Abstract
Recent advances in experimental neuroscience allow non-invasive studies of the white matter tracts in the human central nervous system, thus making available cutting-edge brain anatomical data describing these global connectivity patterns. Through magnetic resonance imaging, this non-invasive technique is able to infer a snapshot of the cortical network within the living human brain. Here, we report on the initial success of a new weighted network communicability measure in distinguishing local and global differences between diseased patients and controls. This approach builds on recent advances in network science, where an underlying connectivity structure is used as a means to measure the ease with which information can flow between nodes. One advantage of our method is that it deals directly with the real-valued connectivity data, thereby avoiding the need to discretize the corresponding adjacency matrix, i.e. to round weights up to 1 or down to 0, depending upon some threshold value. Experimental results indicate that the new approach is able to extract biologically relevant features that are not immediately apparent from the raw connectivity data.
Keywords: matrix functions, network science, neuroscience, unsupervised classification
1. Motivation
In recent years, complex networks have received a significant amount of attention (Strogatz 2001; Albert & Barabasi 2002; Newman 2003). The need to study apparently disparate real-world networks using a single unified language has led to the growth of an interdisciplinary field that involves mathematicians, physicists, computer scientists, engineers and researchers from both the natural and social sciences. In this work, we are interested in nature's most complex system, the human cerebral cortex (Sporns & Zwi 2004). The development of diffusion magnetic resonance imaging (MRI) has enabled neuroscientists to construct connectivity matrices for the human brain and ‘proof-of-principle’ work has shown that existing biological knowledge can be recovered from these connectivity data (Klein et al. 2007).
Our ability to understand and compare different connectivity structures can be greatly facilitated by the introduction of easily computable measures that characterize the network topology. Typically, measures of this type rely heavily on the idea that communication, to be understood here as the ease of information spread between nodes on the network, takes place along geodesics. However, in many real-world networks, information can disseminate along non-shortest paths (Borgatti 2005; Newman 2005), and for such networks any meaningful measure of ‘communicability’ should account not only for the shortest path between two nodes but also for all other possible routes. Motivated by this consideration, Estrada & Hatano (2008) recently advanced a new definition of communicability that takes non-shortest paths into account with an appropriate length-based weighting. This definition applies to networks with unweighted edges. In the case where the connectivity information is real valued, converting this information into the required binary format is undesirable because (i) it requires a cut-off value to be determined and (ii) fine details about connectivity strengths are lost.
This report has two main aims: (i) the introduction of a new computable measure of connectivity for a weighted network, and (ii) the application of this new measure to the case of cutting-edge anatomical connectivity data for the brain. In §2, we develop the new measure by extending the definition of communicability to the case of weighted networks, taking care to deal with the issue of normalization. We then present a comparison of connectivity data for stroke patients and healthy control subjects in §3.
2. Network communicability
Suppose we are given a network consisting of a list of (i) nodes and (ii) edges connecting the nodes. In the language of graph theory, this is an undirected, unweighted graph that could be defined in terms of the adjacency matrix A∈N×N, which has aij=aji=1 if nodes i and j are connected, and aij=aji=0 otherwise. We will always set aii=0, so that self-links, also called loops, are disallowed. Estrada & Hatano (2008) recently put forward the concept of communicability to address the issue that the existence or non-existence of an edge does not necessarily capture the degree of ‘connectedness’ between a pair of nodes. For example, two nodes that are not themselves connected, but have many neighbours in common, should be regarded as closer together than the two unconnected nodes that can only be joined through a long chain of edges. An extremely useful observation is that if we raise the adjacency matrix to the kth power, then its i,jth element
| (2.1) |
counts the number of walks of length k that start at node i and finish at node j. Here, the term walk refers to any possible traversal through the network that follows edges, and length refers to the number of edges involved. Estrada and Hatano argued that a level of communicability between two nodes could be assigned by summing the number of walks of length 1, 2, 3, …. Because short walks are more important than long ones, e.g. in a message-passing scenario shorter walks are faster and cheaper, to arrive at a single real number, walks of length k are penalized by the factor 1/(k!). This leads to a definition of communicability between nodes i and j, for i≠j, given by , or, more compactly, exp(A)ij (Estrada & Hatano 2008). We also note that in addition to giving a neat characterization in terms of the matrix exponential, the choice of scaling factor k! can also be justified from the perspective of statistical mechanics (Estrada & Hatano 2007).
In our context, the connectivity information arises in the form of real-valued, non-negative weights, where a larger weight aij indicates that nodes i and j are more strongly connected. The identity (2.1) remains valid in this more general setting, but now the term does not give a zero/one contribution depending on whether the walk is possible. Instead, it contributes the product of the weights along all the edges in the walk. Down-weighting the contribution of longer walks is especially relevant here, since experimental uncertainty generally increases with length.
Although it is appealing to use exp(A) in this way to define communicability for a weighted network, such a measure is likely to suffer from difficulties if the weights are poorly calibrated. A highly promiscuous node with large weights is liable to have an undue influence. Similar effects have been observed in the context of spectral clustering (Higham et al. 2007), where it has proved successful to judge the size of a cluster not by the number of nodes, but by the total weight of connections that they possess. This results in a natural normalization step in which the weight aij is divided by the product , where is the generalized degree of node i. An example illustrating the benefits of this normalization step can be seen in §3.2. By analogy, we therefore define the communicability between distinct nodes i and j in a weighted network by
| (2.2) |
where the diagonal degree matrix D∈N×N has the form D≔diag(di).
In §3, we show that this new measure extracts useful information from brain connectivity networks.
3. Brain network
3.1 Data and acquisition
As noted by Sporns et al. (2005), a major challenge facing any attempt to model the human brain using complex network theory is that the basic structural units, in terms of network nodes and links, are not well defined. Indeed, at least three levels of description are possible: (i) individual neurons and synapses (microscale), (ii) neuronal groups and populations (mesoscale), and (iii) anatomically distinct brain regions and the corresponding inter-regional pathways (macroscale). In this work, owing to the resolution limits of MRI data, we focus on the macroscale description of the human brain. We define a network using the Harvard-Oxford cortical and subcortical structural atlases as implemented in FSLView, part of FSL (Smith et al. 2004), thereby partitioning the brain into 56 anatomically distinct regions: 48 cortical and 8 subcortical. This produces a weighted, undirected graph with 56 nodes. In our experiments, we have structural diffusion-weighted imaging data for nine stroke patients (at least six months following first, left-hemisphere, subcortical stroke) and 10 age-matched controls.
A more detailed description of the materials and methods is provided in the electronic supplementary material.
3.2 Spectral clustering
We have set ourselves the task of unsupervised clustering of the patients, to check how accurately we can recover the known stroke/control groupings. A patient dataset consists of (562−56)/2=1540 distinct values, giving the connectivity strength between each pair of distinct brain regions. We used each of the 19 patient datasets to create the columns of a matrix W∈1540×19, so that wij gives the connectivity strength for the ith pair of brain regions in patient j. Unsupervised clustering on the 19 columns of this matrix was performed using the singular value decomposition (SVD; Higham et al. 2007). This approach is closely related to many other techniques, such as principal components analysis, support vector machines/kernel-based methods, machine learning and multidimensional scaling (Cox & Cox 1994; MacKay 2003; Skillicorn 2007).
The second right singular vector, v[2]∈19, can be used to assign a value (v[2])j to the jth patient, and the aim is that patients with similar connectivity profiles will be assigned nearby values. This is a classical dimension reduction technique, where a vast amount of information is compressed into a single one-dimensional summary that is much easier to visualize and interpret. In particular, a large gap between successive components, especially a gap that straddles the origin, is an indication that the nodes on either side are the members of distinct subgroups.
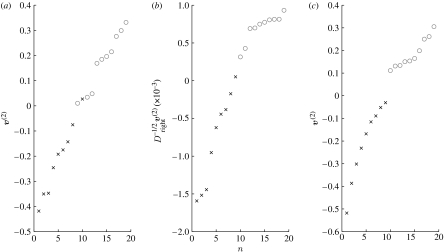
Figure 1a shows the values of v[2], plotted in increasing order. Components corresponding to stroke patients are labelled with crosses, and circles denote controls. We see from figure 1a that although the SVD has placed the strokes and controls approximately in order, a stroke and control (in positions 9 and 10) have been misordered and there is no clear gap separating strokes and controls. Figure 1b shows the corresponding plot when the SVD is applied to the normalized data matrix , with and , and the normalized left singular vector is displayed, as discussed for the case of microarray data in Higham et al. (2007). We see that the classification is improved by the normalization process in the sense that strokes and controls appear sequentially. Closer inspection of the raw data showed that for the two patients who were originally ordered incorrectly, one had unusually large and the other had unusually small overall connectivity weights, (Dright)i; this is precisely the situation where normalization is designed to be beneficial. We note, however, that normalization has not dealt successfully with the separation issue. There is no obvious gap between strokes and controls, and a cut-off at the origin would place a stroke among the controls.
Figure 1.
Components corresponding to stroke patients are labelled with crosses, and circles denote controls. Components of the (a) right singular vector, v[2], of the original data matrix (raw); (b) scaled right singular vector (normalized) and (c) second right singular vector, v[2], of the data matrix after post-processing using communicability.
3.3 Communicability
We motivated the new weighted communicability measure by arguing that the higher order terms in the power series of equation (2.2) contain important additional information. We now provide evidence that weighted communicability does indeed add value to the raw data.
3.3.1 Spectral clustering based on weighted communicability
We now repeat the unsupervised clustering task for the new data matrix, C∈1540×19, whose columns are constructed from the respective communicability networks, so that cij gives the communicability strength for the ith pair of brain regions in patient j. Figure 1c shows the values of the second right singular vector, v[2], plotted in increasing order. We see that post-processing the data using communicability significantly improves the results of the clustering algorithm, giving a correct ordering and a clear separation, with the two groups having opposite signs, negative for strokes and positive for controls. Using the second left singular vector, u[2], we may proceed to identify those connections that enable us to distinguish between stroke and control classes; further details are provided in the electronic supplementary material.
3.3.2 Statistical validation
To quantify the effect of using weighted communicability, we applied the mean-centred partial least-squares (PLS) approach of McIntosh and colleagues (McIntosh & Lobaugh 2004). Through the SVD, PLS analysis returns latent variable pairs (left/right singular vectors containing the connection/group saliences) that describe a particular pattern of connectivity covariance according to the subject. The statistical significance of each latent variable was determined using permutation tests of 500 permutations, while the reliability of saliences of the individual connections in contributing to the pattern of covariance identified by the latent variables was determined using 100 bootstrap analyses.
The PLS analysis returned one significant (p≤0.01) latent variable pair for each of the three datasets described above. In each case, PLS was able to distinguish between stroke and control classes; however, this should not be too surprising since PLS is a supervised method. Perhaps more importantly, the number of connections that returned saliences in the 99th percentile was the greatest for communicability (318), then the normalized data (290) and the lowest in the raw data (266), suggesting that communicability has the effect of reducing the influence of noise in the data.
4. Discussion
Our new network measure extends the concept of communicability in a natural manner to the case of weighted networks. Initial tests reported here on cutting-edge anatomical brain connectivity data show that this measure can give statistically significant enhancement to the performance of standard data analysis tools. In future work, we are planning to study networks relating to a range of brain disorders and investigate the underlying changes in connectivity structure that are revealed through the new measure.
Acknowledgments
We are very grateful to Tim Behrens, Heidi Johansen-Berg, Saad Jbabdi and Rose Bosnell for providing access to the connectivity data and valuable feedback on this work, which was supported by the Medical Research Council under project no. MRC G0601353.
Supplementary Material
References
- Albert R., Barabasi A.L. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47–97. doi: 10.1103/RevModPhys.74.47. [DOI] [Google Scholar]
- Borgatti S.P. Centrality and network flow. Soc. Networks. 2005;27:55–71. doi: 10.1016/j.socnet.2004.11.008. [DOI] [Google Scholar]
- Cox T.F., Cox M.A.A. Chapman and Hall; London, UK: 1994. Multidimensional scaling. [Google Scholar]
- Estrada E., Hatano N. Statistical-mechanical approach to subgraph centrality in complex networks. Chem. Phys. Lett. 2007;439:247–251. doi: 10.1016/j.cplett.2007.03.098. [DOI] [Google Scholar]
- Estrada E., Hatano N. Communicability in complex networks. Phys. Rev. E. 2008;77:036 111. doi: 10.1103/PhysRevE.77.036111. [DOI] [PubMed] [Google Scholar]
- Higham D.J., Kalna G., Kibble M. Spectral clustering and its use in bioinformatics. J. Comput. Appl. Math. 2007;204:25–37. doi: 10.1016/j.cam.2006.04.026. [DOI] [Google Scholar]
- Klein J.C., Behrens T.E.J., Robson M.D., Mackay C.E., Higham D.J., Johansen-Berg H. Connectivity-based parcellation of human cortex using diffusion MRI: establishing reproducability, validity and observer-independence in BA 44/45 and SMA/pre-SMA. NeuroImage. 2007;34:204–211. doi: 10.1016/j.neuroimage.2006.08.022. [DOI] [PubMed] [Google Scholar]
- MacKay D.J.C. Cambridge University Press; Cambridge, UK: 2003. Information theory, inference and learning algorithms. [Google Scholar]
- McIntosh, A. R. & Lobaugh, N. J. 2004 Partial least squares analysis of neuroimaging data: applications and advances. NeuroImage 23(Suppl. 1), 250–263. See http://rotman-baycrest.on.ca/index.php?section=84. ( 10.1016/j.neuroimage.2004.07.020) [DOI] [PubMed]
- Newman M.E.J. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. doi: 10.1137/S003614450342480. [DOI] [Google Scholar]
- Newman M.E.J. A measure of betweenness centrality based on random walks. Soc. Networks. 2005;27:39–54. doi: 10.1016/j.socnet.2004.11.009. [DOI] [Google Scholar]
- Skillicorn D. Chapman and Hall/CRC; Boca Rotan, FL: 2007. Understanding complex datasets. [Google Scholar]
- Smith S.M., et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Suppl. 1):208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Sporns O., Zwi J. The small world of the cerebral cortex. Neuroinformatics. 2004;2:145–162. doi: 10.1385/NI:2:2:145. [DOI] [PubMed] [Google Scholar]
- Sporns O., Tononi G., Kötter R. The human connectome: a structural description of the human brain. PLoS Comput. Biol. 2005;1:245–251. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogatz S.H. Exploring complex networks. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.