Abstract
Tissue morphogenesis and homeostasis are dependent on a complex dialogue between multiple cell types and chemical and physical cues in the surrounding microenvironment. The emergence of engineered three-dimensional (3D) tissue constructs and the development of tractable methods to recapitulate the native tissue microenvironment ex vivo has led to a deeper understanding of tissue-specific behavior. However, much remains unclear about how the microenvironment and aberrations therein directly affect tissue morphogenesis and behavior. Elucidating the role of the microenvironment in directing tissue-specific behavior will aid in the development of surrogate tissues and tractable approaches to diagnose and treat chronic-debilitating diseases such as cancer and atherosclerosis. Toward this goal, 3D organotypic models have been developed to clarify the mechanisms of epithelial morphogenesis and the subsequent maintenance of tissue homeostasis. Here we describe the application of these 3D culture models to illustrate how the microenvironment plays a critical role in regulating mammary tissue function and signaling, and discuss the rationale for applying precisely defined organotypic culture assays to study epithelial cell behavior. Experimental methods are provided to generate and manipulate 3D organotypic cultures to study the effect of matrix stiffness and matrix dimensionality on epithelial tissue morphology and signaling. We end by discussing technical limitations of currently available systems and by presenting opportunities for improvement.
I. Introduction
Tissue development depends on coordinated cycles of transcriptionally regulated cell growth, death, and migration that are controlled by exogenous soluble and physical stimuli and spatially dependent cell–matrix and cell–cell adhesion (Barros et al., 1995; Ingber, 2006; Jacobson et al., 1997; Locascio and Nieto, 2001). Regardless of length scale, understanding the molecular basis of tissue-specific differentiation and homeostasis requires an appreciation of adhesion-dependent cell behavior in the context of a three-dimensional (3D) extracellular microenvironment and a complex adhesion-dependent multicellular tissue. Through genetic and biochemical analysis, we have learned much about cell adhesion, including details of adhesion molecule structure and function, and about how various cell adhesion molecules likely mediate cell–extracellular matrix (ECM) interactions and facilitate cell–cell junctional complexes (Fuchs et al., 1997; Huttenlocher et al., 1998; Springer and Wang, 2004). We have also learned much about how exogenous growth, death, migration, and even mechanical cues activate signaling cascades to influence the fate of individual cells and undifferentiated 2D cell monolayers (Huttenlocher et al., 1995; McBeath et al., 2004; Stegemann et al., 2005; Thornberry and Lazebnik, 1998; Wang et al., 2001). Yet, all too often, experimental conclusions reached from observations of single cells and simplified 2D monolayer cell sheets do not accurately represent how cells behave within 3D tissues in vivo (Green et al., 1999; Sethi et al., 1999). Indeed, developmental models and transgenic animals consistently underscore the importance of studying cell behavior in the correct tissue context. However, live animal experimentation is so inherently complex that systematic assessment of the effect of individual variables, such as cell shape and matrix compliance, on cell behavior is extremely challenging and impractical (Sethi et al., 1999; Wang et al., 2005). At the interface between in vivo studies and 2D culture models are the organotypic culture systems that can faithfully recapitulate various aspects of tissue organization and function ex vivo. These organotypic 3D models have been employed with varying degrees of success to clarify some of the mechanisms, whereby biological processes such as adhesion-dependent survival (Weaver et al., 2002; Zahir and Weaver, 2004), polarity (O’Brien et al., 2002; Wang et al., 1998), proliferation (Zink et al., 2004), and even epigenetics (Bissell et al., 1999; Zink et al., 2004) regulate cell behavior as well as novel feedback/regulatory mechanisms (Bissell et al., 2002). Organotypic culture models have been effectively applied to study tissue-specific differentiation (Bissell et al., 2002), to understand factors controlling stem cell behavior (Hendrix et al., 2001), and even microenvironmental control of malignant transformation and tumor dormancy (Margulis et al., 2005; Weaver et al., 1997). Through the prudent use of organotypic 3D models, critical disparities between the molecular determinants of cell polarity (reviewed in O’Brien et al., 2002), apoptosis resistance (Weaver et al., 2002), and growth factor responsiveness (Wang et al., 1998) in cells incorporated into a 3D tissue and those propagated as 2D monolayers have been revealed (reviewed in O’Brien et al., 2002). Yet, while 3D organotypic models can faithfully recapitulate some aspects of tissue behavior ex vivo, many of the systems routinely used to study tissue-like behaviors employ crudely defined natural ECM molecules that contribute to considerable experimental variance. In addition, many of the approaches used to assemble 3D tissue-like structures in culture operate by simultaneously modifying multiple variables, including restrictions on cell shape, matrix compliance, biochemical cues and metabolites, and even the spatial orientation of the ECM, thereby obscuring definitive experimental conclusions regarding individual experimental parameters (Kleinman et al., 1986; Paszek et al., 2005; Wozniak et al., 2003). Indeed, the engineering of surrogate tissues and the development of tractable approaches to diagnose and treat chronic-debilitating diseases such as cancer and atherosclerosis require both a comprehensive understanding of tissue-specific behavior at the molecular level and highly reproducible systems. Accordingly, considerable effort has been expended toward developing synthetic biomaterials in which individual material properties such as cell shape, matrix presentation (2D vs 3D), ligand density, and elastic modulus can be precisely modulated (Chen et al., 1998; Engler et al., 2004; Tan et al., 2003; Yamada et al., 2003). By applying one of the defined systems in which matrix compliance and ligand density could be rigorously controlled, the critical role of matrix stiffness and integrin adhesions as a key regulator of multicellular mammary epithelial cell (MEC) tissue morphogenesis and malignant transformation has been highlighted (Paszek and Weaver, 2004; Paszek et al., 2005). In this chapter, we discuss the rationale for applying well-defined 3D organotypic culture assays to study adhesion-dependent cell behavior. We describe the use of 3D MEC organotypic cultures to illustrate how matrix compliance plays a critical role in regulating mammary tissue function and signaling. Finally, we outline experimental methods to generate, manipulate, and study the effect of matrix stiffness and matrix dimensionality on epithelial tissue morphology and signaling, and discuss technical limitations of currently available systems and future opportunities for improvement.
II. Rationale
A. Stromal–Epithelial Interactions
During embryogenesis, the epithelium originates from the endoderm and ectoderm and develops into a specialized tissue whose primary functions in the organism are to protect and to control permeation or transport. Unlike skin and esophagus, which are stratified epithelia that provide a critical barrier, the secretory epithelium is composed of a simple layer of epithelial cells lining tubes and ducts, whose principal function is to facilitate secretion and transport of biological materials. In vivo, the secretory epithelium abuts on and is surrounded by a stroma, which consists of cellular and noncellular components, including ECM molecules, soluble factors, and various stromal cells such as fibroblasts, adipocytes, and endothelial cells. Directly interacting with the epithelium is the basement membrane (BM), which is a specialized, highly organized ECM composed primarily of laminins 1, 5, and 10, collagen IV, entactin, and heparin sulfate proteoglycans. The BM in turn intersects with the interstitial matrix, which consists of collagens I and III, fibronectin, tenascin, elastins, and various proteoglycans including lumican, biglycan, and decorin (Kleinman et al., 1986). Collectively, the various components of the ECM and stroma provide biochemical (composition) and biophysical (structural modification and organization) cues to the epithelium and operate in concert with soluble factors released from the resident stromal cells to maintain the epithelium’s organ-specific function. Perturbations in stromal–epithelial interactions and altered epithelial organization are hallmarks of cancer and many chronic degenerative diseases. Moreover, disrupting tissue organization or altering ECM integrity precipitates disease, and restoring tissue structure or proper ECM interactions normalizes tissue behavior (reviewed in Hagios et al., 1998; Jeffery, 2001). Accordingly, the goal of 3D organotypic culture models is to recreate tissue-specific interactions, organizations, functions, and behavior ex vivo through prudent control of the biochemical and biophysical properties of the ECM, in order to understand the role of stromal–epithelial interactions and tissue structures in tissue-specific functions.
Mammary gland organotypic culture models have been used effectively to study the role of stromal–epithelial and ECM interactions in tissue-specific differentiation (Debnath et al., 2003; Petersen et al., 1992; Weaver et al., 1996; Wozniak et al., 2003). Unlike other tissues, the mammary gland undergoes unique developmental cycles in the adult organism and the gland can be readily accessed and manipulated in vivo and in culture. Additionally, reasonable quantities of breast tissue can be isolated and propagated ex vivo for culture experiments. As such, much of what we know regarding ECM-dependent epithelial differentiation has been derived from organotypic cultures of primary and immortalized MECs. Early studies demonstrated that MECs grown as 2D monolayers on rigid tissue culture substrates or within a physically constrained collagen I gel fail to assemble tissue-like structures (acini) and differentiate [no detectable expression of differentiated proteins such as whey acidic protein (WAP) or β-casein], despite the availability of appropriate growth factors and lactogenic hormones (reviewed in Roskelley et al., 1995). Yet, when the same MECs are grown within unconstrained collagen I gels and allowed to deposit and organize their own endogenous BM, or are embedded within a compliant reconstituted BM (rBM), they are able to assemble multicellular tissue-like structures (acini; reminiscent of terminal ductal lobular units in tissues in vivo) and differentiate in response to hormonal cues (expressed β-casein and WAP; reviewed in Roskelley et al., 1995; Fig. 1). Further studies using murine and human MECs have also consistently shown that the composition and spatial context of the ECM profoundly influence the responsiveness of an epithelium to exogenous growth, migration, and death stimuli (Wang et al., 1998, 2005; Weaver et al., 2002). For example, some human luminal epithelial breast tissues in vivo express the estrogen receptor (ER) and proliferate in response to hormonal fluctuations in estrogen. When these MECs are isolated and cultured on tissue culture plastic, they spread to form raised ER-negative, 2D cobblestone monolayer colonies that lack estrogenic responsiveness. However, if the isolated MECs are instead grown in the context of a compliant rBM, they retain their ER expression and maintain their estrogenic responsiveness (Novaro et al., 2003). Likewise, undifferentiated MECs grown on tissue culture plastic are highly sensitive to exogenous death cues, whereas their rBM-differentiated counterparts exhibit extremely high resistance to multiple apoptotic stimuli (Weaver et al., 2002). Analogous observations regarding the importance of biochemical and biophysical ECM cues for epithelial morphogenesis and tissue-specific differentiation have also been reported for thyroid, salivary gland, and kidney epithelia studies (Kadoya and Yamashina, 2005; O’Brien et al., 2001; Yap et al., 1995).
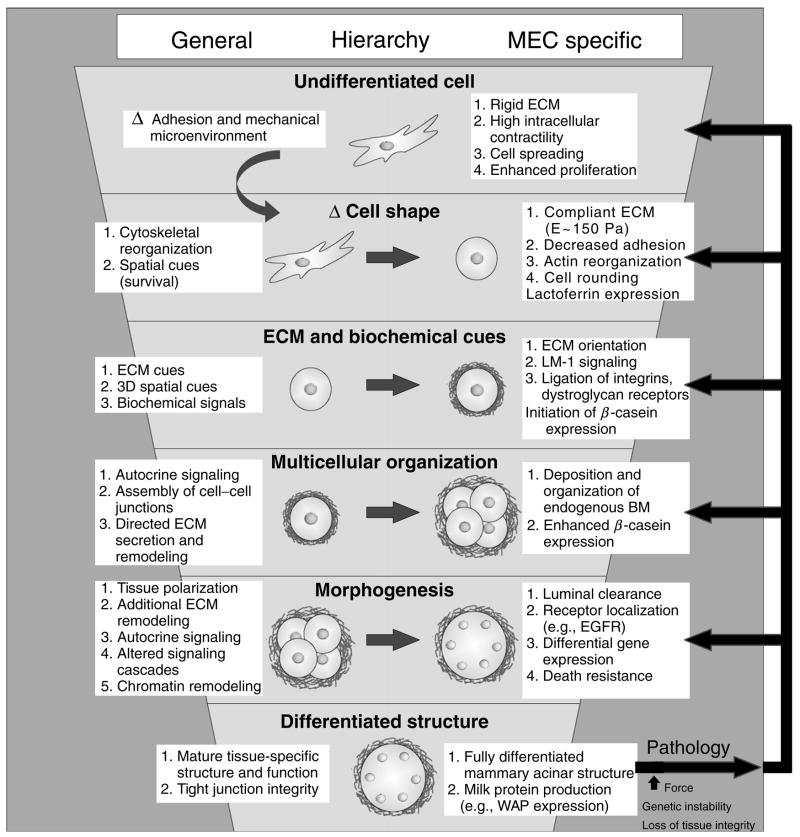
Fig. 1.
Biochemical and biophysical cues from the extracellular matrix regulate tissue-specific epithelial differentiation. Illustration depicting ECM regulation of tissue-specific differentiation through a progressively complex hierarchy of adhesion-regulated events functionally linked to changes in cell shape, receptor-initiated biochemical signaling, assembly of multicellular structures, and reciprocal biochemical and physical modification of the ECM microenvironment adjacent to the epithelial tissue. Undifferentiated cell (top): an undifferentiated cell interacting with a highly rigid 2D ECM substratum, such as matrix-coated tissue culture plastic, will adhere rapidly and, if given sufficient ECM ligand, will spread appreciably using multiple adhesion receptors, including integrins, and assemble mature focal adhesions. Epithelial cells grown on a rigid 2D matrix proliferate readily to form viable polarized cellular monolayers with adherens and tight junctions as well as prominent focal adhesions. Such cells exhibit robust Rho GTPase activation in response to exogenous stimuli, and require activated PI3 kinase or ERK signaling to survive. Under these conditions, epithelial cells do not assemble 3D tissue-like structures or express differentiated proteins in response to “differentiation cues.” Mechanical cues (second tier): an epithelial cell interacting with a highly compliant ECM readily adheres using multiple matrix receptors, including integrins, and assembles small immature focal complexes but fails to spread appreciably. Instead cells interacting with a compliant matrix exhibit profound reorganization of their actin cytoskeleton. MECs grown under these conditions can be induced to express lactoferrin if given the correct exogenous soluble cues. 3D ECM and biochemical cues (third tier): epithelial cells interacting with a highly compliant matrix in three dimensions adhere through multiple adhesion receptors including integrins, syndecans, and DG, and proliferate readily in response to exogenous growth factors. MECs interacting with a highly compliant 3D ECM can be induced to express abundant quantities of the differentiation protein β-casein. Multicellular organization, morphogenesis, and tissue differentiation (fourth to sixth tiers): in response to a 3D compliant ECM, ductal epithelial cells begin to interact with one another and assemble multicellular polarized structures with cell–cell junctions including adherens, scribble, and gap junctions. MECs assembled into multicellular 3D-polarized tissue-like structures begin to deposit and assemble an endogenous basally polarized basement membrane, show enhanced expression of milk protein expression such as β-casein, and exhibit enhanced long-term survival and apoptosis resistance to multiple exogenous stimuli including chemotherapeutics, immune receptor activators, and gamma irradiation. Long term culture of epithelial cells in the context of a 3D compliant ECM permits completion of tissue-like morphogenesis characterized by the assembly of an apically and basally polarized, growth-arrested tissue with a cleared central lumen and spatial restriction of various membrane associated proteins including growth factor receptors. Once a fully polarized and growth-arrested structure has formed, mammary acini can now be induced to express additional milk proteins such as WAP in response to lactogenic hormones. However, in response to an increase in matrix stiffness as occurs following chronic inflammation, injury, or tumorigenesis, or following genetic mutations and oncogene activation, tissue integrity becomes progressively compromised reversing the cell state to a less differentiated condition. In extreme cases, cells can behave analogous to undifferentiated, highly contractile single cells.
B. ECM Mechanics and Epithelial Behavior
Many important discoveries have been made concerning the molecular mechanisms by which the ECM influences epithelial behavior, including the requirement of signaling through laminin-dependent ligation of α3β1 and α6β4 integrins. In addition, cooperative ERK-PI3 kinase and RacGTPase-NFκB signaling through epidermal and insulin growth factor receptors and prolactin-dependent activation of Stat3 have been identified as key biochemical events involved in directing MEC growth, survival, and differentiation (Akhtar and Streuli, 2006; Muschler et al., 1999; Paszek et al., 2005; Zahir et al., 2003). The ECM not only influences epithelial behavior through biochemical signaling but also through the mechanical properties of the microenivornment.
Early studies with constrained versus released collagen gels revealed the importance of ECM mechanics in directing the cell shape of MECs to promote differentiation (Emerman and Pitelka, 1977). MECs plated on constrained collagen gels or gluteraldehyde-crosslinked rBM fail to differentiate in response to lactogenic stimuli and instead spread to form a 2D cell monolayer despite appropriate integrin-ECM ligation and growth factor signaling (reviewed in Roskelley et al., 1995; Weaver and Bissell, 1999). Furthermore, laminin- and proteoglycan-mediated ligation of dystroglycan (DG) has been strongly implicated as the primary mediator of ECM-directed cell shape fate determination in MECs and as a critical component in establishing a continuous BM (Muschler et al., 1999). The hypothesized mechanism seems to depend only on DG’s extracellular domain and to involve DG binding to laminin, which then polymerizes on the cell surface and onto adjacent DG-expressing cells, ultimately establishing a continuous BM. This process of ligation-driven BM assembly is almost certainly in competition with integrin-based and other adhesive processes. The latter seem more mechanosensitive and might dominate on rigid substrates versus soft substrates.
Although the detailed molecular mechanisms of the mechanosensitivity of MEC differentiation remain to be delineated, recent studies using both nontransformed and transformed human MECs suggest that Rho GTPase-dependent cell contractility regulates adhesion-directed, cell shape-dependent, epithelial tissue-specific functions (Paszek et al., 2005). Transformed human mammary epithelial tumor cells propagated on top of constrained collagen I gels assembled aberrant invasive structures with high Rho and ROCK activity, whereas they could form cell aggregates reminiscent of nontransformed tubules when grown in unconstrained collagen I gels (Keely et al., 1995). In concert with these in vitro observations, transformed mammary tumors were recently shown to exhibit enhanced Rho GTPase activity and exert elevated myosin-dependent cell contractility and aberrant integrin adhesions when compared to nontransformed MECs. Normalizing tumor cell contractility through application of pharmacological inhibitors of Rho, ERK signaling, or myosin could phenotypically revert the malignant phenotype (Paszek et al., 2005). Consistent with a critical role for matrix compliance in epithelial behavior, nontransformed MECs grown within highly compliant rBM gels or nonconstrained and compliant collagen I/rBM gels competently assemble polarized, growth-arrested acini-like structures. However, when grown within constrained collagen I/rBM gels or collagen I/rBM gels of higher concentration and stiffness, they form progressively disrupted, disorganized, and continuously proliferating colonies (Paszek et al., 2005; see also Sections III.A.1 and III.A.2).
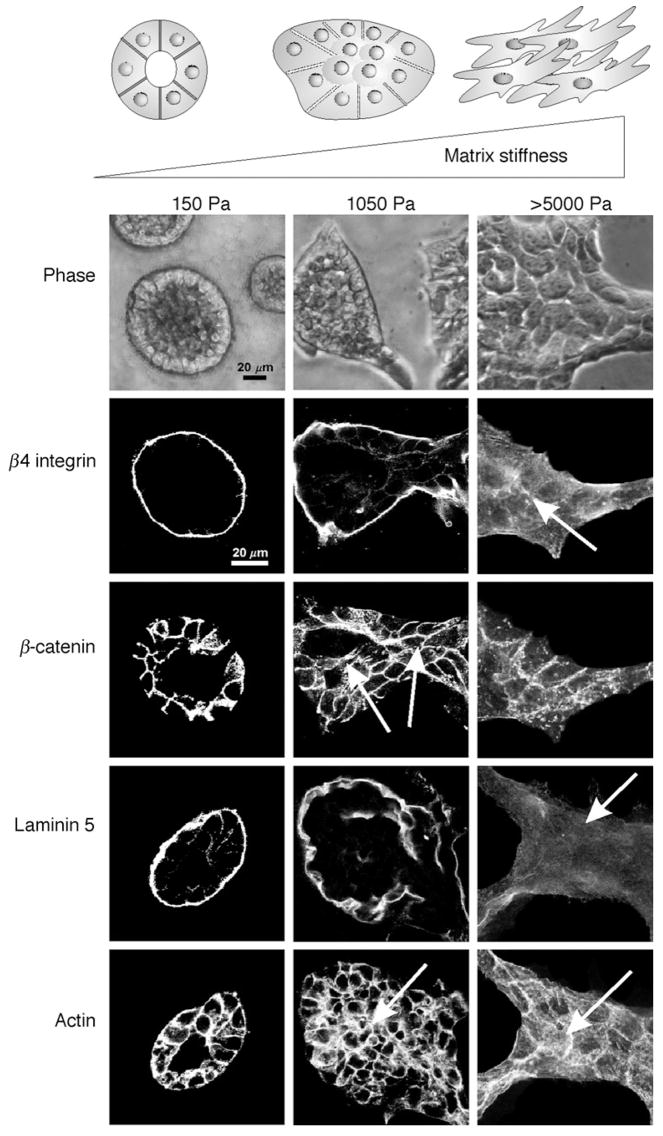
Through the application of defined, synthetic, rBM-crosslinked polyacrylamide gels, it was concluded that matrix stiffness and not matrix density or physical presentation constitutes a critical regulator of multicellular epithelial morphogenesis (Paszek et al., 2005; Fig. 2). These studies clearly emphasize the importance of myosin contractility and integrin adhesion maturation as matrix-regulated cell shape and force regulators. They have also identified altered ERK-dependent cell growth and survival, destabilization of cell–cell adhesions, and perturbed matrix assembly, as central mechanisms for further study. Indeed, the proper assembly of an endogenous cell-derived matrix plays a key role in epithelial differentiation, as has been illustrated by the necessity of proper laminin–nidogen interactions for mammary tissue differentiation and gene expression in culture (Pujuguet et al., 2000) and for multiple epithelial tissues including the kidney in vivo (Willem et al., 2002). Indeed, in lung development, increasing the compliance of the chest wall or decreasing the skeletal muscle fibers that aid in breathing modifies the biophysical properties of the tissue microenvironment by decreasing the applied force to the developing lung, leading to a decrease in lung growth, which further perturbs the tissue ECM and compromises tissue function (reviewed in Liu et al., 1999).
Fig. 2.
Matrix stiffness modulates MEC growth and morphogenesis. Phase-contrast and confocal immunofluorescence images of 3D MEC colonies on 3D rBM-crosslinked PA gels of increasing elastic moduli (E = 150–5000 Pa) after 20 days, showing progressively disrupted colony morphology as matrix stiffness increases (top). Cell–cell adherens junctions are disrupted and luminal clearance is compromised with even a modest increase in the elastic modulus of the matrix (E = 1050 Pa central panel; β-catenin and actin). Basal polarity is perturbed (disorganized β4 integrin and absence of basally deposited laminin-5) once the matrix stiffness stiffens appreciably (E > 5000 Pa; right panel). Scale bar is 20μm. Adapted from Paszek et al. (2005).
C. 3D Organotypic Model Systems
Key to engineering tissue-specific function is the application of an appropriate ECM in which the biochemical, biophysical, and spatial cues can be defined and controlled. An array of natural ECMs and a growing list of synthetic biomaterials, each with advantages and disadvantages, are available to the experimentalist. Ideally, a comprehensive assessment of what constitutes normal ECM composition, mechanical properties, and organization should be taken into consideration. Unfortunately, our comprehension of these variables has lagged behind, due to the complexity, lack of homogeneity, and anisotropy of biological materials.
rBMs isolated from Engelbreth–Holm–Swarm (EHS) mouse sarcomas have been routinely used to assemble tissue-like structures in culture and have been successfully applied to study mammary, thryoid, salivary gland, lung, and kidney epithelial cell morphogenesis and differentiation, and to distinguish between normal and transformed epithelial cells (Azuma and Sato, 1994; Debnath et al., 2003; Nogawa and Ito, 1995; O’Brien et al., 2001; Petersen et al., 1992; Yap et al., 1995). Similarly, fibrin gels have also been successfully used to assemble 3D normal and transformed tissue-like structures in culture (Alford et al., 1998). However, given that rBM is directly isolated from tissues, the matrix is inherently complex, poorly defined, and subject to complications with lot to lot variability and limitations due to the specific nature of the biochemical and biophysical environment associated with sarcomas. Fibrin gels, while attractive, also suffer from preparation variance. Additionally, fibrin gels are easily proteolyzed by cell-derived MMPs and consequently are not viable for long-term culture experiments. While alternative fibronectin sources that are less proteolytically sensitive have proven useful, these matrices have yet to be routinely applied to epithelial organ culture models.
As an alternative, collagen I gels have been extensively used as a 3D tissue matrix. The application of defined collagen gels to replace the more complex and biologically accurate rBM and fibrin gels has several advantages, including the fact that collagen I is a more biologically defined substrate, is relatively inexpensive to prepare or purchase, and is much more readily available. Because collagen I is the most common protein found in vertebrate animals and is structurally highly conserved, it is generally well tolerated for in vivo studies, and multiple cell types readily adhere to this substrate. In addition, the elastic moduli of a collagen I gel can be readily manipulated by varying collagen orientation, fibril crosslinking, concentration, or even biochemical modification or mutation (Christner et al., 2006; Girton et al., 1999; Martin et al., 1996; Roeder et al., 2002), thereby increasing its biological versatility (Elbjeirami et al., 2003; Grinnell, 2003). The magnitude and directional orientation of externally imposed tension can also be easily manipulated with collagen preparations. For example, through the release of collagen gels from the culture vessel, the isometric tension within the gel can be dramatically reduced (Rosenfeldt and Grinnell, 2000). Collagen gels can also be biochemically modified to facilitate epithelial functionality, as for example through the addition of either rBM, purified laminin, or derivatized peptides (Gudjonsson et al., 2002).
Purified, biologically derived materials, such as rBM and collagen I, have an intrinsic amount of biochemical and biophysical variability due to the inherent variability between animals and preparations. This variability leads to inconsistencies between experiments, as well as a high degree of heterogeneity within single gels. Additionally, the dynamic range of elastic moduli that can be reasonably achieved with these systems is limited by biochemical and biophysical constraints of these unique macromolecules. Therefore, although these materials have proven to be useful for clarifying the general influence of matrix on cell and tissue phenotypes, they are not as tractable for defining precise molecular mechanisms mediating mechanotransduction.
To address the issues listed above, especially control over matrix compliance, we and many others use a system first developed by Pelham and Wang (1997) that involves functionalizing synthetic polyacrylamide gels for cell culture (by cross-linking them with precise concentrations of ECM ligands) as 2D model systems for cell spreading, adhesion, and migration. Polyacrylamide gels represent tractable materials to allow studies of molecular pathways and signaling events of cells grown in various mechanical environments. The mechanical properties of these gels, which have been defined using rheology and atomic force microscopy (Engler et al., 2004; Guo et al., 2006; Yeung et al., 2005; Chapter 22 by Engler et al., this volume), can be manipulated by changing the relative concentration of acrylamide and the crosslinker, bis-acrylamide, yielding a system with precisely controlled biochemical and biophysical properties. Polyacrylamide is an exemplary material for studying cell behavior, as it is nonreactive, resistant to nonspecific binding and protein adhesion, and optically clear. The most significant downside to the polyacrylamide gel system is that acrylamide is cytotoxic in its monomeric form, which precludes the extension of its use to 3D cultures in which cells are embedded before polymerization. To overcome this limitation, we have used these polyacrylamide gels to reconstitute 3D conditions by overlaying MECs plated on top of rBM-crosslinked polyacrylamide gels with a blanket layer of rBM. Although the cells undergo normal morphogenesis under these conditions, there are some limitations inherent in this unique technique. Namely, and most importantly, this is neither a true 2D nor a complete 3D system, and the cells behave differently than they do in full 3D cultures (Leight et al., unpublished observations). Although this drawback leads to difficulty in interpretation and definition of these experiments, this system is suitable for approximating the physiological mechanical conditions under which epithelial cells grow and thrive. Alternatively, polyethylene glycol gels combined with bioactive peptides, such as fibronection- and laminin-binding sites, are also attractive biomaterials. However, their 3D organization is significantly different from that found in naturally occurring matrices and in vivo in that they typically have a greater matrix density and altered spatial orientation (reviewed in Zhang, 2004). Furthermore, because matrix remodeling is a critical aspect of epithelial morphogenesis, expensive bioactive peptides that can be proteolytically remodeled need to be incorporated into these synthetic biomaterials to permit proper tissue morphogenesis, migration, and to support long-term cell and tissue viability (reviewed in Lutolf and Hubbell, 2005). As an attractive new strategy in the arsenal of synthetic materials, novel matrices that incorporate recombinant natural and synthetic proteins and biopeptides are currently being developed and offer new hope for future applications.
Progress has been made in recapitulating tissue-specific morphology ex vivo either for tissue transplantation or for the study of tissue-specific function, but the application of these organotypic model systems to dissect the molecular basis of tissue homeostasis and disease has lagged behind significantly. The failure to exploit current 3D model systems for the study of cell behavior and signaling in the context of a tissue-like microenvironment and structure resides primarily in the lack of appropriate, cost-effective, easy, and reproducible strategies to manipulate, analyze, and assess cell function, signaling, and gene expression in these model systems. We have been studying the effect of cell shape, matrix compliance, adhesion, and dimensionality on cell behavior at the molecular levels, and here we provide a detailed description of the methods we have successfully used to do so.
III. Methods
A. Engineered Cell/Tissue Explants
-
Natural matrices: rBM
On ice, evenly coat the bottom of a tissue culture plastic dish with ice-cold rBM and incubate the dishes at 37 °C for 10–20 min to permit gel polymerization (see Table I for volume).
Prepare a single-cell suspension of trypsinized/washed cells in log-phase growth, and adjust the final cell concentration to 1 × 106 cells/ml media.
Aliquot cell suspension into individual tubes, adjusting cell number to desired total gel volume [see step (g)].
Centrifuge individual tubes to pellet cells (5 min, 180 × g rcf).
Aspirate the supernatant from the cell pellet, leaving 5% of the media behind.
Resuspend the cells in the remaining media by vigorously tapping the side of the tube, and place the tube on ice (note: do not vortex).
Add desired volume (Table I) plus an additional 10% of ice-cold rBM, and resuspend cells by gentle pipetting, taking care to avoid bubbles and keep the tube cold.
Transfer the cell/gel solution to the precoated tissue culture chambers, ensuring that the surface is covered with a uniform layer of cell/gel solution. Incubate at 37 °C for 20–30 min to permit gel polymerization.
Gently add complete growth media to the cultures until the gels are fully covered, taking care not to disrupt the gels. Incubate cultures in humidified chambers (37 °C) for desired length of time with media changes every 2–3 days.
-
Natural matrices: collagen I
-
Prepare collagen/rBM solution following an adapted version of the protocol published by BD (BD Biosciences—Discovery Labware, Catalog No. 354236 Product Specification Sheet).
Place the following on ice: acid-solubilized rat tail collagen I, sterile 10× phosphate-buffered saline (PBS), sterile deionized, distilled water (ddH2O), sterile 1-N sodium hydroxide (NaOH), and an empty sterile tube marked “Final Collagen Solution.”
- Calculate the desired volumes required for the experiment. (Note: Prepare 20% extra volume to account for material loss during experimental manipulations. See Table I for suggested total volumes.):
To the tube marked ‘‘Final Collagen Solution,’’ add the desired volumes of sterile ice-cold 10× PBS, 1-N NaOH, and ddH2O, then mix.
To the tube marked ‘‘Final Collagen Solution,’’ add the acid-solubilized collagen [from step (ii)], and mix gently by pipetting several times, taking care to keep the solution ice cold and to minimize air bubble formation. (Note: Do not over mix the gel solution, or the materials properties of the final gel will be altered.)
Neutralize the pH to 7.2–7.6 by titrating, drop-wise with 1-N NaOH until the solution turns a slight shade of reddish-purple indicated by the phenol red dye. Mix gently after the addition of each drop.
If desired, add an appropriate amount of rBM to the gel solution and leave on ice until required. The collagen solution can be used immediately or held on ice for 2–3 h.
Place the desired tissue culture dish on ice, and coat the bottom of each well with a thin layer of the collagen/rBM gel solution. Incubate the plate at 37 °C for 10–20 min to permit gel polymerization (see Table I for volume).
Prepare a single-cell suspension of trypsinized/washed cells in log-phase growth, and adjust the final cell concentration to 1 × 106 cells/ml media.
Aliquot cell suspension into individual tubes, adjusting cell number to the desired gel volume [see step (h)].
Centrifuge individual tubes to pellet cells (5 min, 180 × g rcf).
Aspirate the supernatant from the cell pellet, leaving 5% of the media behind.
Resuspend the cells in the remaining media by vigorously tapping the side of the tube and place the tube on ice (note: do not vortex).
Add desired volume plus an additional 10% of ice-cold collagen/rBM solution, and resuspend cells by gentle pipetting, taking care to avoid bubbles and to maintain the tube cold.
Transfer the cell/gel solution to the precoated tissue culture chambers ensuring that the surface is covered with a uniform layer of cell/gel solution. Incubate at 37 °C for 20–30 min to permit gel polymerization.
After polymerization, release the gel from the sides of the dish by running a small sterile spatula around the edge.
Gently add complete growth media to the cultures until the gels are fully covered, taking care not to disrupt the gels. Incubate cultures in humidified chambers (37 °C) for desired length of time with media changes every 2–3 days.
Anticipated results: Because the cells are seeded as single entities, it is possible to monitor the various stages of morphogenesis. Within 24 h all of the cells should be actively dividing, and by 48 h cell doublets should have formed with detectable polarized deposition of laminin-5 ECM protein and E-cadherin and β-catenin localized at cell–cell junctions. By 72–96 h cell proliferation should approach 60–85% [assessed by 5-bromo-2-deoxyuridine (BrdU) incorporation] and basal/apical tissue polarity should be established (determined by basal localization of β4 integrin and basal deposition of laminin-5). Within 10 days, fully embedded MECs within a compliant 3D rBM or collagen/rBM gel should have assembled small, essentially uniform, growth-arrested, polarized acini (Petersen et al., 1992; Weaver et al., 2002). Tissue morphology can be easily assessed by monitoring morphogenesis using immunofluoresence and morphometric assessment markers (Sections III.E.1 and III.E.2; Debnath et al., 2003; Paszek et al., 2005; Weaver et al., 1997). [Note: Cells grown on top of rBM as opposed to those completely embedded tend to form larger and more heterogeneous spheroids and exhibit slightly delayed lumenal clearance and growth arrest dynamics (Leight et al., unpublished observations). In addition, cells embedded within collagen/rBM gels of increasing elastic modulus (E > 675 Pa) form larger nonpolarized structures that lack a central lumen (Paszek et al., 2005)].
-
-
Synthetic matrices: functionalized polyacrylamide gels [Note: For immunofluorescence techniques, small (18–25 mm) coverslips can be used. For total RNA and protein isolation, larger (50 mm) coverslips should be used. The cell number should be optimized for the experiment performed, based on the degree of cell spreading and proliferation anticipated. For example, at least 750,000 cells are needed for total RNA and protein isolation, while significantly less is required for immunofluorescence visualization.]
Flame coverslip quickly and let cool.
Using a cotton swab, evenly and thoroughly coat the coverslip with 0.1-N NaOH. Air dry the coverslips until a filmy coat appears.
Using a p100 pipette tip, spread an even but thin coat of silane onto the surface of the coverslip (refer to Table II for amount). Allow the silane coating to dry (room temperature, 5–10 min), and place the coated cover-slips silane-side up in a Petri dish. (Note: Do not incubate longer than 20 min.)
Wash the coverslips thoroughly with ddH2O (minimum 3×; 10 min each), tapping the dish vigorously to remove excess liquid.
Incubate the coverslips in 70% glutaraldehyde (1:140; v:v in PBS) at room temperature for 30 min.
Wash the coverslips thoroughly with ddH2O (minimum 3×; 5–10 min each).
Arrange the coverslips face-up to dry.
After fully drying, the activated coverslips can be used immediately or stored for several weeks in a dry place. (Note: If the coverslips turn a rust-brown color, they should not be used as this is indicative of excess silane reacting with glutaraldehyde.)
In a microcentrifuge tube, mix the solutions required for gel preparation (see Table III for recipe).
In another microcentrifuge tube, weigh 5.6 mg of N-succinimidyl ester of acrylamidohexanoic acid (N6 crosslinker) per 1 ml of final desired solution. This compound can incorporate into the polyacrylamide gel, rendering it reactive with amine groups of proteins.
Add 70 μl of 200 proof ethanol and 80 μl ddH2O (per 1 ml of final solution) to the N6 cross-linker. Briefly, sonicate in a sonicating water bath (average peak power = 45 W), or vortex at highest setting until fully dissolved.
Add 844.4 μl of gel solution to the cross-linker/ethanol solution, and vortex.
Degas the gel solution using a vacuum flask or chamber for at least 30 min.
While the solutions are being degassed, evenly coat an additional set of equivalent-sized coverslips with Rain-x. Allow the Rain-x coating to dry at room temperature for 5–10 min, then gently buff the coverslip using a Kimwipe.
Place the activated coverslips face-up on a secured piece of paraffin.
Add 10 μl of freshly made 5% ammonium persulphate (w:v in ddH2O) per 1 ml of fully degassed acrylamide solution, mix well, and quickly dispense the desired volume of solution onto each activated coverslip (refer to Table III for volumes). Carefully place the Rain-x treated coverslip on top without trapping air bubbles and allow the gel solution to polymerize at room temperature for 25–60 min. (Note: Gel polymerization is indicated by retraction of the gel from the edge of the coverslip. Do not allow them to set for longer than 60 min, or the gel will dehydrate.)
While the gels are polymerizing, prepare a surface amenable to placing the gels on ice by covering the top surface of polystyrene tissue culture dishes with parafilm. (Note: The size is dependent on the size of the coverslips.)
Place the parafilm-affixed dishes on ice, and prepare the rBM solution for coating the coverslips. In a prechilled conical tube, prepare a solution of 140-μg/ml rBM, 5-mM EDTA, in 50-mM HEPES buffer, pH 8.0. (Note: Be sure to keep on ice to prevent polymerization of rBM.)
After the polyacrylamide gels have fully polymerized, carefully remove the top Rain-x-coated coverslip using a razorblade, taking care not to scratch the gel surface. Rinse each gel with ice-cold ddH2O. If using 18-and 25 mm coverslips, place the coverslips gel-side up on the parafilm-affixed dishes on ice. Immediately dispense the appropriate amount of rBM solution onto the gel (Table III). For 50 mm coverslips, pipette the appropriate amount of rBM solution (Table III) directly onto the paraffin-affixed dishes, and place the rinsed gels face down on top of the solution. Avoid trapping air bubbles under the coverslip. Incubate cover-slips on ice for 2 h.
While the coverslips are incubating, prepare the ethanolamine solution (1:100 v:v; 50-mM HEPES, pH 8.0) and chill the solution on ice.
Following incubation, individually rinse each gel in ice-cold ddH2O and wipe the parafilm-affixed dishes dry. Following the method in step (s), incubate the coverslips with ethanolamine on ice for 30 min to quench the unreacted N6 crosslinker (refer to Table III for volumes).
Soak the prepared gels in ice-cold PBS. In a sterile tissue culture hood, move the gels to sterile tissue culture dishes and store for up to 3 days in sterile 2-mM sodium azide/PBS at 4 °C.
Prior to cell plating, rinse each gel thoroughly in sterile PBS (minimum 3×), and leave the gels fully immersed in sterile PBS while preparing single-cell suspensions.
Prepare a single-cell suspension of trypsinized/washed cells in log-phase growth, and adjust the final cell concentration to 1 × 106 cells/ml media.
Aliquot cell suspension into individual tubes, adjusting cell number to desired concentration.
Centrifuge individual tubes to pellet cells (5 min, 180 × g rcf).
Aspirate the supernatant from the cell pellet, leaving 5% of the media behind.
Resuspend the cells in the remaining media by vigorously tapping the side of the tube and place the tube on ice (note: do not vortex).
Resuspend the cell suspension in cold growth medium, supplemented with 500-ng/ml fungizone, 50-μg/ml gentamicin sulfate, and 1:100 penicillin/streptomycin (stock concentration: 10,000 units penicillin/ml and 10,000-μg streptomycin/ml).
Pipette the desired cell number onto each gel taking care to ensure an even distribution of cells acrossthe gel surface, and allow the cells to adhere.(Note: The length of time for cells to adhere to the gel surface varies between cell types and needs to be optimized for each experiment.)
To facilitate 3D morphogenesis, after complete cell adhesion (minimum 6–8 h), cover cells in media containing 0.2-mg/ml rBM, and incubate cells for desired number of days. Change the culture media including 0.2-mg/ml rBM, fungizone, gentamicin, and penicillin/streptomycin, the following day and then every other day until termination of experiment.
Table I.
Matrix Volume (μl) per Well Size
| Underlay (μl) | Embed (μl) | |
|---|---|---|
| 60 mm | 1250 | 4250 |
| 35 mm | 450 | 1400 |
| 12-well | 200 | 700 |
| 4-well | 100 | 300 |
| 24-well | 100 | 300 |
| 48-well | 50 | 175 |
| 96-well | 20 | 60 |
Table II.
Amount of Each Solution to Add During Steps (c), (o), (q), and (s) When Using the Stated Coverslip Size
| Silane | Acrylamide solution | rBM solution | Ethanolamine | |
|---|---|---|---|---|
| 18-mm circle | 20 | 20 | 300 | 300 |
| 25-mm circle | 30 | 30 | 450 | 450 |
| 50-mm circle | 100 | 190 | 900 | 900 |
All values listed are in microliters.
Table III.
Recipes of 1-ml Polyacrylamide Gel Solutions for Given Elastic Modulus (Yeung et al., 2005)
| Elastic modulus (Pa) | 140 | 400 | 1050 | 5000 | 60,000 |
| Acrylamide (%) | 3 | 3 | 3 | 5.5 | 10 |
| Bis-acrylamide (%) | 0.04 | 0.05 | 0.1 | 0.15 | 0.5 |
| 40% Acrylamide (μl) | 75 | 75 | 75 | 137.5 | 250 |
| 2% Bis-acrylamide (μl) | 20 | 25 | 50 | 75 | 250 |
| 0.5-M HEPES, pH 4.22 (μl) | 100 | 100 | 100 | 100 | 100 |
| TEMED (μl) | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| ddH2O (μl) | 648.9 | 643.9 | 618.9 | 541.4 | 243.9 |
Anticipated results: After 14 days in culture, MECs plated on top of functionalized polyacrylamide gels with a rBM blanket layer, similar to cells overlaid on rBM gels (Debnath et al., 2003), form larger acini than their counterparts embedded within natural matrices. Analogous to cells grown within a natural matrix or grown on top of rBM with an overlay of rBM, by day 4, cells grown on the rBM PA gels should have acquired detectable cell–cell E-cadherin/β-catenin junctions as well as basal and apical polarity. Thus, by day 4, the cells should be highly proliferative but have acquired basal polarity, detectable by basally localized β4 integrin and deposition of a laminin-5- and collagen IV-rich endogneous BM as well as apically localized cortical actin. Studies have revealed that while the growth rate of MCF10As plated on soft (E = 140 Pa) and stiff (E = 5000 Pa) rBM-functionalized polyacrylamide gels are similar, cells interacting with a matrix stiffer than 1000 Pa fail to fully growth arrest. We have previously shown that MECs plated with a 3D rBM blanket layer on soft rBM-functionalized polyacrylamide gels undergo normal morphogenesis, while morphogenesis is perturbed in those plated on a stiff gel under the same conditions (Paszek et al., 2005; Fig. 2). A protocol outlining basic immunofluorescence techniques for each cell culture method is described in the Section III.E, for the visualization of characteristics indicative of morphogenesis. A comparison detailing morphogenetic characteristics of all of the described methods is currently in progress (Leight et al., unpublished observations).
B. Isolation of Bulk Proteins
-
Natural matrices: rBM (Note: This protocol is for isolating proteins from 1-ml rBM gels. Adjust volumes stated if necessary.)
Prepare 25 ml of ice-cold Dulbecco’s PBS solution (DPBS) containing 5-mM EDTA (EDTA/DPBS).
Supplement 25% of the EDTA/DPBS solution as prepared above with a cocktail of serine and cysteine protease and tyrosine phosphatase inhibitors. (Note: See Section IV for specific reagents and concentrations.)
Place culture on ice and gently aspirate off the medium.
Add 3 ml of the protease inhibitor/EDTA/DPBS solution prepared in step (b) to the culture dish, and pipette up and down using a p1000 pipette until a uniform suspension is obtained, avoiding the formation of insoluble foam.
Transfer the solubilized rBM gel solution to a 15-ml conical tube on ice.
Repeat steps (d) to (f) once, collecting all of the solubilized rBM into the conical tube.
Angle the conical tubes in a box of ice. Secure the tube and box of ice on a rocker and rock at 4 °C for 45–60 min.
Place 24 nonstick microcentrifuge tubes on ice, and aliquot the cell/rBM solution evenly among the tubes.
Centrifuge tubes at 4 °C for 10 min (3200 × g rcf).
Aspirate the supernatant, leaving 5% of the media behind.
Add 500 μl of the EDTA/DPBS solution to one tube, scraping the pellet against the side of the tube to resuspend the pellet. Mix well, and transfer to the next tube. Continue to scrape, mix, and transfer the solution to combine a total of four tubes into one.
Repeat steps (i) to (k) until the original 24 tubes are combined into one.
Centrifuge final tube at 4 °C for 15 min (21,000 × g rcf).
Prepare the lysis buffer, supplementing it with a cocktail of serine and cysteine protease and tyrosine phosphatase inhibitors.
Carefully aspirate the supernatant, and resuspend the pellet in 100–300 μl of lysis buffer (see note in Section IV), depending on the size of the pellet and the amount of protein expected.
Incubate on ice for 30 min.
Sonicate the lysis solution on ice with three pulses of 10 sec each, at an output power of 8 W, pausing for 30 sec between each pulse for the sample to cool down.
Centrifuge final tube at 4 °C for 10 min (10,500 × g rcf).
Transfer the supernatant to a clean microcentrifuge tube and fast freeze on dry ice. Store at −80 °C.
-
Natural matrices: collagen I (Note: This protocol is for isolating proteins from 1-ml collagen/rBM gels. Adjust volumes stated if necessary.)
Prepare 10 ml of ice-cold collagen release solution: 2-mg/ml collagenase, 2-mg/ml trypsin, and 5% fetal bovine serum in DMEM:F12. Keep on ice until needed and warm the amount needed just prior to experimentation. (Note: The trypsin should be EDTA-free or cell cadherin junctions will be disrupted.)
Gently aspirate medium from the 3D culture.
Add 2.5 ml of the collagen release solution and 500 μl of full-strength dispase and incubate at 37 °C for 10–15 min. (Note: The dispase should be prewarmed to 37 °C.)
Pipette up and down vigorously using a p1000 pipette to disrupt the gel. Incubate at 37 °C for 10–15 min.
Repeat step (d) until pipetting is easy and the colonies fall freely.
Transfer the solubilized gel solution to a 15-ml conical tube.
Centrifuge tubes for 5 min at 180 × g rcf.
Aspirate the supernatant, leaving 5% of the media behind.
Resuspend the pellet in 5 ml DMEM:F12 with 10% fetal bovine serum. Pellet by centrifugation for 5 min at 180 × g rcf.
Repeat steps (h) and (i) three times to thoroughly wash the pellet.
Aspirate the supernatant, leaving 5% of the media behind.
Resuspend the pellet in 5 ml ice-cold DMEM:F12. Pellet by centrifugation (4 °C, 5 min; 180 rcf).
Repeat steps (k) and (l) three times to thoroughly wash the pellet.
Aspirate the supernatant.
Prepare lysis buffer, supplementing it with a cocktail of serine and cysteine protease and tyrosine phosphatase inhibitors. (Note: See Section IV for specific reagents and concentrations.)
Carefully aspirate the supernatant, and resuspend the pellet in 100–300 μl of lysis buffer, depending on the size of the pellet and the amount of protein expected.
Incubate on ice for 30 min.
Sonicate the lysis solution on ice with three pulses of 10 sec each at an output power of 8 W, pausing for 30 sec between each pulse for the sample to cool down.
Centrifuge final tube at 4 °C for 10 min (10,500 × g rcf).
Transfer the supernatant to a clean microcentrifuge tube, and fast freeze on dry ice. Store at −80 °C.
-
Synthetic matrices: functionalized polyacrylamide gels (Note: This protocol is for 50-mm polyacrylamide gels.)
Prepare the lysis buffer, supplementing it with a cocktail of serine and cysteine protease and tyrosine phosphatase inhibitors. (Note: See Section IV for specific reagents and concentrations.)
Aspirate the medium from the gels and rinse with ice-cold DPBS.
Aspirate the DPBS from each plate and invert the lids of 60-mm tissue culture dishes onto ice.
Remove the coverslip from the plate, and very carefully wipe the edges clean with a cotton swab and/or Kimwipe to remove any cells adhered to the glass or edges of the gel to eliminate cell variability.
Pipette 250 μl of lysis buffer into the lid of the tissue culture plate.
Place the coverslip gel-side down onto the lysis buffer and incubate on ice for 5 min.
With a cell scraper, push the coverslip down and carefully scrape the coverslip against the lid of the culture plate for at least 5 min.
Squeeze the excess buffer from underneath the glass and remove the coverslip from the lid.
Transfer the solution into a microcentrifuge tube.
Incubate on ice for 30 min.
Sonicate the lysis solution on ice with three pulses of 10 sec each at an output power of 8 W, pausing for 30 sec between each pulse for the sample to cool down.
Centrifuge final tube at 4 °C for 10 min (10,500 × g rcf).
Transfer the supernatant to a clean microcentrifuge tube and fast freeze on dry ice. Store at −80 °C.
C. Isolation of Bulk mRNA
-
Natural matrices: rBM
Prepare a 3-ml solution/ml of rBM of 4-M guanidine thiocyanate; 25-mM sodium citrate–citric acid, pH 7.0; 0.5% (w:v) N-laurylsarcosine, sodium salt; 100-mM 2-mercaptoethanol. (Note: The 2-mercaptoethanol should be added fresh each time.)
Aspirate culture media and add 3 ml guanidine thiocyanate/mercaptoethanol solution as prepared above for each milliliter of rBM.
Using a p1000 pipette, pipette the solution up and down to solubilize the rBM.
Transfer the solubilized solution to an RNase-free polypropylene tube.
Add 1/10 volume {note: for steps (e) through (g), 1 volume refers to the sum of the residual culture volume [from step (b)] and the guanidine thiocyanate/mercaptoethanol volume} of DEPC-treated 2-M acetic acid–sodium acetate, pH 4.0, and mix thoroughly by vortexing at the highest setting. (Note: Be sure to obtain a completely homogeneous solution before proceeding to the next step.)
In a fume hood, add 1 volume of 0.1-M citrate, pH 4.3 buffered/saturated phenol, and mix thoroughly by vortexing at the highest setting. (Note: Protective safety attire should be worn. Be sure to obtain a completely homogeneous solution before proceeding to the next step.)
In a fume hood, add 2/10 volume of 49:1 (v:v) chloroform:isoamyl alcohol. Incubate at room temperature for 5–10 min. Then, briskly shake the tube by hand 8–10 times. Do not vortex here. (Note: A milky solution should form.)
Place the tube on ice for at least 15 min. The emulsion should break into two phases at this point, with a clear aqueous layer forming on top of a milky organic layer.
Centrifuge at 4 °C for 30 min (3200 × g rcf) to clarify the upper aqueous phase.
Transfer the upper aqueous phase to a clean RNase-free tube, taking care to avoid disturbing the interface, and add equal volume (the volume of the aqueous phase) of isopropanol, prechilled to −20 °C. Mix well and incubate at −20 °C for 2 h to overnight to precipitate the RNA.
Pellet RNA by centrifugation at 4 °C for 30 min (21,000 × g rcf).
Carefully aspirate the supernatant, being careful not to disturb the loosely adherent pellet.
Add 500 μl of very cold (−20 °C) 75% (v:v) ethanol in DEPC-treated ddH2O, and immediately pellet the total RNA by centrifugation at 4 °C for 15 min (21,000 × g rcf).
Repeat steps (l) and (m) six times to thoroughly wash the pellet and incubate the last wash in −20 °C 75% ethanol overnight.
Pellet total RNA by centrifugation at 4 °C for 15 min (21,000 × g rcf), and carefully aspirate the supernatant.
Air dry the pellet at room temperature until the pellet appears glassy (usually 10–15 min, depending on the volume of residual ethanol).
Dissolve the RNA pellet in DEPC-treated ddH2O. The volume depends on the expected amount of total RNA. (Note: When starting with 250,000 preformed spheroids, the expected RNA yield usually ranges between 10 and 15 μg.) Incubate on ice for 30 min to 1 h to thoroughly solubilize RNA.
Store at −80 °C until required.
-
Natural matrices: collagen I
Aspirate culture medium. Use a flamed RNase-free razor blade to cut the collagen gel into small pieces for easier RNA extraction, taking care to remove residual medium released prior to gel solubilization.
Add 3 ml of the prepared guanidine thiocyanate/mercaptoethanol solution (as described in C.1) per 1 ml of collagen, and allow to solubilize for 5 min at room temperature on a rocking platform.
Follow steps (C.1.e) to (C.1.r) to extract the total RNA.
-
Synthetic matrices: functionalized polyacrylamide gels
Prepare a solution of 4-M guanidine thiocyanate; 25-mM sodium citrate–citric acid, pH 7.0; 0.5% (w:v) N-laurylsarcosine, sodium salt; 100-mM 2-mercaptoethanol. (Note: The 2-mercaptoethanol should be added fresh each time.)
Invert the lids of 60-mm tissue culture dishes on the bench top. Add 600 μl of the prepared guanidine thiocyanate/mercaptoethanol solution for each 50 mm polyacrylamide gel to each lid.
Remove the coverslip from the culture dish, and very carefully wipe the edges clean with a cotton swab and/or Kimwipe to remove any cells adhered to the glass or edges of the gel to eliminate cell variability.
Place the coverslip gel-side down onto the guanidine thiocyanate/mercaptoethanol solution, and incubate on ice for 5 min.
With a cell scraper, push the coverslip down and carefully scrape the coverslip against the lid of the culture plate for at least 5 min.
Squeeze excess buffer from underneath the glass and remove the coverslip from the lid.
Transfer the solution to an RNase-free polypropylene tube and follow steps outlined in (C.1.e) to (C.1.r) to extract the total RNA.
D. Rapid Protein Isolation Techniques
-
Synthetic matrices: functionalized polyacrylamide gels (Note: This protocol is for isolating Rac-GTP. For rapid isolation of other proteins, the assay will be similar, but may need to be modified and/or optimized. At least 600 μg of total protein is needed. Adjust the number and size of gels to obtain enough protein.)
-
Prepare glutathione-sepharose beads for glutathione-S-transferase-tagged p21-binding domain of Pak1 (GST-PBD) binding.
Centrifuge 1 ml of 50% glutathione-sepharose slurry at 4 °C for 30 sec (21,000 × g rcf).
Aspirate the supernatant and add 500 μl MLB (Section IV. D.I). Pellet by centrifugation at 4 °C for 30 sec (21,000 × g rcf).
Repeat step (ii) three times to wash the beads.
Aspirate the supernatant and resuspend the sepharose beads in an equal volume of MLB (500 μl) to produce a 50% slurry.
Incubate the 20- to 30-μl GST-PBD with 20- to 30-μl sepharose slurry (4 °C; 20 min).
Supplement MLB with a cocktail of serine and cysteine protease and tyrosine phosphatase inhibitors. (Note: See Section IV for specific reagents and concentrations.)
Aspirate the medium from the gels and rinse with ice cold DPBS.
Aspirate the DPBS from each plate and invert the lids of 60-mm tissue culture plates onto ice.
Remove the coverslip from the plate, and very carefully wipe the edges clean with a cotton swab and/or Kimwipe to remove any cells adhered to the glass or edges of the gel to eliminate cell variability.
Pipette 350- to 400-μl MLB into the lid of the tissue culture plate.
Place the coverslip gel-side down onto the lysis buffer and incubate on ice for 5 min.
With a cell scraper, push the coverslip down and carefully scrape the coverslip against the lid of the culture plate for at least 5 min.
Squeeze the excess buffer from underneath the glass and remove the coverslip from the lid.
Pipette the MLB solution into a microcentrifuge tube, combining like samples.
Centrifuge the tubes at 4 °C for 5 min (10,500 × g rcf).
Transfer at least 800 μl of lysate into the tube containing GST-PBD. Leave at least 50 μl of lysate for determining total Rac separately.
Gently rock the solution at 4 °C for 60 min.
Collect the GST-PBD-Rac mixture by centrifugation at 4 °C for 30 sec (21,000 × g rcf).
Carefully aspirate the supernatant and resuspend the beads in 500 μl of ice-cold MLB.
Centrifuge at 4 °C for 30 sec (21,000 × g rcf).
Repeat steps (o) and (p) three times.
Resuspend the beads in 15 μl of sample loading buffer for electrophoresis and vortex briefly. Heat the samples to 95 °C for 10 min. (Note: Visually confirm that the sample loading buffer penetrates the beads before heating.)
Centrifuge the solution at room temperature for 30 sec (21,000 × g rcf).
Fast freeze the samples in a dry ice/ethanol bath and store at −80 °C.
-
E. Immunofluorescence
-
Natural matrices: rBM
Aspirate cell culture medium, and wash in DPBS (containing Ca2+ and Mg2+), if cultures were grown in serum.
Add equal-volume neutralized collagen solution [see step (A.2.a) for directions, omitting ddH2O] and mix thoroughly. (Note: Collagen is added to strengthen the matrix and permits easier cryosectioning.)
Incubate the gels at 37 °C for 30 min to polymerize.
-
If desired, sections can be triton-extracted prior to fixation to facilitate cytoskeletal and nuclear visualization.
Prepare ice-cold cytoskeletal extraction buffer (see Section IV for details) containing Triton X-100 (0.005%; v:v) and 5-mM EGTA, supplemented with protease and phosphatase inhibitors.
Add equal-volume extraction buffer and incubate at room temperature for 30 min.
Fix with 2% paraformaldehyde (pH 7.4) at 4 °C overnight.
Rinse cultures with PBS/glycine at 4 °C (minimum 3×, 5 min each).
Incubate cultures with 18% sucrose-PBS/glycine at 4 °C for 3 h.
Incubate cultures with 30% sucrose-PBS/glycine at 4 °C for 3 h.
Rinse cultures with PBS/glycine at 4 °C for 5 min.
Add OCT Tissue Tek compound for cryosection, and rapidly freeze on a bed of dry ice and ethanol or in liquid nitrogen. Store culture blocks at −80 °C until required.
-
Prepare activated gelatin-coated microscope slides:
Autoclave (121 °C, 30 min) 0.5 g gelatin in 25 ml ddH2O and cool to room temperature.
Add 0.05 g chromium potassium sulfate dissolved in 75 ml of ddH2O to precooled gelatin solution.
Store at 4 °C until required.
-
Prepare microscope slides for tissue culture sections:
To minimize antibody solution requirement, generate a hydrophobic incubation ring on a microscope slide. This can be done using a hydrophobic (wax) pen to draw small rings slightly larger than the tissue samples to be stained. Additionally, this can be achieved by melting paraffin at 95 °C around the circumference of a microcentrifuge tube or the lid of a 15 ml conical tube lid, and then gently, but firmly placing the tube on the slide. Incubate the slide on a heating block at 58 °C for 1 min. Carefully, remove the tube/lid from the slide while the slide is on the heating block, and let the paraffin solidify at room temperature.
Evenly coat the interior of the paraffin ring with activated 0.5% gelatin, and air dry at room temperature overnight.
Using a cryostat, cut frozen sections of 3D tissue blocks (5–20 μm), and transfer sections to the gelatin-coated paraffin ring. Store the slides at −80 °C until required.
For immunostaining, remove sections from freezer, thaw, and air dry at room temperature for 5–20 min.
Rehydrate sections in IF buffer at room temperature for 20 min.
Incubate sections in blocking buffer at room temperature for 1 h or at 4 °C overnight in a humidified chamber.
Incubate sections in primary antibody solution at room temperature for 1–2 h or at 4 °C overnight in a humidified chamber.
Wash sections in IF buffer at room temperature, minimum three times for 15 min each.
Incubate in secondary antibody in IF buffer at room temperature for 45 min. (Note: If the secondary antibody is fluorescent, keep slides under foil.)
Wash sections in IF buffer at room temperature, minimum three times for 10 min each.
To visualize nuclei, counterstain with 1-μg/ml DAPI in PBS/glycine at room temperature for 5 min.
Rinse each gel in PBS/glycine at room temperature, minimum three times for 5 min each.
Aspirate residual liquid from the gels and mount sections with mounting media. Leave under foil to dry at room temperature for 15–30 min. Secure with nail polish when dry.
Store at −20 °C until visualization.
-
Natural matrices: collagen I
Follow the steps outlined in Section III.E.1, omitting steps (b) and (c).
-
Synthetic matrices: functionalized polyacrylamide gels
Aspirate cell culture medium and rinse cells grown in serum with DPBS (containing Ca2+ and Mg2+).
Fix with 2% paraformaldehyde, pH 7.4 at room temperature for 30 min.
Rinse with IF buffer at room temperature, minimum three times for 5 min each.
With fine tip forceps, remove the coverslip from the cell culture dish, and place the coverslips gel-side up on a secured piece of paraffin.
Follow protocol outlined in steps (Section III.E.1.p) to (Section III.E.1.v).
Aspirate residual liquid from the gels and mount gels onto microscope slides with mounting media. Leave under foil to air dry at room temperature for 15–30 min. Secure with nail polish when dry.
Store at −20 °C until visualization.
IV. Materials
A. Engineering Tissue Explants
-
Natural matrices: rBM
Wet ice
rBM, BD Biosciences BD Matrigel™ [Note: As there is inherent lot-to-lot variability, each lot should be tested prior to use. When deciding which lots to choose, we prefer lots that have endotoxin levels less than 2 units/ml and protein concentrations ranging from 9 to 12 mg/ml. Additionally, we have found that MECs behave similarly in Matrigel and Growth Factor Reduced Matrigel (both from BD), although this should be tested for each cell line. Each lot should be tested for compatibility with the various cell lines by looking for changes in morphology in 3D cultures and ensuring low background nucleic acid and IgG levels that would interfere with RNA isolation and immunofluorescence procedures. It should be noted also that the rheological properties of commercially available rBM preparations can also vary from E = 50 to 200 Pa (unpublished observations).]
Cell culture supplies (medium, trypsin, trypsin-inhibiting agent)
-
Natural matrices: collagen I/rBM
Wet ice
Acid-solubilized rat tail collagen I [Note: Although this protocol is designed for acid-solubilized rat tail collagen I, we have previously used acid-solubilized bovine dermal collagen I (ICN Biomedicals Catalog No.152394) and acid-solubilized rat tail collagen I (BD Labware Catalog No. 354236) to embed MECs in 3D ECM gels. As there is inherent lot variability, each lot should be tested prior to use to determine the appropriate concentration and gelling time for each system. Additionally, the elastic modulus of the gels can be manipulated by varying the concentration of collagen I (E = 20–1800 Pa; Leight et al., unpublished observations). We have found that the elastic modulus varies between lots, so each lot should be tested prior to experimentation (unpublished observation).]
10× DPBS containing 1.33-g/ml Ca2+ and 1.0-g/ml Mg2+, supplemented with phenol red.
Deionized, distilled water (ddH2O)
1-N sodium hydroxide (NaOH)
rBM
Cell culture supplies (medium, trypsin, trypsin-inhibiting agent)
Small sterile spatula
-
Synthetic matrices: functionalized polyacrylamide gels (Note: When possible, presterilized materials should be used.)
Bunsen burner
Coverslips (No. 1 thickness, hydrolytic class 1 borosilicate coverslips, Fisher; see note in Section III regarding the size of the coverslip required.)
Cotton swabs
0.1-N NaOH
3-Aminopropyltrimethoxysilane, 97%, Sigma Aldrich
Glutaraldehyde, 70%, Sigma Aldrich
PBS
ddH2O
40% Acrylamide
2% Bis-acrylamide
0.5-M N-(2-hydroxyethyl)-piperazine-N′-2-ethanesulfonic acid (HEPES buffer), pH 4.22
N,N,N′,N′-Tetramethylethylenediamine (TEMED)
N-Succinimidyl ester of acrylamidohexanoic acid (N6 cross-linker) [Note: The N6 cross-linker can be synthesized following the protocol outlined in Pless et al. (1983).]
200-Proof ethanol
Rain-X™ (available at an automobile parts store).
Parafilm™
Ammonium persulfate (APS)
50-mM HEPES buffer, pH 8.0
0.5-M ethylenediaminetetraacetic acid (EDTA), pH 8.0
rBM
Ethanolamine, Sigma Aldrich
Sterile PBS
Sodium azide
Cell culture supplies (medium, trypsin, trypsin-inhibiting agent)
Cell culture antibiotics and antimycotics [fungizone (amphotericin B, Sigma Aldrich), gentamicin sulfate (Gibco™, penicillin G/streptomycin sulfate)]
B. Isolation of Bulk Proteins
-
Natural matrices and synthetic matrices
DPBS (for rBM and functionalized polyacrylamide gels only)
0.5-M EDTA, pH 8.0 (for rBM and functionalized polyacrylamide gels only)
Collagenase, Roche Applied Sciences (for collagen/rBM gels only)
Trypsin (for collagen/rBM gels only)
Dispase, BD Biosciences (for collagen/rBM gels only)
Fetal bovine serum (for collagen/rBM gels only)
DMEM:F12 (for collagen/rBM gels only)
Wet ice
Serine and cysteine protease and tyrosine phosphatase inhibitor cocktail: 2-μg/ml aprotinin (Roche Applied Sciences), 1-μg/ml leupeptin (Sigma Aldrich), 1-μg/ml E-64 (Sigma Aldrich), 50-mM sodium fluoride, 10-μg/ml pepstatin A (Sigma Aldrich), 0.5-mM benzamidine (Sigma Aldrich), 1-mM sodium orthovanadate, 1-mM Pefabloc SC (Roche Applied Sciences). [Note: Activate 125-mM sodium orthovanadate with 100-mM hydrogen peroxide at room temperature for 20 min just prior to use (Zhang et al., 2005). Add activated sodium orthovanadate and Pefabloc SC to solution just prior to use.]
-
Appropriate lysis buffer (Note: The choice of lysis buffer is dependent on the nature of the protein to be studied and should be optimized for each experiment. Radioimmunoprecipitation assay (RIPA) and Laemmli buffers are common choices. RIPA buffer enables suitable extraction of cytoplasmic proteins, while Laemmli buffer is effective for membrane-bound and nuclear proteins.)
RIPA buffer: 50-mM Tris–HCl pH 8.0, 50-mM sodium chloride, 0.5% (w:v) sodium deoxycholate, 1% IGEPAL® CA-630 (Sigma Aldrich), 0.1% (w:v) SDS
Laemmli buffer: 33.3-mM Tris–HCl pH 8.0, 50-mM EDTA, 2% (w:v) SDS
Cell scraper, Sigma Aldrich
Dry ice
C. Isolation of Bulk mRNA
-
Natural matrices and synthetic matrices
Diethyl pyrocarbonate (DEPC)-treated ddH2O (Note: Prepare by adding 1-ml DEPC (Sigma Aldrich) to 500-ml ddH2O in a glass bottle. Mix vigorously, and incubate overnight in a fume hood, leaving the cap slightly loose. Autoclave for 45 min at 121 °C, 20 psig, and store at room temperature.)
4-M guanidine thiocyanate; 25-mM sodium citrate–citric acid, pH 7.0; 0.5% (w:v) N-laurylsarcosine (ICN Biomedicals), sodium salt; in DEPC-treated ddH2O
2-Mercaptoethanol
DEPC-treated 2-M acetic acid–sodium acetate, pH 4.0 (Note: Prepare by adding 4.022-ml glacial acetic acid to 31-ml ddH2O. Adjust the pH to 4.0 with 2-M sodium acetate. Add 2 μl/ml total volume of DEPC. Mix well and incubate at room temperature overnight. Autoclave for 15 min at 121 °C, 18 psig, and store at room temperature.)
0.1-M citrate, pH 4.3 buffered/saturated phenol, Sigma Aldrich
49:1 (v:v) Chloroform:isoamyl alcohol
Wet ice
Isopropanol
75% Ethanol in DEPC-treated ddH2O
D. Rapid Protein Isolation Techniques
-
Synthetic matrices: functionalized polyacrylamide gels
MLB: 25-mM HEPES, pH 7.5; 150-mM sodium chloride; 1% Igepal CA-630; 10-mM MgCl2; 1-mM EDTA; 10% glycerol
Serine and cysteine protease and tyrosine phosphatase inhibitor cocktail: 2-μg/ml aprotinin (Roche Applied Sciences), 1-μg/ml leupeptin (Sigma Aldrich), 1-μg/ml E-64 (Sigma Aldrich), 50-mM sodium fluoride, 10-μg/ml pepstatin A (Sigma Aldrich), 0.5-mM benzamidine (Sigma Aldrich), 1-mM sodium orthovanadate, 1-mM Pefabloc SC (Roche Applied Sciences) [Note: Activate 125-mM sodium orthovanadate with 100-mM hydrogen peroxide at room temperature for 20 min just prior to use (Zhang et al., 2005). Add activated sodium orthovanadate and Pefabloc SC to solution just prior to use.]
Sample loading buffer: 0.25-M Tris, pH 6.8; 50% (v:v) glycerol; 2% (w:v) sodium dodecyl sulfate (SDS); 50-μl/ml 2-mercaptoethanol in ddH2O, supplemented with bromophenol blue
Wet ice
Ice-cold DPBS
Glutathione-sepharose slurry, Sigma Aldrich
E. Immunofluorescence
-
Natural matrices and synthetic matrices
DPBS (containing 1.33-g/ml Ca2+ and 1.0-g/ml Mg2+)
Wet ice
Acid-solubilized rat tail collagen I
10× DPBS with Ca2+ and Mg2+, supplemented with phenol red
ddH2O
1-N NaOH
1.5× Cytoskeletal extraction buffer: 150-mM sodium chloride; 450-mM sucrose; 15-mM piperazine-1,4-bis(2-ethanesulfonic acid) (PIPES) buffer, pH 6.7; 5-mM magnesium chloride in ddH2O
Triton X-100
Serine and cysteine protease and tyrosine phosphatase inhibitor cocktail: 2-μg/ml aprotinin, 1-μg/ml leupeptin, 1-μg/ml E-64, 50-mM sodium fluoride, 10-μg/ml pepstatin, 0.5-mM benzamidine, 1-mM sodium orthovanadate, 1-mM Pefabloc SC (Note: Activate sodium orthovanadate with hydrogen peroxide just prior to use. Add activated sodium orthovanadate and Pefabloc SC to solution just prior to use.)
2% (w:v) Paraformaldehyde, pH 7.4
PBS/glycine (1× PBS, supplemented with 75-mg/ml glycine)
Dry ice
0.5% (w:v) Porcine gelatin (Sigma Aldrich) in ddH2O, supplemented with 0.5-mg/ml chromium potassium sulfate
Microscope slides
Paraffin
IF buffer: 7.6-mg/ml sodium chloride, 1.9-mg/ml sodium phosphate, 0.4-mg/ml potassium phosphate monobasic, 0.5-mg/ml sodium azide, 1-mg/ml bovine serum albumin, 0.2% (v:v) Triton X-100, 0.05% (v:v) Tween 20 in ddH2O
Blocking buffer: 10% normal goat serum, 0.13-mg/ml appropriate Fab fragments, in IF buffer
Primary and secondary antibodies
Aluminum foil
4′,6-Diamidino-2-phenylindole (DAPI), Sigma Aldrich
Mounting medium (e.g., Vectashield Mounting Medium®, Vector)
Nail polish
V. Discussion
Epithelial tissues are highly complex, organized 3D structures that evolve incrementally during development to generate these specialized functional tissues through spatially and temporally controlled stromal–epithelial interactions. The tissue microenvironment of the epithelium is composed of multiple stromal cell types, and these cellular components, together with the epithelium, are embedded within a proteinaceous ECM. It is the combination of cellular and ECM interactions, operating through controlled biochemical and physical cues, that ultimately regulates epithelial cell fate and function. The goal of an epithelial experimentalist is to recreate at least some of the intricate relationships that exist between the various cell types and the ECM in vivo, but in a simple format in culture, such that the recreated system is more amenable to molecular studies without severely compromising the epithelial cell’s normal tissue behavior. The idea is that experimental observations made using such contrived but simplified systems will ultimately be distilled into the critical information that is necessary to systematically engineer surrogate tissues for replacement therapy or to develop tractable treatments to prevent and cure various diseases. Toward this lofty goal, considerable research has been successfully directed at determining how each individual cell variable and microenvironmental component influences epithelial cell behavior (Mostov et al., 2005; Paszek et al., 2005; Petersen et al., 1992).
Despite the efforts, our understanding of what controls the epithelial cells’ behavior within the complex 3D tissue-like structure and how combinations of microenvironmental cues might cooperate to influence epithelial function remains rudimentary at best. Moreover, although we and others have been successful in generating functional data using these “crude” systems, it remains difficult to isolate specific responses to allow the identification of the precise molecular mechanisms linked to the generation of a given tissue phenotype. For example, MECs grown within a 3D rBM simultaneously and acutely change their shape, matrix adhesion, cell contractility, and signaling, as well as growth factor and apoptosis responsiveness, as compared to MECs interacting with a 2D rigid substrate (Debnath et al., 2003; Wang et al., 1998; Weaver et al., 2002). To address this difficulty, tractable culture models that reproducibly reconstruct individual aspects of tissue organization and function and that encompass controllable homotypic and heterotypic cell–cell interactions and ECM cues are needed. Preferably, these newly engineered culture systems will be amenable to precise biochemical and physical manipulation and will be sufficiently robust and versatile for routine experimentation. Additionally, they should be inexpensive and lend themselves to easy and reproducible manipulation.
Conventional organotypic systems are often expensive, labor intensive to generate, and suffer from experimental inconsistencies. It is now feasible to synthesize biocompatible matrices to study the effect of individual parameters, such as matrix binding and ECM orientation, receptor expression and activity, cell shape, matrix compliance, and even ECM dimensionality through a combination of nonreactive hydrogels with cell-adhesive sites. Recombinant synthetic proteins have also been used to promote specific adhesion and to foster cell-specific degradation and remodeling of the matrix by incorporating proteolytically degradable peptide sequences. We and others have applied similar strategies to successfully study the phenotypic behavior of individual cells in response to various physical, architectural, and biochemical cues including issues pertaining to the regulation of cell survival (Buckley et al., 1999; Capello et al., 2006; Chen et al., 1997; Friedland et al., unpublished observations), migration (Gobin and West, 2002; Wong et al., 2003), stem cell fate (McBeath et al., 2004), differentiation (Bokhari et al., 2005; Mauck et al., 2006), and growth regulation (Bokhari et al., 2005; Georges et al., 2006; Paszek et al., 2005). However, such specialized systems have limited application for studying cell behavior in multicellular structures and 3D tissues and have only sparingly been applied to the study of heterotypic cell–cell interactions (Georges et al., 2006). Moreover, many of the currently available synthetic biomaterials exhibit incompatible material properties such as high stiffness, elevated matrix density, and random matrix presentation that render them less than suitable for the study of epithelial tissue morphogenesis (reviewed in Zhang, 2004). To address these concerns, newer generations of biomaterials are currently being developed, including highly compliant synthetic matrices generated using combinations of polyethylene glycol and methylcellulose conjugated with various bioactive peptides and MMP-cleavable proteins (Leach JB, personal communication), polyethylene glycol gels with functionalized recombinant proteins (Rizzi and Hubbell, 2005), electrically spun collagen gels with precisely controlled orientations (Matthews et al., 2002), and synthetic gels with gradients of ECM compliance that recreate durotactic-directed cell migration during development, wound closure, and tumor metastasis (Lo et al., 2000; Wong et al., 2003; Zaari et al., 2004). The application of these novel materials together with the availability of pluripotent and tissue-specific stem cells provide encouragement that we are at least moving closer to our idealized model systems, to begin to elucidate the mechanisms regulating multicellular epithelial tissue-specific structure and function.
In addition to these important considerations, it is recognized that tissues develop progressively and evolve through reciprocal and dynamic dialogues between the cellular and stromal components and tissue milieu, and this temporal relationship must also provide the mature tissue with physiological advantages that need to be identified and assessed. For example, although bioengineers have been able to successfully reconstruct blood vessels that are phenotypically and functionally identical to differentiated arteries in vivo, the engineered vessels rapidly fatigue when transplanted into a host in vivo. One must also consider that our ultimate goal should be the engineering of complex 3D microenvironments that are amenable to dynamic physical and biochemical modification. When seeded with pluripotent and tissue-specific stem cells, they should allow systematic development ex vivo of viable, live tissues to be used for routine and faithful experimentation and for various clinical applications. Clearly, we have our work cut out for us.
Acknowledgments
We thank J. C. Friedland and J. N. Lakins for their contributions. This work was supported by NIH grants CA078731 and BRP HL6438801A1 (to V.M.W.) and T32HL00795404 (to K.R.J.), DOD grants W81XWH-05-1-330 and DAMD17-01-1-0367 (to V.M.W.), and a NSF graduate fellowship (to J.L.L.).
References
- Akhtar N, Streuli CH. Rac1 links integrin-mediated adhesion to the control of lactational differentiation in mammary epithelia. J Cell Biol. 2006;5:781–793. doi: 10.1083/jcb.200601059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford D, Baeckstrom D, Geyp M, Pitha P, Taylor-Papadimitriou J. Integrin-matrix interactions affect the form of the structures developing from human mammary epithelial cells in collagen or fibrin gels. J Cell Sci. 1998;111(Pt 4):521–532. doi: 10.1242/jcs.111.4.521. [DOI] [PubMed] [Google Scholar]
- Azuma M, Sato M. Morphogenesis of normal human salivary gland cells in vitro. Histol Histopathol. 1994;4:781–790. [PubMed] [Google Scholar]
- Barros EJ, Santos OF, Matsumoto K, Nakamura T, Nigam SK. Differential tubulogenic and branching morphogenetic activities of growth factors: Implications for epithelial tissue development. Proc Natl Acad Sci USA. 1995;10:4412–4416. doi: 10.1073/pnas.92.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: Microenvironmental influences in the normal and malignant breast. Differentiation. 2002;9–10:537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Weaver VM, Lelievre SA, Wang F, Petersen OW, Schmeichel KL. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59(7 Suppl):1757–1763s. discussion 1763s–1764s. [PubMed] [Google Scholar]
- Bokhari MA, Akay G, Zhang S, Birch MA. The enhancement of osteoblast growth and differentiation in vitro on a peptide hydrogel-polyHIPE polymer hybrid material. Biomaterials. 2005;25:5198–5208. doi: 10.1016/j.biomaterials.2005.01.040. [DOI] [PubMed] [Google Scholar]
- Buckley S, Driscoll B, Barsky L, Weinberg K, Anderson K, Warburton D. ERK activation protects against DNA damage and apoptosis in hyperoxic rat AEC2. Am J Physiol. 1999;1(Pt 1):L159–L166. doi: 10.1152/ajplung.1999.277.1.L159. [DOI] [PubMed] [Google Scholar]
- Capello A, Krenning EP, Bernard BF, Breeman WA, Erion JL, de Jong M. Anticancer activity of targeted proapoptotic peptides. J Nucl Med. 2006;1:122–129. [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;5317:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Micropatterned surfaces for control of cell shape, position, and function. Biotechnol Prog. 1998;3:356–363. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- Christner PJ, Gentiletti J, Peters J, Ball ST, Yamauchi M, Atsawasuwan P, Beason DP, Soslowsky LJ, Birk DE. Collagen dysregulation in the dermis of the Sagg/+mouse: A loose skin model. J Invest Dermatol. 2006;3:595–602. doi: 10.1038/sj.jid.5700100. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;3:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Elbjeirami WM, Yonter EO, Starcher BC, West JL. Enhancing mechanical properties of tissue-engineered constructs via lysyl oxidase crosslinking activity. J Biomed Mater Res A. 2003;3:513–521. doi: 10.1002/jbm.a.10021. [DOI] [PubMed] [Google Scholar]
- Emerman JT, Pitelka DR. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro. 1977;13:316–328. doi: 10.1007/BF02616178. [DOI] [PubMed] [Google Scholar]
- Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;1(Pt 1):617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Dowling J, Segre J, Lo SH, Yu QC. Integrators of epidermal growth and differentiation: Distinct functions for beta 1 and beta 4 integrins. Curr Opin Genet Dev. 1997;5:672–682. doi: 10.1016/s0959-437x(97)80016-0. [DOI] [PubMed] [Google Scholar]
- Georges PC, Miller WJ, Meaney DF, Sawyer ES, Janmey PA. Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. Biophys J. 2006;8:3012–3018. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girton TS, Oegema TR, Tranquillo RT. Exploiting glycation to stiffen and strengthen tissue equivalents for tissue engineering. J Biomed Mater Res. 1999;1:87–92. doi: 10.1002/(sici)1097-4636(199907)46:1<87::aid-jbm10>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Gobin AS, West JL. Cell migration through defined, synthetic ECM analogs. FASEB J. 2002;7:751–753. doi: 10.1096/fj.01-0759fje. [DOI] [PubMed] [Google Scholar]
- Green SK, Frankel A, Kerbel RS. Adhesion-dependent multicellular drug resistance. Anticancer Drug Des. 1999;2:153–168. [PubMed] [Google Scholar]
- Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends Cell Biol. 2003;5:264–269. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- Gudjonsson T, Ronnov-Jessen L, Villadsen R, Rank F, Bissell MJ, Petersen OW. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115(Pt 1):39–50. doi: 10.1242/jcs.115.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WH, Frey MT, Burnham NA, Wang YL. Substrate rigidity regulates the formation and maintenance of tissues. Biophys J. 2006;6:2213–2220. doi: 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagios C, Lochter A, Bissell MJ. Tissue architecture: The ultimate regulator of epithelial function? Philos Trans R Soc Lond, B Biol Sci. 1998;1370:857–870. doi: 10.1098/rstb.1998.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Meltzer PS, Gardner LM, Hess AR, Kirschmann DA, Schatteman GC, Seftor RE. Expression and functional significance of VE-cadherin in aggressive human melanoma cells: Role in vasculogenic mimicry. Proc Natl Acad Sci USA. 2001;14:8018–8023. doi: 10.1073/pnas.131209798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher A, Lakonishok M, Kinder M, Wu S, Truong T, Knudsen KA, Horwitz AF. Integrin and cadherin synergy regulates contact inhibition of migration and motile activity. J Cell Biol. 1998;2:515–526. doi: 10.1083/jcb.141.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher A, Sandborg RR, Horwitz AF. Adhesion in cell migration. Curr Opin Cell Biol. 1995;5:697–706. doi: 10.1016/0955-0674(95)80112-x. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Mechanical control of tissue morphogenesis during embryological development. Int J Dev Biol. 2006;2–3:255–266. doi: 10.1387/ijdb.052044di. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;3:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Jeffery PK. Remodeling in asthma and chronic obstructive lung disease. Am J Respir Crit Care Med. 2001;10(Pt 2):S28–S38. doi: 10.1164/ajrccm.164.supplement_2.2106061. [DOI] [PubMed] [Google Scholar]
- Kadoya Y, Yamashina S. Salivary gland morphogenesis and basement membranes. Anat Sci Int. 2005;2:71–79. doi: 10.1111/j.1447-073x.2005.00102.x. [DOI] [PubMed] [Google Scholar]
- Keely PJ, Fong AM, Zutter MM, Santoro SA. Alteration of collagen-dependent adhesion, motility, and morphogenesis by the expression of antisense alpha 2 integrin mRNA in mammary cells. J Cell Sci. 1995;108(Pt 2):595–607. doi: 10.1242/jcs.108.2.595. [DOI] [PubMed] [Google Scholar]
- Kleinman HK, McGarvey ML, Hassell JR, Star VL, Cannon FB, Laurie GW, Martin GR. Basement membrane complexes with biological activity. Biochemistry. 1986;2:312–318. doi: 10.1021/bi00350a005. [DOI] [PubMed] [Google Scholar]
- Liu M, Tanswell AK, Post M. Mechanical force-induced signal transduction in lung cells. Am J Physiol. 1999;4(Pt 1):L667–L683. doi: 10.1152/ajplung.1999.277.4.L667. [DOI] [PubMed] [Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;1:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Nieto MA. Cell movements during vertebrate development: Integrated tissue behaviour versus individual cell migration. Curr Opin Genet Dev. 2001;4:464–469. doi: 10.1016/s0959-437x(00)00218-5. [DOI] [PubMed] [Google Scholar]
- Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;1:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- Margulis A, Zhang W, Alt-Holland A, Crawford HC, Fusenig NE, Garlick JA. E-cadherin suppression accelerates squamous cell carcinoma progression in three-dimensional, human tissue constructs. Cancer Res. 2005;5:1783–1791. doi: 10.1158/0008-5472.CAN-04-3399. [DOI] [PubMed] [Google Scholar]
- Martin RB, Lau ST, Mathews PV, Gibson VA, Stover SM. Collagen fiber organization is related to mechanical properties and remodeling in equine bone. A comparison of two methods. J Biomech. 1996;29(12):1515–1521. [PubMed] [Google Scholar]
- Matthews JA, Wnek GE, Simpson DG, Bowlin GL. Electrospinning of collagen nanofibers. Biomacromolecules. 2002;2:232–238. doi: 10.1021/bm015533u. [DOI] [PubMed] [Google Scholar]
- Mauck RL, Yuan X, Tuan RS. Chondrogenic differentiation and functional maturation of bovine mesenchymal stem cells in long-term agarose culture. Osteoarthr Cartil. 2006;2:179–189. doi: 10.1016/j.joca.2005.09.002. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;4:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- Mostov K, Brakeman P, Datta A, Gassama A, Katz L, Kim M, Leroy P, Levin M, Liu K, Martin F, O’Brien LE, Verges M, et al. Formation of multicellular epithelial structures. Novartis Found Symp. 2005;269:193–200. discussion 200–205, 223–230. [PubMed] [Google Scholar]
- Muschler J, Lochter A, Roskelley CD, Yurchenco P, Bissell MJ. Division of labor among the alpha6beta4 integrin, beta1 integrins, and an E3 laminin receptor to signal morphogenesis and beta-casein expression in mammary epithelial cells. Mol Biol Cell. 1999;9:2817–2828. doi: 10.1091/mbc.10.9.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogawa H, Ito T. Branching morphogenesis of embryonic mouse lung epithelium in mesenchyme-free culture. Development. 1995;4:1015–1022. doi: 10.1242/dev.121.4.1015. [DOI] [PubMed] [Google Scholar]
- Novaro V, Roskelley CD, Bissell MJ. Collagen-IV and laminin-1 regulate estrogen receptor alpha expression and function in mouse mammary epithelial cells. J Cell Sci. 2003;116(Pt 14):2975–2986. doi: 10.1242/jcs.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien LE, Jou TS, Pollack AL, Zhang Q, Hansen SH, Yurchenco P, Mostov KE. Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nat Cell Biol. 2001;9:831–838. doi: 10.1038/ncb0901-831. [DOI] [PubMed] [Google Scholar]
- O’Brien LE, Zegers MM, Mostov KE. Opinion: Building epithelial architecture: Insights from three-dimensional culture models. Nat Rev Mol Cell Biol. 2002;7:531–537. doi: 10.1038/nrm859. [DOI] [PubMed] [Google Scholar]
- Paszek MJ, Weaver VM. The tension mounts: Mechanics meets morphogenesis and malignancy. J Mammary Gland Biol Neoplasia. 2004;4:325–342. doi: 10.1007/s10911-004-1404-x. [DOI] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;3:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;25:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;19:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless DD, Lee YC, Roseman S, Schnaar R. Specific cell adhesion to immobilized glycoproteins demonstrated using new reagents for protein and glycoprotein immobilization. J Biol Chem. 1983;258:2340–2349. [PubMed] [Google Scholar]
- Pujuguet P, Simian M, Liaw J, Timpl R, Werb Z, Bissell MJ. Nidogen-1 regulates laminin-1-dependent mammary-specific gene expression. J Cell Sci. 2000;113(Pt 5):849–858. doi: 10.1242/jcs.113.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi SC, Hubbell JA. Recombinant protein-co-PEG networks as cell-adhesive and proteolytically degradable hydrogel matrixes. Part I: Development and physicochemical characteristics. Biomacromolecules. 2005;3:1226–1238. doi: 10.1021/bm049614c. [DOI] [PubMed] [Google Scholar]
- Roeder BA, Kokini K, Sturgis JE, Robinson JP, Voytik-Harbin SL. Tensile mechanical properties of three-dimensional type I collagen extracellular matrices with varied microstructure. J Biomech Eng. 2002;2:214–222. doi: 10.1115/1.1449904. [DOI] [PubMed] [Google Scholar]
- Rosenfeldt H, Grinnell F. Fibroblast quiescence and the disruption of ERK signaling in mechanically unloaded collagen matrices. J Biol Chem. 2000;5:3088–3092. doi: 10.1074/jbc.275.5.3088. [DOI] [PubMed] [Google Scholar]
- Roskelley CD, Srebrow A, Bissell MJ. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr Opin Cell Biol. 1995;5:736–747. doi: 10.1016/0955-0674(95)80117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi T, Rintoul RC, Moore SM, MacKinnon AC, Salter D, Choo C, Chilvers ER, Dransfield I, Donnelly SC, Strieter R, Haslett C. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: A mechanism for small cell lung cancer growth and drug resistance in vivo. Nat Med. 1999;6:662–668. doi: 10.1038/9511. [DOI] [PubMed] [Google Scholar]
- Springer TA, Wang JH. The three-dimensional structure of integrins and their ligands, and conformational regulation of cell adhesion. Adv Protein Chem. 2004;68:29–63. doi: 10.1016/S0065-3233(04)68002-8. [DOI] [PubMed] [Google Scholar]
- Stegemann JP, Hong H, Nerem RM. Mechanical, biochemical, and extracellular matrix effects on vascular smooth muscle cell phenotype. J Appl Physiol. 2005;6:2321–2327. doi: 10.1152/japplphysiol.01114.2004. [DOI] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;4:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry NA, Lazebnik Y. Caspases: Enemies within. Science. 1998;5381:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: A different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;25:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB, Dembo M, Hanks SK, Wang Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci USA. 2001;20:11295–11300. doi: 10.1073/pnas.201201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Goswami S, Sahai E, Wyckoff JB, Segall JE, Condeelis JS. Tumor cells caught in the act of invading: Their strategy for enhanced cell motility. Trends Cell Biol. 2005;3:138–145. doi: 10.1016/j.tcb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Weaver VM, Bissell MJ. Functional culture models to study mechanisms governing apoptosis in normal and malignant mammary epithelial cells. J Mammary Gland Biol Neoplasia. 1999;2:193–201. doi: 10.1023/a:1018781325716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Fischer AH, Peterson OW, Bissell MJ. The importance of the microenvironment in breast cancer progression: Recapitulation of mammary tumorigenesis using a unique human mammary epithelial cell model and a three-dimensional culture assay. Biochem Cell Biol. 1996;6:833–851. doi: 10.1139/o96-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, Werb Z, Bissell MJ. Beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;3:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;1:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willem M, Miosge N, Halfter W, Smyth N, Jannetti I, Burghart E, Timpl R, Mayer U. Specific ablation of the nidogen-binding site in the laminin gamma1 chain interferes with kidney and lung development. Development. 2002;11:2711–2722. doi: 10.1242/dev.129.11.2711. [DOI] [PubMed] [Google Scholar]
- Wong JY, Velasco A, Rajagopalan P, Pham Q. Directed movement of vascular smooth muscle cells on gradient-compliant hydrogels. Langmuir. 2003;19:1908–1913. [Google Scholar]
- Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J Cell Biol. 2003;3:583–595. doi: 10.1083/jcb.200305010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KM, Pankov R, Cukierman E. Dimensions and dynamics in integrin function. Braz J Med Biol Res. 2003;8:959–966. doi: 10.1590/s0100-879x2003000800001. [DOI] [PubMed] [Google Scholar]
- Yap AS, Stevenson BR, Keast JR, Manley SW. Cadherin-mediated adhesion and apical membrane assembly define distinct steps during thyroid epithelial polarization and lumen formation. Endocrinology. 1995;10:4672–4680. doi: 10.1210/endo.136.10.7664688. [DOI] [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;1:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- Zaari N, Rajagopalan P, Kim SK, Engler AJ, Wong JY. Photopolymerization in microfluidic gradient generators: Microscale control of substrate compliance to manipulate cell response. Adv Mat. 2004;23–24:2133–2137. [Google Scholar]
- Zahir N, Lakins JN, Russell A, Ming W, Chatterjee C, Rozenberg GI, Marinkovich MP, Weaver VM. Autocrine laminin-5 ligates alpha6beta4 integrin and activates RAC and NFkappaB to mediate anchorage-independent survival of mammary tumors. J Cell Biol. 2003;6:1397–1407. doi: 10.1083/jcb.200302023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir N, Weaver VM. Death in the third dimension: Apoptosis regulation and tissue architecture. Curr Opin Genet Dev. 2004;1:71–80. doi: 10.1016/j.gde.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Zhang S. Beyond the Petri dish. Nat Biotechnol. 2004;2:151–152. doi: 10.1038/nbt0204-151. [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang J, McNaughton PA. NGF rapidly increases membrane expression of TRPV1 heat-gated ion channels. EMBO J. 2005;24:4211–4223. doi: 10.1038/sj.emboj.7600893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;9:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]