Abstract
Expression microarrays that provide genome-level, transcriptional, high-resolution profiles have been applied successfully to multiple diseases. Although microarrays provide information regarding thousands of genes, many investigators prefer to focus on a single gene and validate its role, an approach often supported by grant and journal reviewers. Only a minority of investigators focus on global changes in gene expression. Here, we describe and contrast two general approaches to the use of microarray data: the reductionist “cherry picking” approach and the more global, quantitative “systems” approach. We describe microarray analysis experiments relevant to idiopathic pulmonary fibrosis (IPF) in the context of these two approaches. Although it seems that the cherry-picking approaches have been successful in identifying new relevant genes in IPF, we suggest that to fulfill the discovery potential of microarrays in IPF and to create a working model of IPF, unbiased integrative systems approaches are required.
Keywords: FIZZ1, matrix metalloprotease, microarrays, osteopontin, systems biology
Since their introduction approximately 10 years ago (1, 2), high-throughput gene expression profiling technologies, such as microarrays, have become a mainstay of biomedical research. Combined with the abundant genomic information available and with the rapid introduction of computational approaches, these methods have greatly improved the ability to create high-resolution expression profiles of distinct disease states. Such detailed profiles improved the ability of investigators to identify key regulatory molecules and to dissect the gene and protein networks that underlie disease. In a classic example, Alizadeh and colleagues identified two classes of diffuse large B-cell lymphoma based on gene expression profiles (3) and zoomed in on a small number of genes that could be used for classification (4); and recently, they identified potential signaling pathways that underlie some of these differences (5). Numerous other examples of the usefulness of these technologies include classification, prediction of and understanding the mechanisms of metastasis in breast cancer (6–10), prognosis determination and mutation detection in lung cancer (11), and prognosis prediction in pediatric leukemias and lymphomas (12–14).
The study of idiopathic pulmonary fibrosis (IPF), a relatively poorly understood progressive and mostly lethal interstitial lung disease (15), seems to be a field that would greatly benefit from application of gene expression profiling. Much of the classical hypotheses of the mechanisms of pulmonary fibrosis were generated in the bleomycin-induced lung injury and fibrosis model in animals. The need to generate and verify new hypotheses seems to be critical to the development of new diagnostic and therapeutic options. Despite this need, the number of articles using expression-profiling technologies to understand lung fibrosis is low. In this review, we introduce the two contrasting approaches to microarray experiments: the “cherry picking” approach and the “systems” (as in “systems biology”) approach. The aim of this review is to describe the few works that applied microarrays to the study of pulmonary fibrosis and highlight the discovery potential in the datasets generated by these works within a general framework of approach to microarray experiments.
A CONCEPTUAL FRAMEWORK FOR ANALYSIS OF MICROARRAY EXPERIMENTS
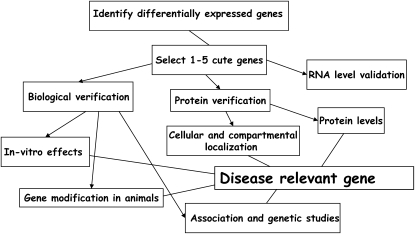
In recent years, there has been a major improvement in our ability to analyze microarray data. Better approaches to normalization, statistical analysis, and pathway analysis greatly simplified the steps from experiment to hypothesis generation. Generally, analytic approaches and their results can be divided into reductionist approaches, where the investigator focuses on a single or a few genes, and global, holistic approaches that try to derive meaningful biological insights from the global changes in gene expression. Historically, the first microarray papers tried to derive global insights; however, very quickly, the reductionist cherry-picking approaches (Figure 1) took precedence, partly because single-gene stories are easier to package and mainly because such papers were better accepted by reviewers, and therefore they were easier to publish. In the cherry-picking approach, a single or a few genes are picked out of the gene expression data, and their relevance is usually validated. The selection is usually based on some statistical criterion and prior biological knowledge. The pathway to validation is outlined in Figure 1. The least important validation is the RNA level validation, which is usually a technical validation. The protein verification suggests that the change in the gene mRNA is meaningful. This can be done in a quantitative manner, verifying the direction of the protein change, and also in a more meaningful way, by demonstrating in which cell and region in the lung the protein is expressed. The biological validation is most important and aimed at answering whether the function of the protein encoded by the gene has a fibrosis relevant function in vivo or in vitro. A final validation is to demonstrate that genetic perturbation in the gene is associated with the disease. Many times it is beyond the ability of a single group or a single article to provide all the details in IPF research and in other fields. The majority of microarray articles published in the general biological literature are cherry-picking articles.
Figure 1.
Cherry-picking approach.
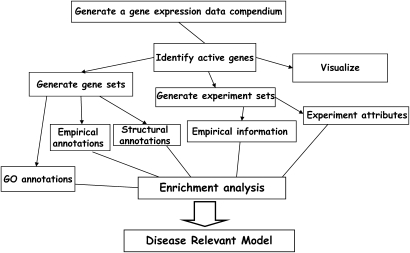
A different and more challenging approach that takes full advantage of the wealth of information in large-scale gene expression data is the global or systems approach (Figure 2). This approach starts with a gene expression compendium that may include only the dataset or multiple datasets that were available. Gene expression data are visualized, and the general trends are observed, usually through some visualization program. This allows the scientist to have a general feeling of the trends in the data. The next step involves identifying all of the active genes (i.e., genes that change in every condition). Then, as much information as possible is collected. A gene can be characterized by the process it is involved in, by its function, and by its cellular localization. All of these are provided at the gene ontology database (www.geneontology.org). A gene can be characterized by the characteristics of the protein it encodes, motifs, and transcription factor binding sites. A gene can be also characterized by everything that is known from the literature. Finally, a gene can be characterized from other microarray datasets (i.e., whether is it induced or inhibited in other experimental systems). In the systems approach, all of these types of information are downloaded and used to create gene attribute files that allow functional characterization of the biological question at hand. This approach has been reviewed previously by us (18), and the software tools for this approach have been described by us (19). What is often neglected is that experiments can also have multiple descriptors (e.g., demographics, dates of experiments, or clinical characteristics). In the global approach, all information is incorporated in the data, and the meaningful or active genes are named and characterized and grouped by a wide variety of functional characterizations that allow identification of functional themes in the data. The algorithms for identifying the clusters are not within the scope of this review and have been previously reviewed (18, 20). The last steps involve using algorithms that allow identification of groups (clusters) of genes and experiments that share common characteristics and then look for enrichment for certain attributes or themes. In the past, investigators applied intuitive approaches and stated that the majority of genes were inflammatory or apoptosis or cell-cycle–related genes; however, such approaches are biased by investigators' lists of favorite topics. To address this problem, we apply methods that look for enrichments that are statistically surprising (e.g., we ask the question whether a certain attribute or characteristic is more abundant within a group of genes or experiments than would be expected by its abundance in the whole genome). In the following sections we characterize microarray experiments by the analytic framework that they belong to and try to define the relative value of the cherries picked and the global insights.
Figure 2.
Systems approach.
MICROARRAY ANALYSIS OF HUMAN SAMPLES
Comparing Normal Control with Diseased Lung
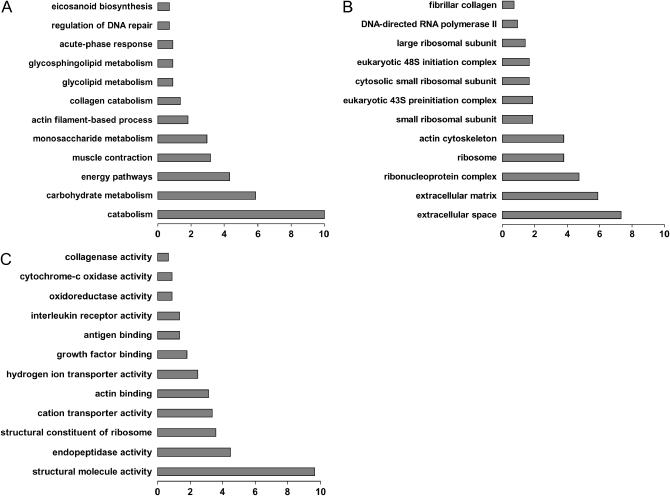
In 2002, two comparisons of gene expression patterns in IPF and normal lungs were published by two different groups, one a conference abstract (21) and the other a full article (17). In their titles, they highlight matrix metalloprotease (MMP) 7 as a potential target for IPF. These findings have previously been reviewed by us (22). Both of these experiments fall into the cherry-picking category. An interesting, statistically significant gene is identified and verified, and its relevance to fibrosis is determined. Our article (17) also contained our first naive experimentation of global analysis. Among the most informative increased genes, we identified genes encoding for proteins expressed in smooth muscle differentiation and muscle contractile machinery, genes that encoded extracellular matrix proteins, and a coordinated increase in the levels of several matrix metalloproteases (MMP1, MMP2, MMP7, and MMP9); however, this analysis was intuitive and not based on an unbiased assessment of enrichment of statically significant annotations in our datasets. For this review and for demonstration, we downloaded the list of genes increased in IPF lungs and analyzed them using NIH DAVID and Ease online tools for functional analysis (www.apps1.niaid.nih.gov/david) (23, 24). We looked for enrichment of GO annotations in the genes up-regulated in IPF (Figure 3). The biological processes profile of IPF (Figure 3A) provides an active picture: IPF lungs are characterized by an increased catabolic state that is also associated with an increase in glucose metabolism, potentially reflecting the active remodeling that occurs in IPF lungs. This is different from the passive view of IPF lungs as end-staged fibrosed organs. Analysis of cellular localization reveals enrichment for genes associated with protein biosynthesis and extracellular matrix as expected (Figure 3B). The most interesting profile is derived from the enrichment in molecular functions (Figure 3C). Although the expected active remodeling is reflected by enrichment for structural constituents and genes with endopeptidase activity, the functional themes contain some unexpected groups, such as antigen binding and hydrogen ion transporter activity. This simple analysis of data generated from five IPF lungs analyzed by the first microarray generation demonstrates the power of the global approach and its discovery potential. Although this dataset has been publicly available for 4 years, we and others have only explored several targets and not necessarily the most interesting ones. Cosgrove and colleagues compared gene expression in five IPF and five normal lungs and focused on angiogenic and angiostatic cytokines in IPF and especially on pigment epithelium–derived factor, a protein with angiostatic properties (25). Their work demonstrates the discovery potential of the cherry-picking approach. Pigment epithelium–derived factor is an excellent example of an unexpected target identified by microarrays. Recently, using an additional dataset of 11 control lungs and 13 IPF lungs, we have again engaged in cherry picking and focused on osteopontin, a phosphoprotein previously shown to be required for development of bleomycin-induced fibrosis in murine lungs (26, 27). Osteopontin was highly expressed in IPF lungs and was expressed mainly in alveolar epithelial cells, and it colocalized with MMP7 in alveolar epithelial cells (16).
Figure 3.
The distribution of genes increased in idiopathic pulmonary fibrosis lungs—functional categories. (A) Biological process. (B) Cellular localization. (C) Molecular function. All enrichments are statistically significant (Fisher's exact score, p < 0.03).
Analyzing Cells Obtained from IPF Lungs
Although whole lungs seem to provide large amounts of information that seems to be highly relevant, at best this information represents an average of gene expression signals generated by multiple cells and in multiple regions. Therefore, the interest in approaches that profile distinct cell populations and regions is high. In recent years, multiple groups have established primary fibroblasts lines from IPF lungs and were able to identify some changes in their biological behavior. Choi and colleagues analyzed chemokine gene expression patterns obtained from fibroblasts isolated from lungs of patients with IPF, nonspecific interstitial pneumonia, and respiratory bronchiolitis–interstitial lung disease and control subjects without interstitial lung disease using expression arrays that contained only probes for chemokines and cytokines (28). All fibroblasts from lungs of patients with idiopathic interstitial pneumonias exhibited increased expression of CCL7. They followed the usual route for verification and demonstrated that CCL7 was overexpressed in IPF lungs. They also demonstrated that the levels of CCL7 were higher in IPF compared with the other interstitial lung diseases. No global observations were made in this report. Renzoni and colleagues analyzed gene expression patterns in normal control fibroblasts and in IPF fibroblasts using oligonucleotide arrays (29). They also analyzed the response to transforming growth factor (TGF)-β. Although they found significant responses to TGF-β, they did not observe any significant difference between fibroblasts obtained from normal control subjects of from patients with fibrotic lung disease (29). Although this article did not shed light on any difference between IPF patients and normal subjects, it provided a detailed description of TGF-β targets in lung fibroblasts, thus providing a useful resource for other investigators. The most important global observation in this article is a negative one; the authors could not find a global difference between IPF and normal control fibroblasts.
Comparing Different Interstitial Lung Diseases
Interstitial lung diseases differ in their clinical course, histologic presentation, radiologic imaging, and prognosis and response to therapy. Therefore, it seems plausible that different interstitial lung diseases express distinct gene expression patterns. Selman and colleagues compared gene expression patterns between IPF and hypersensitivity pneumonitis (HP) (24). Their article is probably the most advanced example of an IPF systems article. Although the authors seem to engage in some cherry picking, this may have been a response to a request by a reviewer. The authors identify multiple genes that distinguish IPF from HP. One interesting aspect of this article is the feature selection. Instead of setting an arbitrary threshold for selecting genes, the authors selected a signature by its use of the highest number of genes that provides the lowest number of classification errors—in other words, by its classificatory efficiency. Using NIH David (http://apps1.niaid.nih.gov/david/), they provide a functional profile of IPF (genes overexpressed in IPF) and of HP (genes overexpressed in HP). One look at these profiles reveals the gene expression parallel to a highly known clinical truth: patients with IPF rarely respond to antiinflammatory treatment (30). The genes that characterize HP are enriched with multiple inflammatory annotations, including T-cell activation, immune response, and defense response, whereas the genes that characterize IPF are enriched with ectoderm development, metalloendopeptidase activity, and extracellular matrix. This view provides an immediate global and unbiased view of the mechanism that underlies the fibrotic lung phenotype in IPF. When these signatures were used to classify a nonspecific interstitial pneumonia, a histologic pattern that is often difficult to differentiate consistently from HP and IPF, we identified three cases that exhibited an IPF-like gene expression and one case that was classified as HP, suggesting that after these gene expression patterns are confirmed on larger cohorts, they could be used to classify disease and potentially to predict response to antiinflammatory therapies.
MICROARRAY ANALYSIS OF ANIMAL MODELS OF DISEASE
Bleomycin-induced Fibrosis
Bleomycin is a common model for induction of fibrosis. We have previously described the two articles that deal with gene expression in bleomycin-induced fibrosis (22). There are also at least two more datasets publicly available at the Gene Expression Omnibus, which is a public microarray database (Table 1). In our first microarray article (31), we applied an intuitive clustering-based approach to obtain global insights regarding the general mechanisms of bleomycin-induced fibrosis using 129 mice homozygous for a null mutation of the integrin β6 subunit gene that develop inflammation, but not fibrosis, in response to bleomycin and wild-type 129 mice (32). Our analysis allowed us to dissect the transcriptional programs associated with inflammation or fibrosis in bleomycin-induced lung injury. Recently, Liu and colleagues analyzed lung gene expression profiles after bleomycin in the rat (33). They identified 628 genes that were changed by bleomycin. The most dramatic increase that they observed was in FIZZ1. They localized FIZZ1 expression to alveolar and airway epithelium. They also identified FIZZ1 in isolated type II alveolar epithelial cells but not in isolated lung fibroblasts. When they transfected FIZZ1 into fibroblasts or cocultured of FIZZ1-expressing type II cells with fibroblasts, the fibroblasts expressed α-smooth muscle actin and type I collagen expression independent of TGF-β stimulation (33).
TABLE 1.
LUNG FIBROSIS–RELEVANT DATASETS
| Description | GEO Accession | Link to Data | Reference |
|---|---|---|---|
| Normal and IPF lungs | N/A | http://www.dom.pitt.edu/paccm/genomics/data.htm | 17 |
| Normal and IPF lungs | GSE2052 | 16 | |
| Normal and IPF fibroblasts | GSE1724 | www.respiratory-research.com/content/5/1/24 | 29 |
| Bleomycin-induced fibrosis in 129/SV (sensitive) and AJ (resistant) mice | GSE452-453 | ||
| Bleomycin-induced fibrosis in C57BL6/J (sensitive) and Balb/c (resistant) mice | GSE485 | www.pepr.cnmcresearch.org/browse.do?action=list_prj_exp&projectId=121 |
Definition of abbreviations: GEO = Gene Expression Omnibus; IPF = idiopathic pulmonary fibrosis; N/A = not applicable.
DO WE IDENTIFY THE RIGHT GENES?
Drug Targets
The studies we described implemented microarrays in the study of human disease and animal models of disease. In most cases, authors have applied cherry-picking approaches and focused on the function of one or two genes and most often determined that these genes (e.g., MMP7, osteopontin, FIZZ1, and CCL7) were important in IPF (16, 17, 25, 28, 33). Although some of the articles made attempts at global observations (17, 32), these observations were mostly qualitative. Only recently have we applied more quantitative methods to better understand the gene expression patterns that characterize IPF (24). Although the relative success of cherry-picking approaches is encouraging, it is also concerning: If so many molecules are involved, how are we ever going to treat IPF? Is blocking any one of these molecules going to be sufficient to halt or reverse lung fibrosis in IPF? Although the results of bleomycin experiments in knockout animals suggest that this is the case, the biological logic does not support this notion because all of these and many other molecules are overexpressed in IPF lungs. An alternative explanation suggests that although each one of these molecules may have isolated profibrotic or antifibrotic effects in healthy animals subjected to a defined injury, in IPF lungs they serve as components of a profibrotic network that determined the lung phenotype in IPF. Therefore, we believe that global approaches that apply system approaches and incorporate multiple levels of information (including the results of cherry-picking experiments) into a robust quantitative model of IPF will allow us to understand and predict the molecular events that lead to the lung phenotype in IPF. Once this model of IPF is created, evaluating the relative role of a specific molecule within this model will allow more accurate predictions about its role as a potential target for therapeutic intervention.
Biomarkers
The mechanistic role of molecules elevated in IPF needs to be validated in complex experiments. The potential role of molecules expressed in IPF lungs as biomarkers seems to be intuitive. This has not been assessed systematically; however, our recent article (24) and the finding of osteopontin, a protein highly expressed in IPF lungs (16) in the peripheral blood of patients with interstitial lung disease (34), suggest that transcriptional profiles may be readily translated into clinical practice by planning studies that will assess their value as markers for disease. The availability of multiple human and mouse datasets (Table 1) should make such analyses possible.
CONCLUSIONS
This article describes and contrasts two general approaches to the use of microarray data: the reductionist cherry-picking approach and the more global, systems approach. We describe microarray experiments relevant to IPF in the context of these approaches and highlight their potential value in understanding the disease. We believe that the increased availability of multiple datasets and the maturation of analytic methods favor the application of systems approaches that will enable us to model and understand IPF in a fashion that will allow better development of new therapeutic and diagnostic modalities.
Supported by NIH grant 1R01 HL073745-01 and by a generous donation from the Simmons family.
Conflict of Interest Statement: N.K. received $5,000 for serving on a Biogen IDEC advisory board in November 2005. I.O.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Lipshutz RJ, Morris D, Chee M, Hubbell E, Kozal MJ, Shah N, Shen N, Yang R, Fodor SP. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques 1995;19:442–447. [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expreshsion patterns with a complementary DNA microarray. Science 1995;270:467–470. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–511. [DOI] [PubMed] [Google Scholar]
- 4.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med 2004;350:1828–1837. [DOI] [PubMed] [Google Scholar]
- 5.Lu X, Nechushtan H, Ding F, Rosado MF, Singal R, Alizadeh AA, Lossos IS. Distinct IL-4-induced gene expression, proliferation, and intracellular signaling in germinal center B-cell-like and activated B-cell-like diffuse large-cell lymphomas. Blood 2005;105:2924–2932. [DOI] [PubMed] [Google Scholar]
- 6.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002;415:530–536. [DOI] [PubMed] [Google Scholar]
- 7.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002;347:1999–2009. [DOI] [PubMed] [Google Scholar]
- 8.Kang Y, He W, Tulley S, Gupta GP, Serganova I, Chen CR, Manova-Todorova K, Blasberg R, Gerald WL, Massague J. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci USA 2005;102:13909–13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature 2005;436:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, Ponomarev V, Gerald WL, Blasberg R, Massague J. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest 2005;115:44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet 2005;37:48–55. [DOI] [PubMed] [Google Scholar]
- 12.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002;8:816–824. [DOI] [PubMed] [Google Scholar]
- 13.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002;346:1937–1947. [DOI] [PubMed] [Google Scholar]
- 14.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell 2002;1:133–143. [DOI] [PubMed] [Google Scholar]
- 15.Costabel U, King TE. International consensus statement on idiopathic pulmonary fibrosis. Eur Respir J 2001;17:163–167. [DOI] [PubMed] [Google Scholar]
- 16.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, et al. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med 2005;2:e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuo F, Kaminski N, Eugui E, Allard J, Yakhini Z, Ben-Dor A, Lollini L, Morris D, Kim Y, DeLustro B, et al. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci USA 2002;99:6292–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: understanding cancer using microarrays. Nat Genet 2005;37:S38–S45. [DOI] [PubMed] [Google Scholar]
- 19.Dave NB, Kaminski N. Analysis of microarray experiments for pulmonary fibrosis. Methods Mol Med 2005;117:333–358. [DOI] [PubMed] [Google Scholar]
- 20.Slonim DK. From patterns to pathways: gene expression data analysis comes of age. Nat Genet 2002;32:502–508. [DOI] [PubMed] [Google Scholar]
- 21.Cosgrove GP, Schwarz MI, Geraci MW, Brown KK, Worthen GS. Overexpression of matrix metalloproteinase-7 in pulmonary fibrosis. Chest 2002;121:25S–26S. [PubMed] [Google Scholar]
- 22.Kaminski N. Microarray analysis of idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol 2003;29:S32–S36. [PubMed] [Google Scholar]
- 23.Hosack DA, Dennis G Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol 2003;4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, Aziz N, Kaminski N, Zlotnik A. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med 2006;173:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosgrove GP, Brown KK, Cool CD, Geraci MW, Schwartz MI, Worthen GS. Angiogenesis in pulmonary fibrosis. Am J Respir Crit Care Med 2002;165:A172. [DOI] [PubMed] [Google Scholar]
- 26.Berman JS, Serlin D, Li X, Whitley G, Hayes J, Rishikof DC, Ricupero DA, Liaw L, Goetschkes M, O'Regan AW. Altered bleomycin-induced lung fibrosis in osteopontin-deficient mice. Am J Physiol Lung Cell Mol Physiol 2004;286:L1311–L1318. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi F, Takahashi K, Okazaki T, Maeda K, Ienaga H, Maeda M, Kon S, Uede T, Fukuchi Y. Role of osteopontin in the pathogenesis of bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol 2001;24:264–271. [DOI] [PubMed] [Google Scholar]
- 28.Choi ES, Jakubzick C, Carpenter KJ, Kunkel SL, Evanoff H, Martinez FJ, Flaherty KR, Toews GB, Colby TV, Kazerooni EA, et al. Enhanced monocyte chemoattractant protein-3/CC chemokine ligand-7 in usual interstitial pneumonia. Am J Respir Crit Care Med 2004;170:508–515. [DOI] [PubMed] [Google Scholar]
- 29.Renzoni EA, Abraham DJ, Howat S, Shi-Wen X, Sestini P, Bou-Gharios G, Wells AU, Veeraraghavan S, Nicholson AG, Denton CP, et al. Gene expression profiling reveals novel TGFbeta targets in adult lung fibroblasts. Respir Res 2004;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selman M, Thannickal VJ, Pardo A, Zisman DA, Martinez FJ, Lynch JP III. Idiopathic pulmonary fibrosis: pathogenesis and therapeutic approaches. Drugs 2004;64:405–430. [DOI] [PubMed] [Google Scholar]
- 31.Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 1999;96:319–328. [DOI] [PubMed] [Google Scholar]
- 32.Kaminski N, Allard JD, Pittet JF, Zuo F, Griffiths MJ, Morris D, Huang X, Sheppard D, Heller RA. Global analysis of gene expression in pulmonary fibrosis reveals distinct programs regulating lung inflammation and fibrosis. Proc Natl Acad Sci USA 2000;97:1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu T, Dhanasekaran SM, Jin H, Hu B, Tomlins SA, Chinnaiyan AM, Phan SH. FIZZ1 stimulation of myofibroblast differentiation. Am J Pathol 2004;164:1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadota J, Mizunoe S, Mito K, Mukae H, Yoshioka S, Kawakami K, Koguchi Y, Fukushima K, Kon S, Kohno S, et al. High plasma concentrations of osteopontin in patients with interstitial pneumonia. Respir Med 2005;99:111–117. [DOI] [PubMed] [Google Scholar]