Abstract
Membrane proteins that transport hydrophobic compounds play important roles in multi-drug resistance1–3 and can cause a number of diseases4,5, underscoring the importance of protein-mediated transport of hydrophobic compounds. Hydrophobic compounds readily partition into regular membrane lipid bilayers6, and their transport through an aqueous protein channel is energetically unfavourable3. Alternative transport models, involving acquisition from the lipid bilayer by lateral diffusion have been proposed for hydrophobic substrates3,4,7–12. To date, all transport proteins for which a lateral diffusion mechanism has been proposed function as efflux pumps. Here we present the first example of a lateral diffusion mechanism for the uptake of hydrophobic substrates, by the Escherichia coli outer membrane long-chain fatty acid (LCFA) transporter FadL. A FadL mutant in which a lateral opening in the barrel wall is constricted, but which is otherwise structurally identical to wild-type FadL, does not transport substrates. A crystal structure of FadL from Pseudomonas aeruginosa shows that the opening in the wall of the β-barrel is conserved and delineates a long, hydrophobic tunnel that could mediate substrate passage from the extracellular environment, through the polar lipopolysaccharide layer and, via the lateral opening in the barrel wall, into the lipid bilayer from where the substrate can diffuse into the periplasm. Since FadL homologues are found in pathogenic and biodegrading bacteria, our results have implications for combating bacterial infections and bioremediating xenobiotics in the environment.
The outer membrane (OM) of gram-negative bacteria provides an efficient barrier for the passage of hydrophobic molecules due to the presence of the polar lipopolysaccharide (LPS) layer on the outside of the cell. The only protein family currently known to be involved in the uptake of hydrophobic molecules is the FadL family, named after the archetypal long-chain fatty acid (LCFA) transporter FadL of E. coli13, 14. The crystal structures of E. coli FadL15 revealed a monomeric 14-stranded β-barrel with an interior occluded by an N-terminal hatch domain, and an inward-pointing kink in β-strand S3 that creates an unusual, lateral opening in the transmembrane barrel15. Based on the FadL structures, two possible LCFA transport mechanisms can be envisioned, both of which are diffusion-based since FadL-mediated transport does not require an energized membrane16. In the “classical” transport model, the hatch undergoes conformational changes creating a transient channel for LCFA transport transversal to the membrane, from the extracellular medium directly to the aqueous periplasm15. The alternative “lateral diffusion” transport model is based on the observation that an LCFA-mimicking LDAO detergent molecule is bound in the lateral opening of E. coli FadL (Fig. 1a,b). In the lateral diffusion model, LCFAs exit FadL laterally through the opening in the barrel wall to move into the OM, from where they could diffuse into the periplasm. To answer the question which transport model is operative for FadL, we designed a number FadL mutants focusing on the hatch domain and the lateral opening.
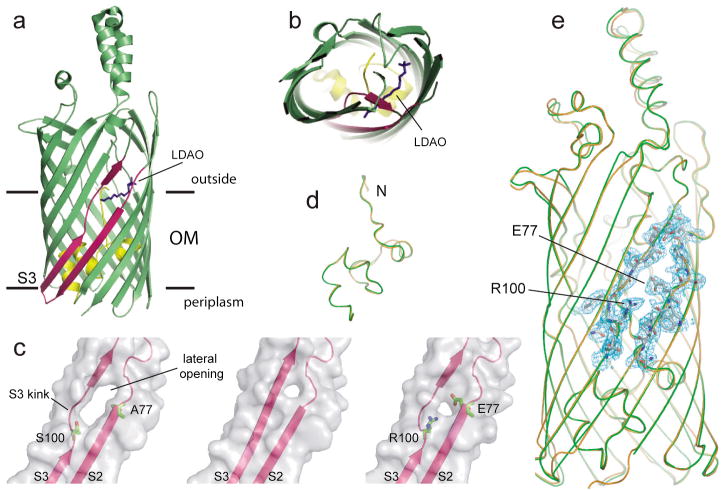
Figure 1. Structural features of FadL mutants.
a,b, Ribbon diagram of wild-type FadL highlighting the S3 kink, viewed laterally (a) and from the extracellular side (b). An LDAO molecule (dark blue) protrudes through the opening in the barrel wall between strands S2 and S3 (dark pink). The hatch domain is colored yellow. The interface boundaries of the OM bilayer are indicated by horizontal lines. c, Surface views of the kink region (with strands S2 and S3 indicated) of wild-type FadL (left), ΔS3 kink (middle) and A77E/S100R (right), showing the smaller lateral opening in ΔS3 kink and A77E/S100R. The side chains for the residues at positions 77 and 100 are shown. d,e, Backbone superpositions of wild-type FadL (orange) and A77E/S100R (green; d, hatch domains only). In e, 2Fo−Fc density (contoured at 1.5σ) is shown as a blue mesh for segments of β-strands S2 and S3 in A77E/S100R, with the side chains of residues E77 and R100 indicated.
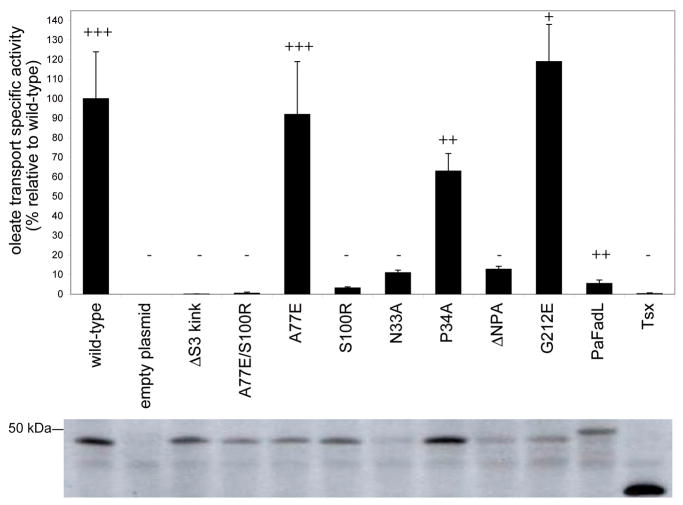
Two FadL mutants, ΔS3 kink and A77E/S100R, were designed to specifically close the barrel wall opening in different ways. In ΔS3 kink, four residues in strand S3 that line the lateral opening (100SNYG103) were replaced with three residues (AND) to bring strand S3 into register with the neighbouring strands and to destabilize the conformation of the S3 kink. In A77E/S100R, residues with long, oppositely charged side chains were introduced for residues A77 and S100, which are located in the barrel wall on different sides of the opening (Fig. 1c) with their side chains pointing towards each other. We anticipated that the introduced glutamic acid and arginine might form a salt bridge, closing the lateral opening. We determined the crystal structures of the ΔS3 kink and A77E/S100R mutants (Supplementary Table 1). The ΔS3 kink mutant, while largely identical to wild-type FadL, lacks density for several extracellular loops (Supplementary Fig. 1). However, the A77E/S100R structure is identical to that of wild-type FadL (Fig. 1d,e), with the sole exception of the lateral opening, which is much smaller in the A77E/S100R mutant (Fig. 1c). Thus, the introduced mutations for residues A77 and S100 are unlikely to affect the interior of the protein, strongly arguing against an inhibitory effect of the mutations on a possible conformational change in the hatch that would form a classical transport channel. Crucially, both the ΔS3 kink and A77E/S100R mutants were completely inactive for in vivo LCFA uptake and growth on palmitate (Fig. 2). Moreover, the low transport activity (~ 3%) of the S100R mutant (Fig. 2) demonstrates that the introduction of a single long side chain is sufficient to inhibit LCFA uptake efficiently. The structural and biochemical data together demonstrate that constricting the lateral opening in the FadL barrel wall is sufficient to block LCFA uptake. While we can not exclude the possibility that subtle changes in barrel structure and dynamics, introduced by the mutations, affect the formation of a lateral opening in other parts of the barrel, our data are most easily explained by a transport model in which LCFAs diffuse laterally from the lumen of the barrel, through the observed opening in the barrel wall, into the OM.
Figure 2. Functional analysis of FadL proteins.
Specific activity expressed relative to wild-type for oleate uptake measured in whole cells at 20°C. Bars represent the average of at least four independent measurements, and the error bars represent the standard deviation. Growth on palmitate minimal medium plates at 37°C, ranging from wild-type levels (+++) to no growth (−), is indicated above the bars. The hatch mutants N33A and ΔNPA do not support growth of E. coli on palmitate at 37°C, since the expression levels of these two mutants are below detection limits at this temperature. The bottom panel shows representative western immunoblots indicating the expression levels of the various proteins in the OM at 20°C. Tsx is the E. coli OM nucleoside transporter, which is included as an additional negative control.
If transport by lateral diffusion is a general feature of FadL channels, then the opening in the barrel wall should be structurally conserved. To test this notion we have determined the crystal structure (Supplementary Table 2) of a FadL homologue from Pseudomonas aeruginosa (PaFadL), which has low (20%) sequence identity to E. coli FadL (EcFadL). Despite the modest sequence identity, PaFadL is structurally similar to EcFadL, with the exception of a number of extracellular loops (Fig. 3a,b). PaFadL has a pronounced lateral opening in the barrel wall at the same location as EcFadL (Fig. 3a), suggesting that the lateral opening is conserved in FadL family members. The most striking feature of the PaFadL structure is the presence of well-defined density for three complete C8E4 detergent molecules inside the barrel lumen (Fig. 3c). The first two detergent molecules (1, 2) are present at positions that are analogous to those occupied by detergent molecules in EcFadL15. The third C8E4 molecule (3) is located in the lateral opening, analogous to the LDAO molecule in wild-type FadL (Fig. 1a,b). The presence of a detergent molecule in the lateral opening suggests that this part of the potential substrate passageway has a substantial affinity for LCFA substrates. This is borne out by the fact that the environment of the detergent molecule in the opening is largely hydrophobic (Supplementary Fig. 2), like the other LCFA binding sites in FadL15.
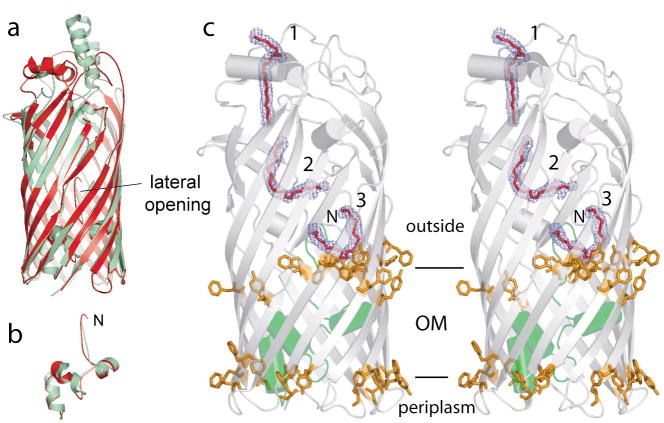
Figure 3. A hydrophobic passageway for substrate diffusion in PaFadL.
a, Superposition of EcFadL (green) and PaFadL (red), showing the conservation of the lateral opening. b, Superposition of the hatch domains. c, Stereo side view of PaFadL, with the three bound C8E4 detergent molecules indicated in red. 2Fo−Fc density is shown as a blue mesh, contoured at 2.0 σ. The hatch domain is colored green. The belts of aromatic residues that delineate the polar-apolar interfaces of the OM are shown as orange stick models.
The three detergent molecules in PaFadL clearly delineate a long (~55 Å) passageway that runs from the extracellular surface all the way to the lateral opening in the barrel wall. Of the ~ 50 residues that are located within 4.5 Å of the detergent molecules, more than 80% are hydrophobic (Supplementary Fig. 2). Moreover, structural and sequence alignments show that the hydrophobic character of these residues is conserved. The hydrophobic channel is continuous, and is interrupted only by the first three N-terminal residues of the hatch, most notably F3. Remarkably, a conformational change of the N-terminus as previously observed in EcFadL15, would generate an uninterrupted, hydrophobic passageway for substrate diffusion all the way from the extracellular medium to the lateral opening (Supplementary Fig. 3). As in EcFadL (Fig. 1a), the lateral opening in PaFadL is situated in the region of the polar-apolar interface of the outer leaflet of the OM (Fig. 3c). This location makes sense, since it likely provides a favourable environment for both the carboxylate head group and the hydrocarbon chain as the LCFA emerges from the FadL lumen. Our observation is in agreement with recent data obtained for multidrug efflux pumps, where a range of hydrophobic substrates was shown to be preferentially localized in the interface region of the lipid bilayer6. Despite the structural similarity to EcFadL, PaFadL has very low activity for LCFA transport (Fig. 2), confirming that FadL channels are substrate specific17.
To support our lateral diffusion model we also asked whether the hatch domain of EcFadL is flexible. The hatch NPA sequence (residues 33–35) is absolutely conserved and is a signature of FadL channels. The N33 amide side chain of the NPA sequence forms a hydrogen bond with the G21 backbone carbonyl, bringing distant parts of the hatch together (Supplementary Fig. 4). The removal of this hydrogen bond would seem a good candidate to probe if the hatch domain can undergo conformational changes. We determined the X-ray crystal structures of the mutants N33A, P34A and ΔNPA (N33G/P34G/A35G; Supplementary Table 1), and found that they are very similar to wild-type FadL (Supplementary Fig. 1 and Supplementary Fig. 4) and do not have a hatch channel. The same is true for the G212E mutant (Supplementary Fig. 1 and Supplementary Fig. 4), which previously was proposed to have an open channel18. Consistent with the structural data, all hatch mutants are active in oleate transport (Fig. 2). The combined structural and biochemical data suggest that the NPA sequence is not directly involved in substrate transport, but it may be important for proper folding or OM targeting of FadL. In addition, it appears that the hatch domain is rigid, providing support for the lateral diffusion transport model. It should also be noted that although the hatch domain of E. coli FadL has a number of hydrophobic residues, these are interspersed with many polar residues (Supplementary Fig. 5). Therefore, even if a hatch channel in E. coli FadL could form by spontaneous conformational changes, it would not provide a suitably hydrophobic conduit for LCFA transport.
We now propose a general, lateral diffusion mechanism for the uptake of hydrophobic substrates by the FadL outer membrane protein family (Fig. 4). According to this mechanism, hydrophobic substrates diffuse via a long hydrophobic passageway laterally into the outer leaflet of the OM, most likely via a stable, lateral opening in the barrel wall. From the outer leaflet the substrate can move to the inner leaflet of the OM and diffuse into the periplasm. By using this mechanism, the hydrophobic substrates bypass the hydrophilic LPS layer of the OM without having to move through an aqueous channel, which would be energetically unfavourable due to the extremely low aqueous solubilities of FadL substrates (< 0.1 nM for palmitate19). Intriguingly, medium-chain fatty acids do not require FadL for uptake in E. coli20. Apparently these relatively water-soluble substrates (> 0.5 mM for laurate19) utilize other OM channels for uptake, most likely porins. Recently, crystal structures of TodX and TbuX were determined, two members of a subfamily of FadL proteins from biodegrading bacteria involved in the uptake of mono-aromatic hydrocarbons21,22. As expected, TodX and TbuX both have a kink in strand S3 as well as a lateral opening at the same position as in FadL17, providing additional support for the generality of the lateral diffusion mechanism in FadL proteins. However, TodX and TbuX also have a narrow, continuous channel through the hatch domain that might serve as a classical channel. The TodX/TbuX hatch channel is relatively polar17, and may have evolved for the uptake of relatively water-soluble (~1–5 mM)23 mono-aromatic hydrocarbons in bacteria lacking porins. Future experiments will be required to establish whether the hatch channel in TodX/TbuX contributes to transport.
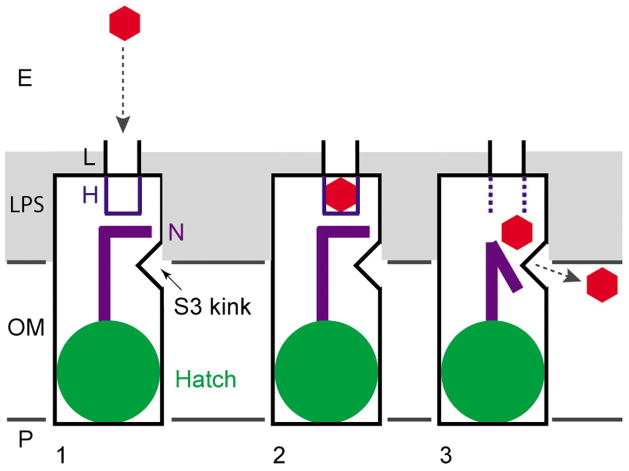
Figure 4. Proposed lateral diffusion model for the uptake of hydrophobic substrates by FadL proteins.
(1) substrate (red hexagon) capture from the extracellular medium by a low-affinity binding site (L)15; (2) diffusion of the substrate into an adjacent high-affinity binding site H (blue)15; and (3) spontaneous conformational changes in the N-terminus (purple) result in substrate release and create a continuous passageway to the barrel wall opening formed by the kink in strand S3. The substrate diffuses laterally through the opening into the OM. The polar part of the LPS, constituting the principal barrier in the transport process, is shown in gray. The extracellular milieu (E) is at the top and the periplasm (P) is at the bottom.
Structural features resembling the lateral opening of FadL channels have been observed in two other, unrelated OM proteins, PagP and OmpW. The lateral opening in the lipid A palmitoyl-transferase PagP may allow access of the lipid substrate to the active site of the enzyme24. OmpW belongs to a widespread family of 8-stranded β-barrels25, with members present in operons dedicated to the degradation of hydrophobic molecules such as naphthalene (NahQ)26 and alkanes (AlkL)27, suggesting that they may form uptake channels for these compounds. Such uptake would have to occur by lateral diffusion, since the 8-stranded barrel lumen is too narrow to form a classical channel25. Thus, bacterial substrate uptake by lateral diffusion into the OM may be a widespread phenomenon. Taking into account the known occurrence of lateral diffusion in multidrug transporters, this mode of transport likely represents a universal mechanism for membrane proteins that modify and transport hydrophobic substrates.
METHODS SUMMARY
LCFA functional assays were performed using a modified approach from that described previously18. FadL mutant proteins were overexpressed and purified from E. coli C43(DE3) as described for the wild-type protein15. Crystals were obtained by the hanging drop method, and structures were solved by molecular replacement using as a search model the monoclinic FadL structure (PDB 1T16) in which the hatch domain and the kink were deleted. Pseudomonas aeruginosa FadL (PaFadL) was overexpressed in native form in E. coli C43(DE3), and purified as EcFadL. The PaFadL crystal structure was determined by the multiple isomorphous replacement with anomalous scattering (MIRAS) method, using a gold and osmium derivative obtained by the quick soaking method28.
METHODS
FadL mutagenesis
The Escherichia coli fadL gene, including the signal sequence and a C-terminal hexahistidine tag, was cloned as previously described15 into the pBAD22 vector, which is under the control of the arabinose-inducible promoter29. Mutations were introduced into the fadL gene by using the QuikChange® Site-directed Mutagenesis kit (Stratagene), and the mutations were verified by nucleotide sequencing.
LCFA functional analyses
For the functional analyses, E. coli LS6164 ΔfadR ΔfadL30 was transformed with the pBAD22 plasmids carrying the mutant fadL genes and FadL homologues. As a negative control, the pBAD22 plasmid carrying the OM nucleoside transporter Tsx31 was introduced into E. coli LS6164. The ability of the mutant proteins to support growth on LCFAs was measured by plating cells (5×106 cfu mL−1) on agar plates containing 5 mM sodium palmitate (Sigma), 0.5% (w/v) Brij™ 58, M9 minimal medium, and 1.5% (w/v) Noble agar (Difco). Growth was scored after 96 h incubation at 37°C.
Transport assays with radiolabeled oleic acid were performed with modifications to the method described by Kumar and Black18. E. coli LS6164 ΔfadR ΔfadL cells with the FadL mutant and pBAD22 plasmids were grown in LB medium to mid-log phase, and FadL expression was induced with 0.01% (w/v) arabinose at 20°C for 5 h. Cells were harvested, washed in EB1 buffer (10 mM citric acid, 0.8 mM magnesium sulfate, 20 mM sodium ammonium phosphate, 60 mM potassium phosphate, 0.02 mM thiamine), and resuspended to an OD600 of 1 in EB1 buffer with 0.5% (w/v) Brij™ 58. After 30 min starvation at 37°C, samples were taken for western immunoblotting analysis and also for measurement of oleic acid transport activity. The transport assay was performed by diluting the cells in EB1 buffer containing 20 mM glucose, 0.5% (w/v) Brij™ 58, and 0.02 μCi [3H-9,10]-oleic acid (Sigma, specific activity 30.0–50.5 μCi mmol−1. At 25 minutes, samples were removed and filtered through 0.45μm membrane filters (Metricel GN-6, Pall Life Sciences). Filters were washed with EB1 buffer containing 0.5% Brij™ 58, and the radioactivity retained on the filters was counted. Counts for the cells expressing the FadL protein were corrected for the background radioactivity associated with cells containing an empty pBAD22 plasmid. To determine the amount of FadL protein expressed in the outer membrane, cells were incubated with BugBuster (Pierce) while shaking for 30 min at 37°C, and centrifuged for 10 min at 14,000 rpm. Proteins were detected by western immunoblotting using the Penta-His-horseradish peroxidise conjugate antibody (Qiagen) and the ECL chemiluminescence detection kit (GE Healthcare). Proteins were quantitated against FadL standards using the ImageJ program32. The transport activities of the cells expressing mutant FadL proteins were corrected for FadL expression levels, and the rates were expressed as a percentage of wild-type.
Purification, crystallization and structure determination of FadL mutants
For crystallization, the FadL mutant proteins were expressed from the pBAD22 plasmids in E. coli C43(DE3) cells33 grown in 2×YT medium by induction with 0.2% (w/v) arabinose for 6 h at 30°C. Cells were harvested, and the proteins were purified from the total membrane fraction as previously described for wild-type FadL15 by Ni affinity chromatography in LDAO, gel filtration chromatography in LDAO (Anatrace), and a final gel filtration chromatography step in C8E4 (Sigma). The purified proteins were concentrated to 5–10 mg mL−1 and flash frozen in liquid nitrogen.
Crystals were obtained by the hanging drop method at 22°C using commercially available screens (The Classics and MB Class II, Qiagen) or in-house screens. The mutant FadL proteins crystallized under the following conditions: N33A, 0.1 M cadmium chloride, 0.1 M sodium acetate (pH 4.6), 30% (v/v) PEG 400; P34A, 0.2 M ammonium acetate, 0.1 M sodium acetate (pH 4.6), 30% (w/v) PEG 4000; ΔNPA, 0.05 M magnesium acetate, 0.05 M cacodylate (pH 5.5), 35% (w/v) PEG 2000; G212E, 0.2 M zinc acetate, 0.1 M cacodylate (pH 6.5), 18% (w/v) PEG 8000; ΔS3 kink, 0.2 M ammonium sulfate, 0.1 M MES (pH 6.5), 30% (w/v) PEG 5000 MME; and A77E/S100R, 0.1 M sodium chloride, 0.1 M citrate (pH 5.6), 16% PEG 4000. The crystals were flash frozen (100 K) in the reservoir solution containing C8E4 and 15–25% (v/v) glycerol by plunging in liquid nitrogen.
Diffraction data were obtained on beamline X6A at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY). Data sets were integrated and scaled using HKL200034. The structures of the mutant proteins were solved by molecular replacement using Phaser35; the monoclinic FadL structure (PDB 1T16) without the N-terminal 40 amino acids and the S3 kink residues 99–108 was used as the search model. Model building was done using Coot36, and refinement was done with CNS37. Data collection and refinement statistics for the mutant FadL proteins are summarized in Supplementary Table 1.
Cloning of PaFadL
The signal sequence cleavage site for PaFadL was predicted to occur between residues 20 and 21 using the SignalP program38. For expression and outer membrane localization of PaFadL, the mature gene (lacking the endogenous signal sequences and the first two residues, and additionally modified with a C-terminal hexahistidine tag) was amplified by PCR from P. aeruginosa PAO1 (ATCC 47085) genomic DNA. The mature gene fragment was cloned in-frame with the E. coli FadL signal sequence (plus the first two residues of the mature FadL sequence) into the pBAD22 vector29.
Purification, crystallization and structure determination of PaFadL
PaFadL was expressed in E. coli C43(DE3) cells33 and purified using the detergents LDAO and C8E4 as described for E. coli FadL15. Crystallization trials of PaFadL were set up using the hanging drop technique with commercially available crystallization screens (The Classics and MB Class II screens, Qiagen). Crystals for the PaFadL were obtained at 22°C in 0.1 M lithium sulfate, 0.1 M MES buffer (pH 6.5), and 28–32% (w/v) PEG 400. The crystals typically appeared after 10 days and grew to their full size (~15 × 30 × 100 μm) in 3–4 weeks. They belonged to space group C2221, diffracted to a resolution of 2.1 Å, and contained one molecule in the asymmetric unit (Matthews coefficient VM ~ 4.2 Å3/Da, corresponding to ~55% (v/v) solvent content39). The crystals were flash frozen (100 K) directly from the drop by plunging in liquid nitrogen.
A native dataset was collected, as well as datasets for crystals that were soaked for ~40 min in 10 mM OsCl3 or KAuCl4. The datasets were processed using HKL200034, showing that the osmium and gold crystals were isomorphous with the native crystals (Supplementary Table 2). Two osmium and two gold sites were found by multiple isomorphous replacement in SOLVE (Z-score 10.2; FOM 0.39 for data between 20 and 2.5 Å)40. Five additional gold and three osmium sites were found in isomorphous/anomalous difference maps and refined using SHARP41. Electron density maps obtained from SHARP were used for automatic model building in RESOLVE40, which resulted in ~ 60% of the model being built. Further model building was done manually in COOT36, followed by refinement in CNS 1.237. Data collection and refinement statistics for PaFadL are summarized in Supplementary Table 2.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank the personnel of the National Synchrotron Light Source (NSLS) beamlines X6A and X29 for beam time and beamline support. We are grateful to P. Black and C. Petteys (Ordway Research Institute and Albany Medical College) for the strain D10 and for their technical advice on the fatty acid transport assays. This work was supported by a training grant from the National Insitutes of Health (E.M.H) and by a NIH research grant (1R01GM074824 to B.v.d.B.).
Footnotes
Author Contributions E.M.H. cloned, purified and crystallized FadL mutants, performed activity assays, and wrote the paper, D.R.P. cloned, purified and crystallized FadL mutants, B.W.L. performed activity assays, M.I. purified and crystallized PaFadL, B.v.d.B. determined crystal structures, designed research and wrote the paper.
Author Information Coordinates and structure factors have been deposited in the Protein Data Bank: PaFadL, 3DWO; ΔS3 kink, 2R88; A77E/S100R, 3DWN; P34A, 2R4L; N33A, 2R4N; ΔNPA, 2R4O; and G212E, 2R4P. The authors declare no competing financial interests.
References
- 1.Saier MH, Jr, Paulsen IT. Phylogeny of multidrug transporters. Semin Cell Dev Biol. 2001;12:205–213. doi: 10.1006/scdb.2000.0246. [DOI] [PubMed] [Google Scholar]
- 2.Poole K. Efflux pumps as antimicrobial resistance mechanisms. Ann Med. 2007;39:162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 3.Sharom FJ. Shedding light on drug transport: structure and function of the P-glycoprotein multidrug transporter (ABCB1) Biochem Cell Biol. 2006;84:979–992. doi: 10.1139/o06-199. [DOI] [PubMed] [Google Scholar]
- 4.van Meer G, Halter D, Sprong H, Somerharju P, Egmond MR. ABC lipid transporters: extruders, flippases or flopless activators? FEBS Lett. 2006;580:1171–1177. doi: 10.1016/j.febslet.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 5.Borst P, Zelcer N, van Helvoort A. ABC transporters in lipid transport. Biochim Biophys Acta. 2000;1486:128–144. doi: 10.1016/s1388-1981(00)00053-6. [DOI] [PubMed] [Google Scholar]
- 6.Siarheyeva A, Lopez JJ, Glaubitz C. Localization of multidrug transporter substrates within model membranes. Biochemistry. 2006;45:6203–6211. doi: 10.1021/bi0524870. [DOI] [PubMed] [Google Scholar]
- 7.Higgins CF, Gottesman MM. Is the multidrug transporter a flippase? Trends Biochem Sci. 1992;17:18–21. doi: 10.1016/0968-0004(92)90419-a. [DOI] [PubMed] [Google Scholar]
- 8.Higgins CF. Multiple molecular mechanisms for multidrug resistance transporters. Nature. 2007;446:749–757. doi: 10.1038/nature05630. [DOI] [PubMed] [Google Scholar]
- 9.van Veen HW, Putman M, Margolles A, Sakamoto K, Konings WN. Structure-function analysis of multidrug transporters in Lactococcus lactis. Biochim Biophys Acta. 1999;1461:201–206. doi: 10.1016/s0005-2736(99)00172-8. [DOI] [PubMed] [Google Scholar]
- 10.Bolhuis H, et al. Multidrug resistance in Lactococcus lactis: evidence for ATP-dependent drug extrusion from the inner leaflet of the cytoplasmic membrane. EMBO J. 1996;15:4239–4245. [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro AB, Ling V. Extraction of Hoechst 33342 from the cytoplasmic leaflet of the plasma membrane by P-glycoprotein. Eur J Biochem. 1997;250:122–129. doi: 10.1111/j.1432-1033.1997.00122.x. [DOI] [PubMed] [Google Scholar]
- 12.Shapiro AB, Ling V. Transport of LDS-751 from the cytoplasmic leaflet of the plasma membrane by the rhodamine-123-selective site of P-glycoprotein. Eur J Biochem. 1998;254:181–188. doi: 10.1046/j.1432-1327.1998.2540181.x. [DOI] [PubMed] [Google Scholar]
- 13.Nunn WD, Simons RW. Transport of long-chain fatty acids by Escherichia coli: mapping and characterization of mutants in the fadL gene. Proc Natl Acad Sci. 1978;75:3377–3381. doi: 10.1073/pnas.75.7.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Black PN, Said B, Ghosn CR, Beach JV, Nunn WD. Purification and characterization of an outer membrane-bound protein involved in long-chain fatty acid transport in Escherichia coli. J Biol Chem. 1987;262:1412–1419. [PubMed] [Google Scholar]
- 15.van den Berg B, Black PN, Clemons WM, Jr, Rapoport TM. Crystal structure of the long-chain fatty acid transporter FadL. Science. 2004;304:1506–1509. doi: 10.1126/science.1097524. [DOI] [PubMed] [Google Scholar]
- 16.DiRusso CC, Black PN. Bacterial long-chain fatty acid transport: gateway to a fatty acid-responsive signalling system. J Biol Chem. 2004;279:49563–49566. doi: 10.1074/jbc.R400026200. [DOI] [PubMed] [Google Scholar]
- 17.Hearn EM, Patel DR, van den Berg B. Outer-membrane transport of aromatic hydrocarbons as a first step in biodegradation. Proc Natl Acad Sci. 2008;105:8601–8606. doi: 10.1073/pnas.0801264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar GB, Black PN. Linker mutagenesis of a bacterial fatty acid transport protein: identification of domains with functional importance. J Biol Chem. 1991;266:1348–1353. [PubMed] [Google Scholar]
- 19.Vorum H, Brodersen R, Kragh-Hansen U, Pedersen AO. Solubility of long-chain fatty acids in phosphate buffer at pH 7.4. Biochim Biophys Acta. 1992;1126:135–142. doi: 10.1016/0005-2760(92)90283-2. [DOI] [PubMed] [Google Scholar]
- 20.Black PN. Characterization of FadL-specific fatty acid binding in Escherichia coli. Biochim Biophys Acta. 1990;1046:97–105. doi: 10.1016/0005-2760(90)90099-j. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, et al. Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol Gen Genet. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- 22.Kahng HY, Byrne AM, Olsen RH, Kukor JJ. Characterization and role of tbuX in utilization of toluene in Ralstonia pickettii PKO1. J Bacteriol. 2000;182:1232–1242. doi: 10.1128/jb.182.5.1232-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eastcott L, Shiu WY, Mackay D. Environmentally relevant physical-chemical properties of hydrocarbons: a review of data and development of simple correlations. Oil and Chemical Pollution. 1988;4:191–216. [Google Scholar]
- 24.Ahn VE, et al. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 2004;23:2931–2941. doi: 10.1038/sj.emboj.7600320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong H, Patel DR, Tamm LK, van den Berg B. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J Biol Chem. 2006;281:7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- 26.Eaton RW. Organization and evolution of naphthalene catabolic pathways: sequence of the DNA encoding 2-hydroxychromene-2-carboxylate isomerase and trans-o-hydroxybenzylidenepyruvate hydratase-aldolase from the NAH7 plasmid. J Bacteriol. 1994;176:7757–7762. doi: 10.1128/jb.176.24.7757-7762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Beilen JB, Eggink G, Enequist H, Bos R, Witholt B. DNA sequence determination and functional characterization of the OCT-plasmid-encoded alkJKL genes of Pseudomonas oleovorans. Mol Microbiol. 1992;6:3121–3136. doi: 10.1111/j.1365-2958.1992.tb01769.x. [DOI] [PubMed] [Google Scholar]
- 28.Sun PD, Radaev S, Kattah M. Generating isomorphous heavy-atom derivatives by a quick-soak method. Part I: test cases. Acta Cryst. 2002;D58:1092–1098. doi: 10.1107/s0907444902006510. [DOI] [PubMed] [Google Scholar]
- 29.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ginsburgh CL, Black PN, Nunn WD. Transport of long-chain fatty acids in Escherichia coli: identification of a membrane protein associated with the fadL gene. J Biol Chem. 1984;259:8437–8443. [PubMed] [Google Scholar]
- 31.Ye J, van den Berg B. Crystal structure of the bacterial nucleoside transporter Tsx. EMBO J. 2004;23:3187–3195. doi: 10.1038/sj.emboj.7600330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 33.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Cryst. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 39.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Cryst. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multi-wavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.