62. doi: 10.1074/jbc.274.10.6754. [DOI] [PubMed] [Google Scholar]
- 245.Kamoun P, Aral B, Saudubray JM. A new inherited metabolic disease: delta1-pyrroline 5-carboxylate synthetase deficiency. Bull Acad Natl Med. 1998;182:131–7. [PubMed] [Google Scholar]
- 246.Baumgartner MR, Rabier D, Nassogne MC, et al. Delta1-pyrroline-5-carboxylate synthase deficiency: neurodegeneration, cataracts and connective tissue manifestations combined with hyperammonaemia and reduced ornithine, citrulline, arginine and proline. Eur J Pediatr. 2005;164:31–6. doi: 10.1007/s00431-004-1545-3. [DOI] [PubMed] [Google Scholar]
- 247.Tadros SF, D’Souza M, Zettel ML, et al. Glutamate-related gene expression changes with age in the mouse auditory midbrain. Brain Res. 2007;1127:1–9. doi: 10.1016/j.brainres.2006.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]