Abstract
Neuroscience increasingly uses computational models to assist in the exploration and interpretation of complex phenomena. As a result, considerable effort is invested in the development of software tools and technologies for numerical simulations and for the creation and publication of models. The diversity of related tools leads to the duplication of effort and hinders model reuse. Development practices and technologies that support interoperability between software systems therefore play an important role in making the modeling process more efficient and in ensuring that published models can be reliably and easily reused. Various forms of interoperability are possible including the development of portable model description standards, the adoption of common simulation languages or the use of standardized middleware. Each of these approaches finds applications within the broad range of current modeling activity. However more effort is required in many areas to enable new scientific questions to be addressed. Here we present the conclusions of the “Neuro-IT Interoperability of Simulators” workshop, held at the 11th computational neuroscience meeting in Edinburgh (July 19-20 2006; http://www.cnsorg.org). We assess the current state of interoperability of neural simulation software and explore the future directions that will enable the field to advance.
Keywords: Neural simulation software, Simulation language, Standards, XML, Model publication
Introduction
Computational neuroscience tries to understand or reverse-engineer neural systems using mathematical or algorithmic models. Most of these are too complex to be treated analytically and must be evaluated numerically on a computer. However writing the appropriate simulation software presents a formidable challenge, because it requires knowledge from many disciplines, ranging from neuroscience, over mathematics, to computer science and simulation theory.
The driving force for most simulator development comes from scientific questions that can be addressed with a single modeling system. Over time, different simulation systems have evolved and in some areas, such as the construction of morphologically detailed neurons models, a small number of programs have even emerged as de-facto standards. Here, Neuron (Hines and Carnevale 1997; Carnevale and Hines 2006) and Genesis (Bower and Beeman 1998) have by far the largest user base for whole-cell and small network modeling, and MCell (Stiles and Bartoll 2001) is the tool of choice for stochastic modeling of individual particles in complex geometries. In other domains where concepts and terminology are not as well developed, researchers still write new simulation programs from scratch. Examples are large networks of point neurons, large networks of detailed model neurons (e.g., Brunel and Wang 2001; Traub et al. 2005) or fine-scale simulation of synapses (Roth et al. 2000).
The plethora of simulation programs (e.g. see Brette et al. 2007 for a recent review of eight systems for simulating networks of spiking neurons) poses a number of problems to the scientific community. First, when models are published, only a specification of the model, rather than the complete implementation is presented. As with many applications of computational modeling, such specifications are frequently incomplete (Schwab et al. 2000), so the reader of the publication may be unable to access particular details of the model. Second, even if the simulation code is published, it can still be tedious and error-prone to extract the model. Most new implementations of models are the result of incremental exploratory development and consequently mix model description, data analysis, visualization, and other peripheral code, causing the model description to be hopelessly obfuscated by a much larger body of ancillary material. The result is that the scientific value of publications with results from simulations is often questionable. Standardization on a single well-supported simulation system might solve some of the problems, but it is not a practical solution for most applications, since research inevitably involves frequent modification of models that require changes to the implementation. A more realistic approach is to encourage and facilitate interoperability between different simulators of the same domain, and maybe even across domains. Interoperability comprises all mechanisms that allow two or more simulators to use the same model description or to collaborate by evaluating different parts of a large neural model.
More generally, the interest in interoperability arises from the expectation that various tasks in modeling and software development can be made more efficient and productive by a relatively modest investment in interoperability work. Given the existing investment in disparate software systems, some form of interoperability may yield a valuable return at modest cost. One extreme case where the return is clear would be the possibility to take a complex whole-cell model developed within one system and run it as is, within another system that provides additional features such as large-scale parallelization or parameter searching. However, as discussed later, effortless gains of this nature are rarely achievable. This shifts the focus onto questions of what is technically feasible and what the real benefits would be of different possible strategies. The main goals are that interoperability work should either enable new scientific questions to be addressed or should reduce the time and effort required to develop a system to the level where a particular problem can be tackled.
Here we present the conclusions of the “Neuro-IT Interoperability of Simulators” workshop, held at the 15th computational neuroscience meeting in Edinburgh (July 19-20 2006; http://www.cnsorg.org). The aim of the workshop was to assess the current state of interoperability of neural simulation software and to explore the future directions that will enable the field to advance.
Interoperability can be achieved in a variety of ways that fall into two broad categories. These categories are addressed in turn in the following two sections. The first case corresponds to the example above, where different systems support the same portable model format, so that models built for one simulator can be run on another. In the second flavor, different simulators, working on different domains, interoperate at run-time. Here, we discuss two sub-cases. In the first, a model stays within the simulator for which it was built, but structures are provided to build a ‘super-model’ where different components running in separate systems can be made to interact. The second case involves a much tighter integration where separate software components no longer need to cover all the functions of a stand-alone system and only work within an enclosing environment that provides the necessary infrastructure for them to do their specific task. The third section summarizes open issues, and the final section discusses where further effort can most effectively be applied.
Standardizing Model Descriptions
What is a Model Description Anyway?
Most simulators start out as a monolithic program where the model is defined in the same programming language as the simulator and is interwoven with a large body of code that supports the simulation but is not part of the actual model. As a consequence, each change of the simulation or a parameter requires recompilation of the entire simulator. The second step of maturation introduces a simple interface to set and inspect parameters. This can be a light-weight graphical interface, or may be text-based using a simple configuration language. The third step of maturation introduces a domain-specific programming language that is usually hand-written, but in some sense complete. These languages are usually interpreted and can, in most cases, be used interactively. In the fourth and final step of maturation, developers encapsulate their model implementations within well-defined interfaces and make them accessible from an existing programming language like Matlab, Python, or Java. This shifts responsibility for the development and maintenance of the language front-end, that most users will work with, onto a strong specialized community. It also brings the additional benefit of a large body of libraries that is often provided by the community (Python calls this “batteries included”).
This maturation process separates the code that is specific to a model from the code that is needed to execute it. To date almost all publicly available simulation systems provide an interpreted language interface to set up a simulation. Neuron even supports two different specification languages (Hoc and Modl) (Hines and Carnevale 2000). Conversely, with the continued development of interpreted high-level languages like Matlab, IDL and Python, it is increasingly possible to tackle some modeling problems in these languages alone, possibly with the addition of compiled modules to improve performance in core parts of the simulation. Examples of the scripting languages used by Neuron and Genesis are shown in Fig. 1. Although the concepts are the same, the language details are quite different.
Fig. 1.
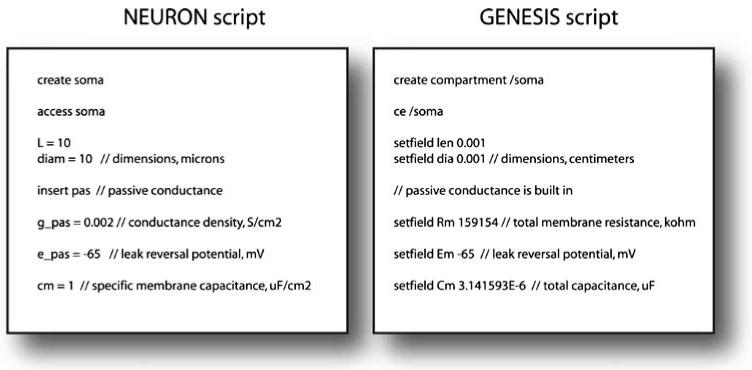
Examples of script files to create a simple single compartment model neuron in NEURON and GENESIS. Although the commands to create the compartment and set the parameters are similar, there are subtle differences in the way the elements of the model are created and addressed, and in the naming of internal variables. The set of units used in each simulator is different also (many GENESIS models use SI units as opposed to the physiological units used here). It is clear though that there is scope to define a standard for describing the elements of such models which can then be mapped to the script particular for a given simulator
Separating model specific code from peripheral simulator code offers the possibility of agreeing on a standard for model descriptions whose appeal is easy to appreciate: the same model will run without modification on a number of different simulators. The standardized model description would give researchers a simple tool to reproduce and validate models published by others. Models are then no longer tied to a particular software system and they gain a degree of durability and universality that models tied to specific systems cannot hope to achieve. In the best cases it would enable truly incremental science (rather than the widespread “rebuild-from-scratch” methodology currently practiced) where reference models can be reused at will, independent of the user’s and original author’s preferred modeling systems. The current situation, however, is rather disenchanting as the examples of Genesis and Neuron illustrate. Although these simulators have an extensive overlap in their application domain, they differ greatly in their specification languages.
There are two main routes towards a standardized model description. The first tries to replace the many different scripting languages by a single standard language that should be supported by all simulators. The second tries to introduce a new portable model description format that is distinct from the scripting languages currently used by any of the simulators. Although at first, the two routes may appear rather similar, they are in fact complementary. While the first tries to introduce a common description language in which users formulate their models, the second introduces a machine readable model exchange format in addition to the scripting languages used by any of the simulators. To date, most activities follow the second route.
Imperative and Declarative Model Specifications
Both Neuron and Genesis use imperative model descriptions, where the definition consists of a precise sequence of instructions specifying how to create and combine the parts of a model. Although imperative model descriptions require a full-featured programming language, they have the obvious advantage that the user can choose the algorithms that create the model. But, because imperative model descriptions specify how models should be constructed using the internal data structures of a simulator, they tend to be simulator-specific. Even where two simulators operate on exactly the same problem domain, they are unlikely to use similar internal structures and the procedures required for setting up a model may be quite different.
The second, declarative, approach is to describe the model that is required and let the simulator determine how it is actually created and mapped to the simulator’s data structures. Such descriptions should be as compact as possible, for example by specifying a parameterized projection pattern for a large network rather than actual connections of each cell. The latter is also declarative, but is typically not something a user would create by hand. The focus here is on declarative descriptions of comparable scale and complexity to the more familiar imperative methods used for setting up models.
An example of the distinction is shown in Fig. 2 where distinct imperative scripts used for setting up a section of cable in Neuron and Genesis have both been derived from a single, declarative, fragment of NeuroML as described below.
Fig. 2.
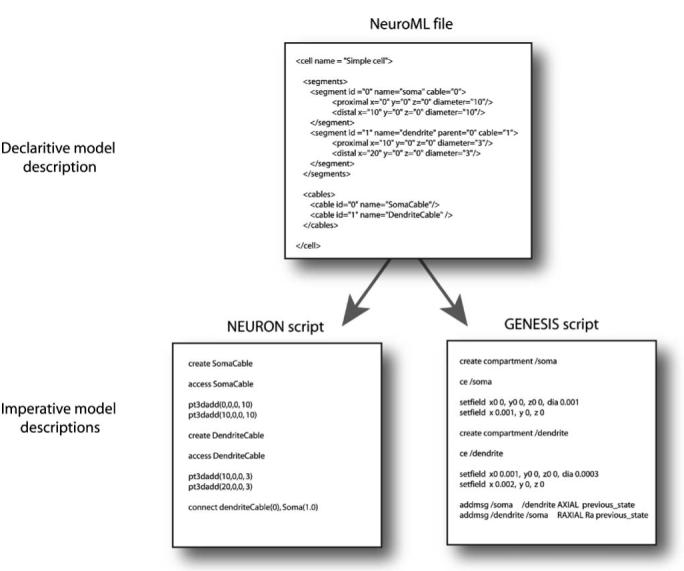
Examples of declarative and imperative model descriptions. The fragment of a NeuroML file above describes two cables, each with a single segment. This declarative specification does not detail the method of creating these, just contains information on the parameters describing them, in a structured format. The NEURON and GENESIS script files, on the other hand, outline the steps needed to create the model, in the native language of the simulator
Declarative descriptions encapsulate the simulator dependent mapping between model and data structures and are, thus, promising candidates for a standard model description format. In computer science, declarative programming is of interest for its readability, because it defines what a program is doing instead of how it is doing it (Lloyd 1994), leaving the problem of how the desired function is achieved to the implementation of the language. For a certain class of declarative languages the correctness of algorithms can be proven mathematically. But, for the case of general purpose declarative languages, readability and formal correctness come at the price that such languages are notoriously inefficient and difficult to implement. This motivates the development of domain-specific declarative specification languages that cover only the constructional parts of a model (such as which channels are present on which compartments) and assume that functional knowledge (such as how channels affect membrane potential) is built into the simulator. The distinction is clearly exemplified by the approaches used with CellML (Cuellar et al. 2003) and SBML (Systems Biology Marup Language) (Hucka et al. 2003). The CellML specification aims to allow complete declarative specification of a model. It uses MathML for specifying equations and the Resource Description Format (RDF, (http://www.w3.org/RDF/). for relations. But the focus of CellML is on document creation and validation, where the document is a complete, portable electronic specification of the model. However in general simulation systems working with CellML only support a small subset of what can be expressed. With SBML, there is a stronger interest in runnable models and the specification assumes that a lot of domain specific knowledge is built into the simulation environment. Nevertheless, most SBML-aware software systems still only support part of the specification. In neuroscience, the main interest to date has been on even higher levels where declarative specifications are only developed to encapsulate models for which implementations already exist.
XML Based Standards
In the domain of data formats, eXtensible Markup Language (XML) is a successful example of a declarative format that was developed to meet the challenges of largescale electronic publishing (http://www.w3.org/XML/). The big appeal of XML based languages is their combination of a standardized low-level structure, with easily extended domain-specific structures. The low-level standardization allows XML documents to be processed automatically without domain-specific knowledge while the extensions allow complex structures to be cleanly represented. XML provides a powerful framework for creating declarative model description formats and the XML community has created a wide range of tools that help in creating domain specific XML variants. Thus, it is not surprising that numerous projects in computational neuroscience and bioinformatics use XML based description formats (e.g. NeuroML (Goddard et al. 2002; Crook et al. 2005), CellML (Cuellar et al. 2003), ChannelML (http://www.neuroconstruct.org/), MorphML (Crook et al. 2007), and SBML (Hucka et al. 2003). It is somewhat ironic that XML is the most successful representative of declarative languages as it misses out on the initial goal of human readability. XML was designed as a machine readable format to be processed automatically and even though it is declarative, in most production applications it is too verbose to be read or written by humans. This gives it a good chance as portable model exchange format, but writing standards-compliant XML directly is not a serious contender to replace the use of imperative scripting languages that researchers currently rely on to formulate their models in the first place. Likewise, RDF provides an XML based ontology system for the semantic web, but the need to create RDF documents has motivated the development of more compact and readable notations such as N3 (http://www.w3.org/DesignIssues/Notation3).
How Much Can a Standardized Model Description Cover?
It is almost inevitable that a complete declarative specification standard for models that are themselves the objects of active research cannot be laid down (with the exception of complete, but impractical specifications as discussed above). The focus here is, instead on highly domain-specific specifications where exact meanings and parameterizations are attached to terms already used in the modeling literature (Ion Channel, Hodgkin-Huxley gate, Spine etc). For models at the level of networks or larger systems, the range of possible models is much broader and no suitable set of terms and concepts is currently in sight. The situation is similar at much smaller scales although standards for sub-cellular morphology specifications are being developed.
A natural starting point for any standardization effort of this nature is the set of declarative specifications already supported by different software systems. Where two systems address the same class of models, this immediately raises the question of whether the standard should cover the intersection, only the parameterizations common to both systems; the union, everything that either of them can express; or something else entirely, based on some new notion of what ought to be modelable, as illustrated in Fig. 3. To date, different emerging standards have adopted different positions on the matter. For example, within the NeuroML framework MorphML (Crook et al. 2007) combines the second and third courses, driven partly by the desire of modelers to be able to use the same morphology specifications with different simulators and partly by the desire of experimental neuroscientists to accurately represent the structures they are tracing. But ChannelML tends towards an intersection of what Neuron and Genesis can support, although it is also under pressure to support some specification styles, such as channel specifications via kinetic schemes that only one of the two supports. At present, the main application of ChannelML is within NeuroConstruct (http://www.neuroconstruct.org) which facilitates the creation of declarative models that can subsequently be converted to input scripts both for Neuron and for Genesis. This leads to a natural tendency towards intersection based specifications and also to an inevitable tension between the desires to be able to represent as many existing models as possible while still being able to export to both systems.
Fig. 3.

Different options for the concepts to be included in standardized specifications when two simulators already support some of the concepts in the domain. Restricting the scope to the intersection ensures that new models defined using the specification can be run in either simulator. Covering the union ensures that models already defined for the simulators can be converted to the standard. A third option is to focus on the domain itself, without regard to existing software support
The case of ChannelML is also instructive from the perspective of channel modeling systems for which it was not originally defined. For example, can models built by biophysicists be represented in ChannelML as is? The answer, in general, is that they cannot, for two main reasons: first, biophysicists prefer to use kinetic schemes (e.g. Chen and Hess 1990; Vandenberg and Bezanilla 1991) for channels rather than Hodgkin-Huxley style models, and second they are concerned with single-channel conductance rather than conductance densities as in ChannelML. Although the specification could readily be extended to support these cases, the result would be that channels could be specified that could not be run in either Neuron or Genesis. This example is by no means an isolated case. In many respects, since they have been so well studied, ion channels could be expected to be one of the easier cases for the creation of a comprehensive specification language.
The channel example illustrates the core issue with the standardization of model descriptions: intersection standards are valuable and initially increase in value as their scope grows, but at some point the extension should be stopped and the remaining cases should be tackled by a different approach. For different levels of maturity and different types of problems the cutoff point can be expected to vary widely, from getting the majority of models within the standard with just a few edge cases to areas, like network specification, where almost everything is an edge case. Here, the default strategy is to use high-level languages like Matlab or Python as the modeling system.
Declarative standards development therefore breaks down into two distinct problems: the development of core standards and the handling of edge cases. The first is well understood and well supported by a wide range of tools and technologies. The second is relatively new area of research and one where neuroscience, with its tendency for almost everything to be an edge case, has great potential to drive the development of new approaches to the handling of deeply heterogeneous models. If computational neuroscience is to exploit this potential, it must come up with a modeling approach which is more attractive than the current de-facto standard of NEURON/HOC or GENESIS/SLI and Matlab for off-line analysis.
Models That Do Not Fit Within a Standard
Even where no specification standard has been laid out, declarative model specifications promise significant benefits in terms of their readability and at least partial system independence. They are also an excellent starting point for any rationalization of models with overlapping domains. Freed from the constraints of existing standards, such models are also at liberty to use recent ideas and technologies for achieving the most readable and expressive formats.
Where independent specifications have been used for separate models that occupy the same domain and could usefully be interconverted, a range of options are open to the researcher wishing to port a model. It can be done by hand, benefiting from the readability of declarative specification languages and avoiding the need to learn the independent scripting languages. In some cases relatively straightforward approaches such as XSL transforms (http://www.w3.org/TR/xslt) may work for systematizing at least part of the process. But in general model reuse will require the injection of additional knowledge about both the biological system being modeled and about the software systems to be used. A relatively simple case would be the mapping of a Hodgkin-Huxley ion channel model onto an equivalent kinetic scheme (Hille 2001) for use within a biophysical modeling package. A slightly more complex example is the replacement of an enzymatic reaction expressed with a reduced approximate equation such as the Michaelis Menten rule by two separate mass-action reactions (Bhalla 2001). The more detailed reactions require quantities to be supplied that are not derivable from the original model. In this case an exact equivalent is not possible, but one which is functionally equivalent to within the original experimental error bounds almost certainly is. A similar problem arises in mapping ion channels onto a system that only supports a few possible parameterizations of transition rates. Again, exact equivalents may not exist, but functional ones are likely to exist and can be found by a combination of model screening and model fitting within the new system.
These examples, where mathematically exact equivalents do not exist in the target system, but where the original modeler could nevertheless perfectly well have achieved their scientific objectives within that system, are likely to make up the majority of cases, rather than being rare exceptions. They call for novel and imaginative approaches to model transformations and knowledge representation. The observation that existing model implementations force the modeler to go beyond their initial intentions and specify quantities that the system needs but that the data does not supply, also hints at the possibility of further levels of model specification that lie somewhere between the empirical observations or hypotheses and the very precise specifications needed to make a working model within a particular system. Whether such forms of specification are achievable, and how they might prove useful remains an open question.
Comparison with Systems Biology Markup Language
In the context of the previous section, SBML takes the approach that for the domain of biochemical reaction systems in a single mixed pool, everything should fit within the standard and there should be no edge cases. The standard has therefore grown with every new version (despite occasional pruning such as the elimination of Fahrenheit as a unit of temperature). This has several notable consequences: no software system supports all of SBML; the conversion of models between systems is inevitably lossy; many systems support SBML in write-only mode. Despite these apparent disadvantages, the SBML movement brings the huge benefit that now the large majority of systems biology models can be represented within a well defined, tightly controlled specification. Although it does not make model portability simple, it certainly facilitates it, and most importantly, it enables the community to move towards a situation in which the provision of a standardized SBML version of a model can be made a condition of publication.
Do the same costs and benefits apply in neuroinformatics, or is the balance subtly different in such a way that we should not aim for the same goals? The most striking difference is in the breadth of the domain to be covered: neuroinformatics increasingly touches on systems biology as one small subset, but approaches it from a direction where detailed 3-D geometry is an essential prerequisite (Stiles and Bartol 2001). This immediately makes any hypothetical catch-all standard substantially larger before even considering the many layers above through neurons and networks to large-scale connectivity. As such, and given the effort already required for the domain covered by SBML, the prospects of a similar program for the whole of neuroinformatics seem at best very remote. However, it may be possible to standardize specifications for models in certain more restricted areas. For example, considerable effort is currently devoted to modeling single neurons and there is a broad consensus about what is needed. Although a standard that catches all cases is unlikely, one that catches a large majority of them should be achievable. For other areas, the benefits of moving towards a system of complete and correct model publication still apply just as much in neuroinformatics as in systems biology, and raises the question of what alternative strategies can be adopted to achieve the same goal. This is considered further in “Discussion and Open Issues.”
Run-time Interoperability
The second approach to interoperability abandons the requirement that a model specification alone should be portable and requires instead that the specification plus a specific implementation should interoperate with other systems. This loses out on the hypothetical benefits of durability and universality that come with standardized and purely declarative model specifications, but it has the large pragmatic advantage of being achievable in many cases.
Examples where run-time communication has an obvious benefit come from cases where there is a big investment in complex, simulator specific models, such as the De Schutter Purkinje cell model running in Genesis (De Schutter and Bower 1994) or Traub’s CA3 pyramidal cell models running in a proprietary system (Traub et al. 1994). Re-implementing the Purkinje cell model in Neuron with the goal of quantitative identity proved very difficult (Arnd Roth, private communication), so for non-Genesis users who nevertheless wish to study the model or include it within a network simulation, the natural solution is to take both the model and Genesis itself and connect that to whatever system they are using. In practical terms, the biology makes this rather easier than it might seem because the communication can often be reduced to sending and receiving discrete spikes with relatively large (several ms) latencies (although, gap junctions, spillover or other local spatial interactions, if present, turn it back into a very hard problem).
The implementation of run-time interoperability can be subdivided into horizontal communication, between simulators operating at the same level, and vertical communication, where they address different levels of detail or different spatial scales. The two cases present rather different challenges. The first is primarily a technical and engineering problem. The second has similar technical requirements but also introduces new conceptual and scientific problems as to how different spatial and temporal scales can be successfully combined within a single system.
Multi-level Interaction
The potential for multi-level modeling has been extensively discussed (Kötter at al. 2002; De Schutter et al. 2005)but there are still very few concrete examples of such approaches in practice. On a technical level, Python based “glue” or its equivalent should be adequate to connect, for example a detailed biophysical model of a synapse run in MCell with a whole cell model run in Neuron or Genesis. The bigger question is how this capability can be scientifically exploited, or, put another way, “who really wants to do multi-level modeling anyway?” Even if it were computationally tractable to model every synapse in a large cell with a detailed biophysical model, the problem would be seriously under-constrained because of the lack of detailed morphology. However, this difficulty rarely arises because of computational limits on what can be modeled. The greater challenge, therefore, and indeed the “multi-level” buzzword itself, come from the need to be very selective about which parts of a model are tackled at the finest scale and how the results of such models can be exploited. The situation can be contrasted with astrophysics where numerical techniques have been developed to compute behavior simultaneously across a wide range of scales, such as collisions between stars with diameters differing by a factor of a million or more. The key difference is that in the astrophysical case, it is a priori clear that the finest scales only matter in very restricted areas. In biology, however, it is rarely so clear that fine-scale behavior in some region is more important than fine-scale behavior in another.
These considerations effectively preclude the simplest interpretation of multi-level modeling (“let’s just replace this synapse by a biophysical model and leave all the rest as they are”) and shift the focus towards the use of representative cases and statistical applications of behavior at different scales.
Technology for Run-time Interoperability
Technically, run-time interoperability between simulators can be achieved in different ways that fall into three broad categories. The first two can be used to couple independent simulators during run-time. The third category introduces a general simulation kernel that orchestrates different simulator modules that are no longer independent.
Direct Coupling
The first category directly connects two or more simulators using the devices of the operating system. Instead of designing a dedicated communication protocol, it is often possible to exploit the extensive scripting interfaces that most simulators have. For two simulators, A and B, each one only needs to be able to generate, rather than understand, the language used by the other. Simulator A talks to simulator B in B’s native language and simulator B responds by sending commands in A’s language. Thus, communication can be established with no more than a few functions on each side. This approach is taken by PyNEST, which connects the NEST (Diesmann and Gewaltig 2002) simulator to extension modules written in Python and has the big pragmatic advantage that it is quick and easy to implement.
Indirect Coupling Via Interpreted Languages
The second category uses an interpreted high-level language to couple different simulators. Popular choices are languages with strong development communities, like Python, Perl, or Guile, that provide powerful mechanisms to glue applications together. Python has a number of features that make it an interesting candidate. First, Python has a clear and concise syntax that supports declarative and functional programming styles. Second, Python’s support for data analysis and visualization is almost on a par with Matlab. Indeed there is now a broad consensus that Python is a good choice for use with neuroscience simulators, the debate being almost completely free from the style of dogmatic language preferences that have surfaced in the past. This may reflect the growing familiarity of software engineers with many different languages and the fact that most imperative interpreted languages look much the same anyway. It may also reflect the recognition that Python is already the de-facto communication standard in several areas, with NEST and Topographica (Bednar et al. 2003) already providing Python interfaces, and Neuron rapidly catching up. The PyNN project (Brette et al. 2007) is developing an API (Application Programming Interface) to unify many of these interfaces so that the same Python code can be used with different systems. Simulator-specific bindings translate the API calls into the appropriate instructions for the system that the model is to be run on.
Coupling Via Object Oriented Frameworks
The final form of interoperability provides an object oriented framework that consists of a general simulation kernel, which handles the overall management of a simulation and delegates specific tasks to modules, plus an open set of modules, that correspond to entities in the neural model. The modules must conform to a carefully designed interface, which supports common operations on the modules including creation and communication. An example of this type of interface for the case of kinetic scheme ion channels is shown in Fig. 4.
Fig. 4.

Software interfaces for constructing and using kinetic scheme ion channel models. A channel implementation that supports these interfaces can be used from an object orientated framework without the internal model of either being exposed to the other. Channel construction is performed by a builder object that must keep track of channels and states as they are mentioned. The “channel” and “state” arguments can be any identifiers convenient to the framework. When requested, the builder returns an object that is able to compute the behavior of a collection of channels (e.g. for an isopotential compartment). The “advance” and getter methods here are specific to single step calculation: further methods would be required to support more complicated numerical algorithms in the container
This modularization enables the subdivision of tasks into smaller modules, such as the solution of the voltage diffusion equation over a branched structure or the update of the state of a single group of ion channels. It has the benefit that the many smaller modules are easier for human engineers to work on. An efficiency advantage of this approach is that the existence of a strictly defined interface makes it possible to design highly optimized numerical engines that can invisibly ‘take over’ calculations nfrom many smaller modules (De Schutter and Beeman 1998). Furthermore, different numerical engines can be written for the same set of modules, allowing different kinds of calculations to be done on a given model description as embodied in the simulation entities. For example, the same model could be simulated using adaptive or fixed time-step algorithms provided both numerical engines were available. It is hoped that careful development to strictly-defined interfaces allows such a system to work efficiently at higher inter-module communication requirements than would be the case for a looser assembly of components linked by an interpreted language like Python. This approach is exemplified by a number of object oriented simulation systems, including the Messaging Object Oriented Simulation Environment (MOOSE, http://sourceforge.net/projects/moose-g3) being developed as a successor to the Genesis framework, Catacomb2 (Cannon et al. 2003), or the discontinued Neosim project (Goddard et al. 2001; http://www.neosim.org).
From a development perspective, however, modular architectures impose much stricter constraints on the implementation of models than simply the need to provide an interface in a specified scripting language such as Python. A module must implement the interfaces specified by the framework for its particular functionality. And if the module is to be developed for simultaneous use in other environments, the interface requirements are likely to propagate through and exert architectural constraints on those environments too. For these reasons, the software interfaces are subject to many of the same pressures and requirements as the development of cross-systems model specification formats, and it is just as important that they achieve community buy-in if they are to be widely adopted. The big advantage for the framework developer over the standards developer, however, is that they have a great deal to offer up-front. Whereas supporting a standard typically involves extra work for a developer, using an existing domain-specific framework dramatically reduces the effort involved in developing a usable implementation of a novel algorithm compared with building a stand-alone application. Even with extensive use of existing libraries, the peripheral code necessary to provide persistence, error reporting, visualization, etc. can substantially outweigh the code required to actually implement the core algorithm. By writing within an existing framework, much, if not all, this ancillary work can be eliminated. Experience will show whether it is possible to design object frameworks that are universal enough to embrace all modeling domains (without coming to the point that it is as general as the operating system kernel) or whether we need different frameworks for the various domains in computational neuroscience.
Discussion and Open Issues
The increasing use of computational models in neuroscience research calls for significant improvements in both technology and in modeling practice in order for the field to progress effectively. The key requirement is for the transferability of models so that new work can build on prior results. Except for the simplest models, paper publication alone is rarely adequate to communicate a model since it gives only description of the model and its behavior, not the model itself. To turn a model into a durable electronic entity that has the same permanence and accessibility as the paper published about it is a very challenging task, but solutions are gradually emerging. Our focus has been on the technical approaches that can improve accessibility of models within an environment where many different simulations systems are in use. All the approaches considered are under active development and it is too early to say which will be most fruitful. Indeed, no one approach is able to solve all the problems, so a flexible combination of techniques is called for according to the needs of different modeling problems.
Beyond the mainly technical concerns of interaction between software systems, the effective use of models also requires changes to the way they are used and published. Certain problems can be solved relatively easily by the adoption of tools and practices that are already routine among software developers (Baxter et al. 2006). In particular the use of version control systems, and the extensive support for documentation standards are as necessary for effective model development as for software development and exactly the same tools can be used. Other problems are more specific to neuroscience and to the decentralized way in which the modeling community operates where changes that could benefit the community at large are still in the hands of individual researchers. We first consider the problem of how to motivate the adoption of standards, followed by issues of model publication and we finally explore the next step of making not just the model itself, but the data on which the model is based readily available for reanalysis and reuse.
Adoption of Proposed Standards
Whatever the solutions to the problems of simulator interoperability may be, it is crucial that they are accepted and used by the prospective users as early as possible. From a technical perspective, the minimal criteria for acceptance are clear. A standard (whether it is a model description language, software interfaces, or a policy on the use of common languages) must be clear and concise, since otherwise prospective users cannot easily decide whether their model is covered by the standard or not and may be put-off by its complexity. In addition, the software implementing the standard must be stable, light-weight, and easy to use. Without this, researchers may fear the work involved in installing and using it, and may prefer to write their own simulator over which they have control.
The sociological aspects are far more difficult. Science is a competitive business and experience tells us that research is driven by tight schedules, scarce resources, and correspondingly pragmatic approaches and decisions. Thus, adhering to standards with the only benefit of helping others may not be at the top of the research agenda. Where the use of standards is primarily a technical issue to be resolved between the developers of different simulators, the benefits to the user comes at little extra cost so there are few barriers to adoption. But in most cases, the use of standards requires changes in the way researchers build models, whether in writing new software or using existing simulators. To be accepted, standards for interoperability must first help the users in their daily modeling work.
Both developing the standards themselves and persuading users to adopt them are slow iterative processes during which the benefits are not fully realizable. It is therefore important to find other possible drivers for the process. One promising route towards this end arises from the strong user preference for developing models using graphical user interface tools rather than by writing code. Graphical tools fit most easily on a declarative data model which is a good starting point for standardization. Another clear motivation would be a standardized notation which is so concise, expressive, and clear that models can be set up and simulated with much less effort than today. Such a notation would both serve interoperability and be a direct advantage for the researchers who use it. Unfortunately, research in this area has hardly started. Although current work on XML based standards will address many of the technical and conceptual issues of such a format, the difficulty of working directly with XML calls for an extra layer between the standard and the user. The notational problems could be addressed by using more readable equivalents of XML such as N3 or YAML, but the conceptual issues remain.
Another possibility is to change the environment in which the individual works so as to promote outcomes, such as portable models, that benefit the community. Obligatory model publication, as discussed below, is one such strategy. Others could include conditions on the award of grants, analogous to existing data sharing requirements (Koslow 2002; Gardner et al. 2003; Insel et al. 2003), or a shift of funding emphasis to reward technologies that allow the easy reuse of existing models as well as the creation of new ones.
Model Publication
Despite significant progress on XML formats and Python interfaces in certain areas, at present no complete standards for model descriptions or simulation middle-ware are in sight and there may be considerable work required to reach a solution that meets all the criteria mentioned here. Until then pragmatic intermediate solutions have to be found. One approach that would bring significant short-term benefits would be to require the publication of source code of a model along with its description in a journal. This at least would ensure that there is a complete and correct, although often hard to read, version of the system described in the paper. A first and encouraging project in this direction is the SenseLab’s ModelDB database (http://senselab.med.yale.edu/senselab/modeldb/) (Migliore et al. 2003) where researchers can submit their models once the publication is accepted. ModelDB already hosts a large number of models and is becoming a valued resource for researchers. Placing source code in ModelDB is actively promoted by the Journal of Computational Neuroscience (JCNS) where the review process involves specifying whether the authors should be asked to make their code available. Although not a strict requirement, this strategy proves quite effective in having source code made available where appropriate.
Inclusive repositories, such as ModelDB are well placed to support emerging standards as they develop. Although a model can be deposited in any format, it is easy to envisage views of the repository that provide better access to and information about models that use recognized standards.
Data Sources
Many models are based primarily on empirical data. Even in the ideal case of a widely used portable standard for model specification, and successful data sharing infrastructure making the data easily available, the process whereby a model was created could still remain something of a black art. Increased compatibility and interoperability between simulators could even exacerbate the problem by allowing more diverse processes to be used in building models and more eclectic mixtures of software to be hooked up together. This goes well beyond issues of model specification, but in principle many of the same concepts are applicable. Instead of specifying only the structure of a final model, the specification could include references to the sources on which the model is based and how each one has been incorporated. The biomodels database (Le Novère et al. 2006) provides examples of how this could work, although these documents are typically created after the model has been published rather than as an integral part of the modeling process itself.
Conclusion
The workshop on which this discussion is based provided a valuable opportunity for developers of a range of neuroscience simulation systems to discuss their problems, objectives, and how their varied projects relate to one another. It was characterized by an unexpected degree of agreement on many key issues: certainly in terms of what the important issues are, and even in many cases on how they should be addressed. Given the scope and scale of the modeling work already undertaken in neuroscience, and the ever-increasing demand for new and better software tools, any activity that increases the productivity of the development community is to be welcomed. In particular, events such as this workshop that bring active developers together provide a very valuable forum for the synchronization and planning of future work, and should certainly be repeated in the future.
Acknowledgments
The workshop was supported by the European Commission IST-2001-35498 Neuro-IT-Net Thematic Network. Robert Cannon was supported by NIH NIDA grant DA16454 as part of the CRCNS program. Michael Hines is funded by grants NINDS NS11613 (NEURON) and NIH DC04732 (ModelDB). Padraig Gleeson is in receipt of a Special Research Training Fellowship from the United Kingdom Medical Research Council.
Contributor Information
Robert C. Cannon, Textensor Limited, Edinburgh, UK
Marc-Oliver Gewaltig, Honda Research Institute Europe GmbH, Offenbach, Germany.
Padraig Gleeson, Department of Physiology, University College London, London, UK.
Upinder S. Bhalla, National Centre for Biological Sciences, Tata Institute of Fundamental Research, Bangalore, India
Hugo Cornelis, University of Texas San Antonio, San Antonio, TX, USA.
Michael L. Hines, Section of Neurobiology, Yale University School of New Haven, CT, USA
Fredrick W. Howell, Textensor Limited, Edinburgh, UK
Eilif Muller, Kirchhoff Institute for Physics, University of Heidelberg, Heidelberg, Germany.
Joel R. Stiles, Center for Quantitative Biological Simulation, Pittsburgh Supercomputing Center, Pittsburg, PA, USA
Stefan Wils, Theoretical Neurobiology, University of Antwerp, Antwerpen, Belgium.
Erik De Schutter, Theoretical Neurobiology, University of Antwerp, Antwerpen, Belgium.
References
- Baxter SM, Day SW, Fetrow JS, Reisinger SJ. Scientific software development is not an oxymoron. PLoS Comput Biol. 2006;2:e87. doi: 10.1371/journal.pcbi.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar JA, Choe Y, De Paula J, Miikkulainen R, Provost J, Tversky T. Modeling cortical maps with topographica. Neurocomputing. 2003;58:1129–1135. [Google Scholar]
- Bhalla US. Modeling networks of signaling pathways. In: De Schutter E, editor. Computational neuroscience: Realistic modeling for experimentalists. CRC; Boca Raton: 2001. pp. 25–48. [Google Scholar]
- Bower JM, Beeman D. The book of Genesis: Exploring realistic neural models with the GEneral NEural SImulation System. 2nd ed. Springer; New York: 1998. [Google Scholar]
- Brette R, Rudolph M, Carnevale T, Hines M, Beeman D, Bower JM, et al. Simulation of networks of spiking neurons: A review of tools and strategies. 2007 doi: 10.1007/s10827-007-0038-6. http://arxiv.org/abs/q-bio.NC/0611089. [DOI] [PMC free article] [PubMed]
- Brunel N, Wang X-J. Effects of neuromodulation in a cortical network model of object working memory dominated by recurrent inhibition. Journal of Computational Neuroscience. 2001;11:63–85. doi: 10.1023/a:1011204814320. [DOI] [PubMed] [Google Scholar]
- Cannon RC, Hasselmo ME, Koene RA. From biophysics to behavior: Catacomb2 and the design of biologically plausible models for spatial navigation. Neuroinformatics. 2003;1:3–42. doi: 10.1385/NI:1:1:003. [DOI] [PubMed] [Google Scholar]
- Carnevale NT, Hines ML. The NEURON book. Cambridge University Press; UK: 2006. [Google Scholar]
- Chen C, Hess P. Mechanism of gating of T-type calcium channels. Journal of General Physiology. 1990;96:603–630. doi: 10.1085/jgp.96.3.603. DOI 10.1085/jgp.96.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook S, Beeman D, Gleeson P, Howell F. XML for model specification in neuroscience: An introduction and workshop summary. Brains, Minds, and Media. 2005;1:bmm228. urn:nbn:de:0009-3-2282. [Google Scholar]
- Crook S, Gleeson P, Howell F, Svitak J, Silver RA. MorphML: Level 1 of the NeuroML standards for neuronal morphology data and model specification. Neuroinformatics. 2007;5 doi: 10.1007/s12021-007-0003-6. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar AA, Lloyd CM, Nielsen PF, Bullivant DP, Nickerson DP, Hunter PJ. An overview of CellML 1.1, a biological model description language. Simulation. 2003;79:740–747. [Google Scholar]
- De Schutter E, Beeman D. Speeding up GENESIS simulations. In: Bower JM, Beeman D, editors. The book of GENESIS: Exploring realistic neural models with the GEneral NEural SImulation System. 2nd ed. Telos; Springer New York: 1998. pp. 329–347. [Google Scholar]
- De Schutter E, Bower JM. An active membrane model of the cerebellar Purkinje-cell .2. Simulation of synaptic responses. Journal of Neurophysiology. 1994;71:401–419. doi: 10.1152/jn.1994.71.1.401. [DOI] [PubMed] [Google Scholar]
- De Schutter E, Ekeberg Ö, Kotaleski JH, Achard P, Lansner A. Biophysically detailed modelling of microcircuits and beyond. Trends in Neurosciences. 2005;28:562–569. doi: 10.1016/j.tins.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Diesmann M, Gewaltig M-O. In: NEST: An environment for neural systems simulations in Forschung und wissenschaftliches Rechnen, GWDG-Bericht. Plesser T, Macho V, editors. Ges. fuer Wissenschaftliche Datenverarbeitung; Göttingen (D): 2002. pp. 43–70. [Google Scholar]
- Gardner D, Toga AW, Ascoli GA, Beatty J, Brinkley JF, Dale AM, et al. Towards effective and rewarding data sharing. Neuroinformatics. 2003;3:286–289. doi: 10.1385/NI:1:3:289. [DOI] [PubMed] [Google Scholar]
- Goddard NH, Beeman D, Cannon RC, Cornelis H, Gewaltig M-O, Hood G, et al. NeuroML for plug and play neuronal modeling. Neurocomputing. 2002;44:1077–1081. [Google Scholar]
- Goddard N, Hood G, Howell F, Hines M, De Schutter E. NEOSIM: Portable large-scale plug and play modelling. Neurocomputing. 2001;38:1657–1661. [Google Scholar]
- Hille B. Ionic channels of excitable membranes. Sinauer Associates INC.; Sunderland, MA: 2001. [Google Scholar]
- Hines ML, Carnevale NT. The NEURON simulation environment. Neural Computation. 1997;9:1179–1209. doi: 10.1162/neco.1997.9.6.1179. [DOI] [PubMed] [Google Scholar]
- Hines ML, Carnevale NT. Expanding NEURON’s repertoire of mechanisms with NMODL. Neural Computation. 2000;12:995–1007. doi: 10.1162/089976600300015475. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, et al. The systems biology markup language (SBML): A medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. DOI 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Insel TR, Volkow ND, Li TK, Battey JF, Landis SC. Neuroscience networks: Data-sharing in an information age. PLoS Biol. 2003;1:e17. doi: 10.1371/journal.pbio.0000017. DOI 10.1371/journal.pbio.0000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslow SH. Sharing primary data: A threat or asset to discovery? Nature reviews. Neuroscience. 2002;3:311–313. doi: 10.1038/nrn787. [DOI] [PubMed] [Google Scholar]
- Kötter R, Nielse P, Dyhrfjeld-Johnsen J, Sommer FT, Northoff G. Multi-level neuron and network modeling in computational neuroanatomy. In: Ascoli G, editor. Computational neuroanatomy: Principles and methods. Humana; Totowa, NJ: 2002. [Google Scholar]
- Le Novere N, Bornstein B, Broicher A, Courtot M, Donizelli M, Dharuri H, et al. BioModels database: A free, centralized database of curated, published, quantitative kinetic models of biochemical and cellular systems. Nucleic Acids Research. 2006;34:D689–D691. doi: 10.1093/nar/gkj092. DOI 10.1093/nar/gkj092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd WJ. Practical advantages of declarative programming; Proc. Joint Conference on Declarative Programming; GULP-PRODE. 1994. [Google Scholar]
- Migliore M, Morse TM, Davison AP, Marenco L, Shepherd GM, Hines ML, et al. ModelDB: Making models publicly accessible to support computational neuroscience. Neuroinformatics. 2003;1:135–139. doi: 10.1385/NI:1:1:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Nusser Z, Häusser M. Monte Carlo simulations of synaptic transmission in detailed three-dimensional reconstructions of cerebellar neuropil. European Journal of Neuroscience. 2000;12(Suppl 11):14. [Google Scholar]
- Schwab M, Karrenbach M, Claerbout J. Making scientific computations reproducible. Computing in Science & Engineering. 2000;2:61–67. [Google Scholar]
- Stiles JR, Bartol TM. Methods for simulating realistic synaptic microphysiology using MCell. In: De Schutter E, editor. Computational neuroscience: Realistic modeling for experimentalists. CRC; Boca Raton: 2001. pp. 87–127. [Google Scholar]
- Traub RD, Contreras D, Cunningham MO, Murray H, LeBeau FEN, Roopun A, et al. Single-column thalamocortical network model exhibiting gamma oscillations, sleep spindles and epileptogenic bursts. Journal of Neurophysiology. 2005;93:2194–2232. doi: 10.1152/jn.00983.2004. [DOI] [PubMed] [Google Scholar]
- Traub RD, Jefferys JGR, Miles R, Whittington MA, Toth K. A branching dendritic model of a rodent CA3 pyramidal neuron. Journal of Physiology (London. Print) 1994;481:7995. doi: 10.1113/jphysiol.1994.sp020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg CA, Bezanilla F. A sodium-channel gating model based on single channel, macroscopic ionic, and gating currents in the squid giant-axon. Biophysical Journal. 1991;60:1511–1533. doi: 10.1016/S0006-3495(91)82186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


