Introduction
The increase in adult type diabetes is becoming a global epidemic and calls for swift actions to better understand the disease mechanisms, thus leading to improved targeted therapies. Emerging knowledge surrounding the role of microRNAs (miRNAs) in the regulation of post-transcriptional protein expression has dramatically altered the view of how target genes are regulated and how they are involved also in controlling glucose homeostasis. In addition to an improved understanding of miRNA functions, epigenetic control mechanisms are becoming better known. Thus, for example the effect of prenatal nutritional deficiencies and hereditary epigenetic changes, including DNA methylation and histone modifications are emerging as important players in the finely tuned balance of factors ultimately yielding the altered functions under various pathologic conditions.
In this article, we review the current understanding of the major epigenetic and post-transcriptional regulatory mechanisms with particular emphasis on podocytes during their injury associated with diabetic kidney damage. It is foreseen that this research line will bring major advances in diagnostics and understanding of pathomechanisms of both type 1 and 2 diabetes and will lead to identification of novel biomarkers and therapeutic targets.
Epigenetic regulation
The term epigenetics is typically defined as heritable changes in gene expression that are not encoded directly within the DNA sequence of genes. Epigenetic changes are crucial for the development and differentiation of the various cell types in an organism. However, epigenetic states can become disrupted by environmental influences or during ageing, and the importance of epigenetic changes in the development of cancer and other diseases is increasingly being discovered.
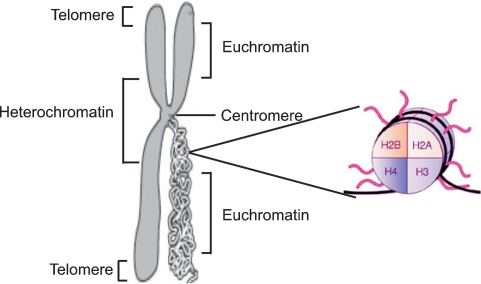
Eukaryotic genomes are packaged in two general varieties of chromatin: gene-rich euchromatin and genetically inactive heterochromatin. Heterochromatin is a tightly packaged form of DNA, and its major characteristic is that DNA transcription is limited. Centromeres and telomeres are both heterochromatic (Figure 1). The euchromatin, in contrast, contains ‘active’ chromatin: DNA sequences that are being transcribed into RNA [1]. Heterochromatin replicates in the S phase (synthesis phase) of the cell cycle later than euchromatin, most likely preserving DNA structure during replication. Heterochromatin also maintains a compact and visible structure during mitosis therefore differing from euchromatin, which undergoes a typical cycle of condensation and unravelling during this process [2].
Fig. 1.
Schematic figure of chromatin and histone structure. Chromatin is the complex of DNA and protein that makes up chromosomes. Heterochromatin locates in the centromeric region and telomeres whereas transcritpionally active euchromatin is located in the less condensed region. The functions of chromatin are to package DNA into a smaller volume to fit in the cell, to strengthen the DNA to allow mitosis and meiosis and to serve as a mechanism to control expression. The major proteins involved in chromatin are histone proteins. The flexible N-terminal tails of the four core histones H2A, H2B, H3 and H4 may undergo a range of post-translational modifications, including acetylation, methylation, O-GlcNac modification, phosphorylation and ubiquitination, all leading to changes in the gene expression level.
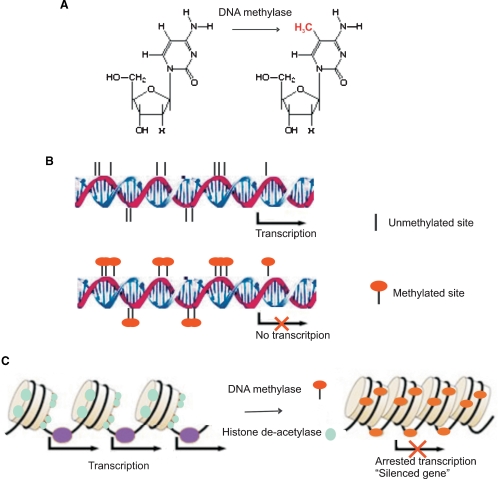
The DNA packaging densities of these two chromatin types vary along the length of the chromosome. High-density heterochromatin regions surround the centromeric region of the chromosome and have a low amount of expressed genes but a high content of repetitive DNA sequences including transposons, which are mobile genetic elements, and highly repetitive DNA regions called satellite DNAs [3]. These two chromatin states are controlled by reversible epigenetic patterns of DNA methylation and histone modifications that changes gene expression without involving changes in the DNA sequence (Figure 2A and B).
Fig. 2.
Schematic figure of epigenetic regulation mechanism. DNA methylase catalyze the transfer of a methyl group to the cytosine residues in CpG dinucleotide sequences (A). Methylation of the CpG islands in the gene promoter region inhibits gene expression (B). Histone acetylation plays an important role in the regulation of gene expression. Hyperacetylated chromatin is transcriptionally active whereas hypoacetylated chromatin is silent. Methyl-CpG-binding proteins interact with histone deacetylase causing gene silencing (C).
Heterochromatin has diverse known and postulated functions: maintenance of genome stability, proper chromosomal segregation during mitosis and prevention of telomere fusion [4]. Heterochromatin also regulates gene expression by both repression and activation, by directly silencing genes or by removal of silencing hallmarks of gene expression [5]. These mechanisms result in considerable regulatory functions during development and in different physiologic states of cells.
In addition to the heterochromatin diversity, the mammalian genome contains well-defined sequences, which notably consist of repeats of cytocine-phosphate-guanidine (CpG) motifs. These areas were earlier considered as non-functional redundant domains of the chromosomes during evolution while recent research has revealed strong regulatory functions to these elements mainly in influencing histone modifications and gene-silencing networks within the cells [6].
In general, the epigenetic regulation of gene transcription–translation machinery consists of now known multiple mechanisms for modifying the readout of the genetic code and repression of the chromatin state. In the context of DNA methylation, sequences within the genome can be classified into two different groups: CpG-deprived regions and CpG-rich regions called ‘CpG islands’ (see Figure 2). CpG islands are defined as being longer than 500 bp and having a GC base content >55% [7]. Considering their regulatory role, CpG islands are often found in promoter regions and about half of all genes contain CpG islands that are situated at the end of the 5′ region [8]. Altered methylation status of GC-rich regions may alter the chromatin structure and typically modulates the finely tuned promoter-transcription factor interactions with the transcriptional machinery [9]. This alteration leads to repressed gene expression associated with hypermethylated CpG islands. In contrast, genomic instability and aberrant gene expression have been associated with hypomethylated islands [10]. It has been proposed that more than half of the repeating 5′-CpG island sequences ultimately participate in transcription regulation as a result of methylation [11].
Histones are core proteins involved in DNA assembly and yielding chromatin structure. Histones may dynamically undergo post-translational modifications, which alter their interaction with DNA and nuclear proteins. The flexible N-terminal tails of the four core histones (H2A, H2B, H3 and H4, see Figure 1) may undergo a range of post-translational modifications, including acetylation, methylation, O-GlcNac modification, phosphorylation and ubiquitination [12,13] all leading to changes of the genetic readout. Histone modifications are indicators of active or repressed chromatin, and the ‘histone code’ hypothesis proposes that combinations of specific histone modifications create a complex defining the functional hierarchy for chromatin regulation.
Key enzyme families involved in histone modification include histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases and the methyl-binding domain protein MECP2 [6]. Whereas methylation of lysine-9 of the histone structure defined as (H3-K9) is a hallmark of ‘silenced’ genomic area throughout heterochromatic regions, the methylation of lysine 4 of histone H3 (H3-K4) denotes gain of activity and is found predominantly at the promoter areas of active genes [14]. The evidence that several methyl-CpG-binding proteins interact with histone deacetylase supports a mechanism linking DNA methylation and histone modifications together (Figure 2C) [15]. In addition, DNA methylation of CpG-rich regions has recently been connected to histone deacetylation, methylation of histone H3 at a specific site of lysine 9 as well as to direct interference by small RNAs [16].
Taken together, remarkable progress has been made in the understanding of the epigenetic mechanisms by which the gene readout can be modulated and changes in gene expression patterns achieved without assuming changes in the gene coding sequence. While the present understanding may already explain a substantial level of epigenetic regulation, a comprehensive understanding bringing all the elements together in understanding distinct pathophysiologic entities still remains to be established.
MicroRNA biogenesis
MicroRNAs are single-stranded transcribed RNAs of 19–25 nucleotides in length that are generated from endogenous hairpin structured transcripts throughout the genome [17]. Recent studies have shown that miRNAs have pivotal roles in diverse gene regulatory pathways, including e.g. control of timing of developmental processes [18], haematopoietic cell differentiation [19], apoptosis [20], cell proliferation and organ development [21]. MicroRNAs also represent one of the largest gene families, accounting for ∼2% of the whole genome [17]. Efforts to identify the specific miRNA targets have lead to speculations that miRNAs can directly regulate the imprint of >90% of human genes [22]. Their defined functions as key post-transcriptional regulatory mediators are rapidly emerging and have shown to be involved in the pathogenesis of a variety of diseases [23], including type 2 diabetes (see Table 1). Even now little is known of direct miRNA effects in the regulation of the functional kidney filtration barrier [24].
Table 1.
Validated and hypothetic miRNA targets
| Validated miRNA targets in type 2 diabetes and in the endocrine system | |||
|---|---|---|---|
| miRNA | Target | Function | Reference |
| miR-375 | Myotrophin | Inhibition of insulin secretion | [69] |
| miR-9 | OneCut2 transcription factor | Inhibition of glucose-stimulated insulin release | [70] |
| miR-192 | E-box repressors | TGF-beta-induced matrix protein collagen Col1a1 and -2 | [24] |
| miR-143 | GLUT4, HSL, fatty acid-binding protein aP2, PPAR-μ2 | Adipocyte differentiation | [49] |
| Hypothetic miRNA targets associated with type 2 diabetes | |||
| miR-30 family | Receptor for advanced glycation end product, immediate early response 3, vimentin, heat-shock protein-20 | Podocyte apoptosis and cytoskeletal structure | [79,80] |
| miR-23b | Hairy/enhancer of the split protein (Hes1) | Downstream target of activated Notch signalling and expressed in nephron segments during development | [81,84] |
| miR-15, miR-16 | Wnt and β-catenin | Nephron induction during embryo development | [75,76] |
| miR-7 | GY-box | Notch signalling | [73,74] |
| miR-4, miR-79 | Brd-box | ||
| miR-2, miR-11 | K-box | ||
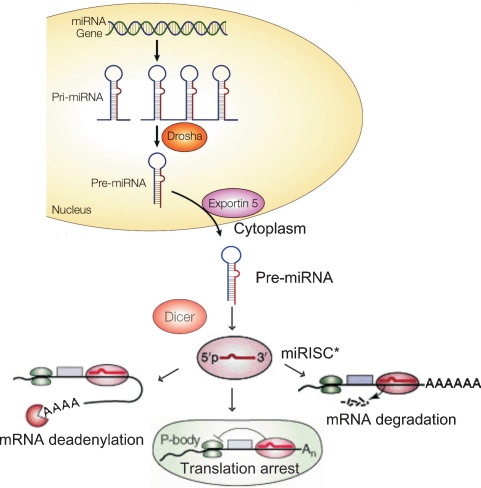
Biogenesis of miRNAs starts in the nucleus (see Figure 3) where stretches of several kilobases long primary miRNAs (pri-miRNAs) are transcribed from the genome, 3′-polyadenylated [covalent linkage poly (A) tail of 3′ end of miRNA], 5′-capped [7-methyl guanylate (m7G) cap of 5′ end of miRNA]. Pri-miRNAs are further modified by a microprocessor that consists of Drosha, a nuclear RNase-III enzyme, and its essential cofactor called Pasha [25]. This resulting protein complex processes pri-miRNAs into ∼70 nucleotide (nt)-long hairpin-shaped premature miRNAs (pre-miRNAs), and the subsequent cleavage at approximately two helical turns from the loop structure. Pre-miRNAs bear a two nt 3′-overhang that contributes to pre-miRNA export out of the nucleus to the cytoplasm by a RanGTP/exportin-5-dependent mechanism [26]. Dicer, a cytoplasmic RNaseIII enzyme, then subjects pre-miRNA to a second cleavage step in the cytoplasm. Dicer cleaves near the hairpin loop to release a short, ∼22 base-pair (bp)-long incomplete RNA duplex with characteristic two nt-long 3′-overhangs at both ends that anchor miRNA molecules to the miRNA-induced silencing complex miRISC [27]. The miRNA strand having lower thermodynamic stability at its 5′-end is selectively identified by miRISC and incorporated into miRISC that contains an Argonaute protein that is the catalytic component of RISC, and other protein factors [28]. When activated by miRNA, miRISC acts as an effector complex of the miRNA pathway interacting with messenger RNA (mRNA). There are several proposed and verified mechanisms regarding how these genome-originated miRNAs can modulate gene and protein expression [29]. It has been shown that miRNAs exhibiting full complementarity with target mRNA elicit mRNA degradation via a mechanism shared with short interfering RNAs (siRNAs). However, in cases of non-perfect complementary binding to the target mRNA 3′-untranslated region (3′-UTR), miRISC may hinder protein translation by several different mechanisms (Figure 3). These mechanisms include translational inhibition at the level of initiation and elongation, degradation of mRNA or the immature protein products or mRNA segregation into P bodies for translational inhibition and/or by mRNA deadenylation that causes destabilization of mRNA [30–32]. The outcome of all these mechanisms either singularly or acting in combination is an effective regulation of the final mRNA yielding protein expression.
Fig. 3.
MicroRNA genesis. Long stretches of immature pri-miRNAs are transcribed from the genome, Drosha and its microprocessor partner Pasha processes pri-miRNAs into ∼70 nt long hairpin-shaped pre-miRNAs. Pre-miRNAs are exported out of the nucleus to the cytoplasm by the RanGTP/exportin-5-dependent mechanism. Dicer cleaves pre-miRNAs near the hairpin loop to release a short ∼22 base-pair (bp) long imperfect RNA duplexes that are further incorporated into the miRISC complex. MiRNAs exhibiting full complementarity with target messenger RNA (mRNA) are shown to prompt mRNA degradation. MiRISC may also hinder protein translation by several different distinct mechanism including translational inhibition at the level of initiation and elongation, degradation of mRNA or the immature protein products or mRNA are segregated into P bodies for translational inhibition and/or mRNA deadenylation that causes destabilization of mRNA.
Tools to generate epigenetic and post-transcriptional profiles
Individual gene expression outcomes in complex diseases, typically in metabolic disorders such as type 2 diabetes, are unlikely to completely explain their impact on disease pathogenesis [33]. Multiple factors are normally needed to manifest any polygenic human disease. In addition to DNA mutations and protein post-translational modifications, well-known factors typically include a complexity of environmental, dietary and exercise factors while the epigenetic regulatory mechanisms and any combinations of the convoluted mechanisms involved appear to be responsible in an increasing number of disease outcomes [34]. Present day DNA analytics using robust techniques of, for example, genome-wide sequencing and mapping of global single nucleotide polymorphisms (SNPs), may thus not ultimately yield sufficient knowledge to understand disease pathogenesis. In combination with the new high-capacity array technologies [35], a new balance between global genomics, epigenetic gene regulation and post-transcriptional effects on protein landscape integrating the distinct data streams is needed to build a comprehensive understanding of underlying disease mechanisms in detail [36].
The huge screening capacity provided by modern microarrays enable the study of individual factors involved in disease pathogenesis on a genome-wide scale simultaneously. A wide range of approaches is rapidly emerging to track the gene targets under epigenetic regulation and to determine the global changes in genomic DNA methylation. Customized combinations of these approaches, based on individual requirements, pave the way to an in-depth understanding of the processes involved and, hopefully, will yield better understanding for improved diagnostics and for example new targets for better therapies.
Computational public database analysis [available e.g. at National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/), UCSC Genome Bioinformatics group, University of California, Santa Cruz, http:// genome.ucsc.edu/] is a key step in analysing and locating CpG islands in the promoter region of genes of interest. Considerable advances have been made in hybridization-based microarray technologies for genome-wide analysis for DNA methylation that is designed to discriminate between methylated and unmethylated sequences in gene promoters [36]. A variety of other tools (see Table 2) for genome-wide discovery of differentially methylated sites include restriction mapping, bisulfite nucleotide sequencing, PCR amplification and chromatin immunoprecipitation on DNA microarray (ChIP-on-chip; for an excellent recent review of the techniques see Ho and Tang [37]).
Table 2.
Techniques to analyse DNA methylation
| Method | Description | Reference |
|---|---|---|
| Methylation-sensitive restriction mapping (MSRF) | PCR-based method for genomic DNA after BstU1 (CG specific, methylation sensitive) and/or MseI (non-CG specific) digestion | [85] |
| Restriction landmark genomic scanning (RLGS) | Genome-wide screening method based on two dimensional separation of genomic DNA fragments containing radiolabel at the NotI restriction sites (GC-rich regions) | [86] |
| Methylated CG island amplification (MCA) | Amplification of DNA sequences with closely spaced (<1 kb) methylated SmaI sites that are commonly found in CG islands | [87] |
| Differential methylation hybridization (DMH) | Array-based method allows genome-wide screening of differentially methylated CG islands between two samples | [88] |
| Bisulphite sequencing | Sequencing-based method. Bisulfite treatment of genomic DNA converts cytosine to uracil, but 5-methylcytosine remains nonreactive | [89] |
| Methylation-specific oligonucleotide array (MSO) | Array-based high throughput method. Detection of methylation status of GC-rich genomic DNA fragments by comparison of signal intensities between the paired ‘methylated’ and ‘unmethylated’ oligonucleotide probes | [90] |
| DNA demethylating agents and gene expression analysis | Detection of reactivated gene expression after treatment of DNA demethylating agent (e.g. 5-aza-2′deoxycytosine) by gene expression microarrays | [38] |
| Genome-wide methylation array/promoter array | Human and mouse CpG island/promoter arrays are constructed based on the CG island library containing CG-rich DNA fragments. University Health Network Microarray Center (UHNMC, Toronto, www.microarray.ca) | [91] |
| Chromatin immunoprecipitation on DNA microarray (ChIP-chip) | The DNA fragments isolated from chromatin immunoprecipitation assay are used as targets in a microarray | [92] |
| Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) | Method is based on analysis and precise quantification of methylation on CpG positions and patterns in genomic DNA by MALDI-TOF-MS | [93] |
With current microarray methods, it is also possible to monitor the effects of DNA demethylating agents (e.g. 5-aza-2′-deoxycytidine) or histone deacetylation inhibitors (HDACs) by exploring transcription profiles of reactivated genes [38]. The approaches taken to distinguish miRNA expression signatures associated with kidney disorders differ from approaches used to assay DNA methylation levels because miRNAs interfere at the post-transcriptional level while DNA methylation influences the transcription of the genome expression. In addition, it is a challenge to validate the miRNA target sequences because several different miRNAs may bind to and cooperatively control a single mRNA target and, conversely, a single miRNA can bind to and regulate many different mRNA targets [39]. Therefore, high-throughput methods supplemented with extensive in silico data analysis and the subsequent investigation of the regulation of miRNA-mediated protein translation typically consist of comparison of protein expression levels [e.g. difference gel eletrophoresis (DiGE), mass spectrometry (MS) and western Blot] alongside with microRNA expression levels (miRNA arrays, qPCR) [40]. Hybridization-based microarrays to detect miRNAs are based on miRBase Sequence database (http://microrna. sanger.ac.uk/) that is a searchable database of published miRNA sequences and their annotations. MiRBase Sequence database consists of over 5000 miRNA loci from 58 species, ranging from vertebrates and invertebrates to prokaryotes including viruses [41]. Recently, next-generation sequencing (NGS) technology has been shown to be capable of producing millions of DNA sequence reads ranging from 35 to 250 base pairs in a single run (commercially available sequencers: Roche (454) GS FLX sequencer http://www.454.com/enabling-technology/ the-system.asp; Illumina/Solexa Genome analyser http:// www.illumina.com/; and the Applied Biosystems SOLiD system http://marketing.appliedbiosystems.com/mk/get/ SOLID_KNOWLEDGE_LANDING). This new technology provides multiple advantages when compared to gene expression microarray platforms, not least in discovering non-coding RNA sequences, especially novel microRNAs [42].
The final in vitro miRNA-target validation should be brought together by using pre-miRNA structures or miRNA inhibitors as demonstrated by Tian et al. [42] for example. Only a few in vitro validated miRNA targets have thus far been identified using the pre-miRNA hairpin structures that mimic natural miRNA molecules [43]. These pre-miRNAs are computationally predicted sequences emerging from the miRBase Sequence databases and typically increase the natural miRNA effect. Distinct pre-miRNA vector constructs as well as synthetic miRNA mimics are available commercially for miRNA functional analyses and miRNA-target site validation (e.g. Systems Biosciences, Mountain View, CA, USA: pre-miRNA clones; Dharmacon, Inc. Lafayette, CO, USA: miRIDIAN miRNA Mimic Library; Ambion, Inc. Austin TX, USA: Pre-miRTM miRNA Precursor Molecules) for extensive miRNA target validation for in vitro and in vivo purposes. Recently, it was shown that synthetic single stranded mature miRNAs or hairpin pre-miRNA structures cannot replace endogenous miRNAs already present in RISC [44]. This affects the validation of miRNA targets by exogenous miRNA mimic structures.
Antisense DNA oligonucleotides (ASOs) and anti-miRNA oligonucleotides (AMOs, antagomirs) are considered as a practical approach for specific pharmacological inhibition of miRNA function [45]. There are two suggested strategies to target miRNA function in vitro and in vivo. First, ASOs are designed for RNaseH-mediated targeted degradation of pri-miRNA molecules in nucleus [46]. The stereochemistry at the 2′-position of the ribose has been shown to be a major determinant in the target RNA-binding affinity and the activation of RNaseH [47]. Most common chemical modifications are phosphorothioate (PS) backbone modification combined with 2′-O-methyl (2′-O-Me) or with 2′-O-methoxyethyl (2′-O-MOE) antisense RNA oligomers or mixed locked nucleic acid (LNA)-DNA ASOs [46]. The base-pair interaction between mature miRNA and mRNA is another logical target for an inhibitor. AMOs are targeting mature miRNA molecules in cytoplasm and designed to block its function in miRISC [46]. Whereas pre-miRNA structures and unmodified single stranded mature miRNA molecules were not able to dissociate endogenous miRNAs from RISC, chemically modified ribo-oligonucleotides have been shown to present with this potential [44]. AMOs consist of chemical modifications of 2′-ribose and/or phosphate backbone (including 2′-O-Me, 2′Fluoro, 2′-O-MOE, LNAs and PS backbone modifications) essential in protecting AMOs against endonuclease-mediated degradation and in improving affinity to target miRNA molecules [46]. MiRNA–mRNA base-pair interaction has successfully been interrupted by transfecting modified AMOs complementary to miRNAs leading to blockage of the miRNA–RISC complex [48]. Esau et al. applied [49] 2′-O-MOE phosphorothioate-modified antisense RNA oligonucleotides targeting miRNAs and demonstrated the potential inhibition of adipocyte differentiation by a mir-143 antagomir.
Epigenetic regulation in diabetes
Epigenetic modulating mechanisms have recently been established as a massive regulatory machine that cannot be ignored in searching for a new mechanistic understanding of metabolic syndrome, obesity and type 2 diabetes [50]. Type 2 diabetes is typically characterized by a combination of peripheral insulin resistance with an insulin secretion defect that varies in severity. The known mechanisms for disrupted insulin secretion in type 2 diabetes include accumulated damage caused by hyperglycaemia, hyperlipidaemia and oxidative stress in combination [51,52]. Oxidative stress and reactive oxygen species have been shown to directly affect DNA methylation pattern and histone organization, therefore tuning the expression of multiple genes [53]. In addition, early nutritional status and possible intrauterine deprivation of nutrients have been shown to strongly impact the long-lasting DNA methylation effects directed at the genome readout [54] with immediate effects on development. A proposed mechanism includes effects by dietary methyl and cofactor sources, such as vitamin B12 and folic acid [55] linked to single-carbon metabolism providing the methyl groups for all biological methylation reactions. During embryonic development, the genome undergoes extensive demethylation followed by remethylation, and appropriate DNA methylation status must be maintained over the rounds of rapid cellular proliferation steps [56]. Unbalanced methyl donors from nutrition during embryonic development may thus irreversibly affect the DNA methylation patterns [57]. Therefore, extremes of nutritional availability and distinct environmental factors have been suggested to influence the epigenetic modifications in human genome and to enhance susceptibility to altered epigenetic pattern and later to metabolic disorders and adult chronic diseases [58]. Epidemiological studies have indicated that sub- or super-optimal nutrient provision in utero may lead to specific chromatin modifications as described above and have been proposed as mechanisms associated with the world-wide epidemic of type 2 diabetes [59]. To support this hypothesis, dietary protein restriction of pregnant rats typically associated with hypomethylation of e.g. the glucocorticoid receptor (GR) and peroxisome proliferator-activated receptor gamma (PPARγ) [60]. PPARγ coactivator 1 α (protein PGC-1α, gene PPARGC1A) is a well-established node in the pathogenesis of diabetes and a master transcriptional coactivator of mitochondrial genes [61]. Its expression is decreased and related to impaired ATP production as a consequence of reduced oxidative phosphorylation in patients with type 2 diabetes. It has also been reported that DNA methylation of the PPARGC1A promoter is increased in diabetic islets, demonstrating a plausible mechanism for reduced PPARGC1A mRNA expression and insulin secretion in pancreatic islets of patients with type 2 diabetes [62]. Consequently, super-optimal methyl donors from nutrition may influence DNA methylation in the PPARGG1A promoter region and therefore represent a potential epigenetic cause for type 2 diabetes. Heterochromatin silencing and histone H3-K9 methylation of heterochromatin transcription and generation of heterochromatin-originated short interfering RNA (siRNA) molecules have been shown in fission yeasts [63]. This epigenetic regulation has specifically been timed during the S-phase of the cell cycle in a temperature sensitive manner. In mammals, levels of DNA methylation correlate with age [64]. This suggests that gene expression may be repressed over time. It has been shown that in the glomerulus ageing podocytes exhibit changed gene expression patterns [65]. In addition, it has been shown that the developmental regulator Pax2 promotes assembly of an H3K4 methyltransferease complex through nuclear factor PTIP (Pax Transcription activation domain Interacting Protein) [66]. The PTIP complex localizes to a Pax2 DNA-binding sequence and recruits the methyltransferease complex. In the embryo, together, Pax2 and Pax8 are important to regulators inducing nephric structures in the intermediate mesoderm and urogenital epithelium [67]. Therefore, Pax2 provides a kidney-specific locus for epigenetic modifications during development.
In addition, it has been shown that some genes that become DNA hypermethylated and silenced in cancer cells require intact DICER function for the maintenance of their epigenetic status [68]. These results support the hypothesis that RNA interference-mediated DNA methylation in cooperation with environmental responses modify the genetic risk factors and the epigenetic inheritance of heterochromatin.
MicroRNAs in insulin release regulation and kidney actions
A subset of miRNAs has shown to be involved in metabolic regulation of glucose homeostasis (Table 1). However, only very few miRNA species have been fully characterized experimentally and most of their functions remain unknown. Pancreatic islet-specific miR-375 inhibits insulin secretion in mouse pancreatic β-cells by inhibiting the expression of the protein myotrophin [69]. In addition, myotrophin, also known as V-1, has been shown to interact with an actin-capping protein inhibiting F-actin assembly, and to induce exocytosis of insulin granules [69]. In another study, increasing the level of miR-9 resulted in a severe defect in glucose-stimulated insulin release by down-regulating the transcription factor Onecut2 (OC2) [70]. OC2 represses the activity and expression of Rab27a effector granuphilin, which is a key component of the machinery controlling insulin release [71]. Also miR-192 levels have been shown to be increased in glomeruli isolated from streptozotocin-injected diabetic mice as well as diabetic mice db/db when compared to non-diabetic mice [24]. MiR-192 was shown to regulate E-box repressors that are responsible for controlling TGF-β induced extracellular matrix proteins collagen 1-α 1 and 2 (Col1a1 and 2) expression [24]. Col1a1 and 2 were shown to accumulate during diabetic nephropathy; therefore, these results suggest an potential role of miR-192 in kidney diseases.
Recently, a correlation between elevated Notch signalling pathway gene expression and diabetic nephropathy has been shown, in concert with Gremlin, the gene associated with tubulointerstinal fibrosis in diabetic nephropathy [72], suggesting the presence of miRNAs and CpG islands as a potential regulatory strategy in this disease [73]. Interestingly, distinct miRNAs appear to modify Notch pathways in Drosophila melanogaster with an effect in signalling cascades determining cell specification and development [74]. Lai et al. showed that specific miRNAs regulated negative regulatory sequence motifs known as the GY box, the Brd box and the K box in the 3′ UTR of the Notch target genes [74]. These regulatory boxes have been shown to serve as binding sites for miRNAs: GY-box is inhibited by miR-7, Brd-box by miR-4 and miR-79, K-box by miR-2 and miR-11; however, it remains to be determined whether these miRNAs regulate Notch signalling in diabetic nephropathy. MiR-143 has been experimentally shown to regulate genes that are crucial for adipocyte differentiation, (including GLUT4, HSL, fatty acid-binding protein aP2 and PPAR-γ2) demonstrating a role for miRNAs in fat metabolism and in endocrine function in humans [49]. In addition, miR-15 and miR-16 have been proposed to control the Wnt/β-catenin signalling pathway during the embryonal stage [75]. Wnt/β-catenin signalling regulates the early events of nephrogenic induction during mouse kidney development [76]; however, a regulatory role for miR-15 or -16 has not yet been demonstrated during this process.
In the human kidney cortex, the renal glomerulus forms the biological sieve that allows the passage of water and small molecules while macromolecules e.g. albumin cannot normally pass beyond the filtration barrier [77]. Podocytes surround the basement membrane of glomerular capillaries from the outside and present specialized structures, foot processes linked to each other forming the slit diaphragms [78]. Therefore podocytes are considered to be critical for maintaining the functional filtration barrier and preventing albuminuria: alterations in slit diaphragm-associated genes result in severe proteinuria [77].
While the association of podocytic miRNA and DNA methylation profiles in health and disease is an important topic to study, little is still known of the role of miRNAs for this complex. General differences in miRNA expression as well as in the proteome profile have been shown in the rat renal medulla and in the renal cortex region using the microRNA microarray [40]. Very recent findings of podocyte-specific deletion of Dicer demonstrated a critical role for miRNA regulation in the progression of glomerular and tubular damage, and therefore the development of proteinuria [79,80]. Dicer deletion in podocytes led to podocyte apoptosis and depletion; proteinuria was significant 3 weeks after birth in mouse models. Also the rapid progression of glomerular and tubular injury was prominent at week 3, and culminated in death several weeks later. Based on altered gene expression profile in podocyte-specific Dicer knock-down glomeruli, especially the miR-30 family has been highlighted as candidates participating in podocyte homestasis and pathogenesis of kidney diseases of podocyte origin. In another study with the podocyte-specific Dicer knock-out mice, it was noticed that expression of slit diaphragm proteins nephrin and podocin was decreased [81–83]. In this study, mmu-miR-23b, mmu-miR24 and mmu-miR26a were implicated as critical to maintain the glomerular filtration barrier [81].
The rapid emergence and advances in high throughput screening techniques facilitate focused research of both genome-wide and cell-specific epigenetic and post-transcriptional imprints in diabetes and kidney disease. Improved understanding and identification of new epigenetic and post-transcriptional regulatory mechanisms involved in the regulation of the key molecular complexes of podocytes will be of utmost importance in advancing diagnosis and novel biomarker development, and in future therapeutics.
Acknowledgments
This study was supported by European Union FP6-Lifescihealth-7 (DiaNa), The Sigrid Juselius Foundation of Finland and Science Foundation Ireland.
Conflict of interest statement. None declared.
References
- 1.Dimitri P, Corradini N, Rossi F, et al. The paradox of functional heterochromatin. Bioessays. 2005;27:29–41. doi: 10.1002/bies.20158. [DOI] [PubMed] [Google Scholar]
- 2.Heitz E. Das heterochromatin der Moose. Jehrb Wiss Botanik. 1928;69:762–818. [Google Scholar]
- 3.Elgin SC, Grewal SI. Heterochromatin: silence is golden. Curr Biol. 2003;13:R895–R898. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Bailis JM, Forsburg SL. S phase assembly of centromeric heterochromatin and cohesion. Cell Cycle. 2004;3:416–418. [PubMed] [Google Scholar]
- 5.Piacentini L, Fanti L, Berloco M, et al. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- 8.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 9.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 10.Eden A, Gaudet F, Waghmare A, et al. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 11.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 13.Kaleem A, Hoessli DC, Ahmad I, et al. Immediate-early gene regulation by interplay between different post-translational modifications on human histone H3. J Cell Biochem. 2008;103:835–851. doi: 10.1002/jcb.21454. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein BE, Humphrey EL, Erlich RL, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–217. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Morita K, Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 2006;25:5794–5804. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgantas RW, 3rd, Hildreth R, Morisot S, et al. CD34 +hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 23.Perera RJ, Ray A. MicroRNAs in the search for understanding human diseases. BioDrugs. 2007;21:97–104. doi: 10.2165/00063030-200721020-00004. [DOI] [PubMed] [Google Scholar]
- 24.Kato M, Zhang J, Wang M, et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macrae IJ, Zhou K, Li F, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 28.Tomari Y, Matranga C, Haley B, et al. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 29.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 30.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 31.Humphreys DT, Westman BJ, Martin DI, et al. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Valencia-Sanchez MA, Hannon GJ, et al. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romao I, Roth J. Genetic and environmental interactions in obesity and type 2 diabetes. J Am Diet Assoc. 2008;108:S24–S28. doi: 10.1016/j.jada.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 34.Hanson MA, Gluckman PD. Developmental origins of health and disease: new insights. Basic Clin Pharmacol Toxicol. 2008;102:90–93. doi: 10.1111/j.1742-7843.2007.00186.x. [DOI] [PubMed] [Google Scholar]
- 35.Gresham D, Dunham MJ, Botstein D. Comparing whole genomes using DNA microarrays. Nat Rev Genet. 2008;9:291–302. doi: 10.1038/nrg2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi H, Maier S, Nimmrich I, et al. Oligonucleotide-based microarray for DNA methylation analysis: principles and applications. J Cell Biochem. 2003;88:138–143. doi: 10.1002/jcb.10313. [DOI] [PubMed] [Google Scholar]
- 37.Ho SM, Tang WY. Techniques used in studies of epigenome dysregulation due to aberrant DNA methylation: an emphasis on fetal-based adult diseases. Reprod Toxicol. 2007;23:267–282. doi: 10.1016/j.reprotox.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dannenberg LO, Edenberg HJ. Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genomics. 2006;7:181. doi: 10.1186/1471-2164-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 40.Tian Z, Greene AS, Pietrusz JL, et al. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 43.Kuhn DE, Martin MM, Feldman DS, et al. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang F, Hajkova P, O’Carroll D, et al. MicroRNAs are tightly associated with RNA-induced gene silencing complexes in vivo. Biochem Biophys Res Commun. 2008;372:24–29. doi: 10.1016/j.bbrc.2008.04.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esau CC. Inhibition of microRNA with antisense oligonucleotides. Methods. 2008;44:55–60. doi: 10.1016/j.ymeth.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Davis S, Lollo B, Freier S, et al. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamaratski E, Pradeepkumar PI, Chattopadhyaya J. A critical survey of the structure-function of the antisense oligo/RNA heteroduplex as substrate for RNase H. J Biochem Biophys Methods. 2001;48:189–208. doi: 10.1016/s0165-022x(01)00149-x. [DOI] [PubMed] [Google Scholar]
- 48.Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 49.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 50.Junien C, Nathanielsz P. Report on the IASO Stock Conference 2006: early and lifelong environmental epigenomic programming of metabolic syndrome, obesity and type II diabetes. Obes Rev. 2007;8:487–502. doi: 10.1111/j.1467-789X.2007.00371.x. [DOI] [PubMed] [Google Scholar]
- 51.Wallace TM, Matthews DR. Coefficient of failure: a methodology for examining longitudinal beta-cell function in type 2 diabetes. Diabet Med. 2002;19:465–469. doi: 10.1046/j.1464-5491.2002.00718.x. [DOI] [PubMed] [Google Scholar]
- 52.Robertson RP, Harmon J, Tran PO, et al. Glucose toxicity in beta-cells: type 2 diabetes, good radicals gone bad, and the glutathione connection. Diabetes. 2003;52:581–587. doi: 10.2337/diabetes.52.3.581. [DOI] [PubMed] [Google Scholar]
- 53.Cerda S, Weitzman SA. Influence of oxygen radical injury on DNA methylation. Mutat Res. 1997;386:141–152. doi: 10.1016/s1383-5742(96)00050-6. [DOI] [PubMed] [Google Scholar]
- 54.Gallou-Kabani C, Junien C. Nutritional epigenomics of metabolic syndrome: new perspective against the epidemic. Diabetes. 2005;54:1899–1906. doi: 10.2337/diabetes.54.7.1899. [DOI] [PubMed] [Google Scholar]
- 55.Van Den Veyver IB. Genetic effects of methylation diets. Annu Rev Nutr. 2002;22:255–282. doi: 10.1146/annurev.nutr.22.010402.102932. [DOI] [PubMed] [Google Scholar]
- 56.Morgan HD, Santos F, Green K, et al. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 57.Waterland RA, Garza C. Potential mechanisms of metabolic imprinting that lead to chronic disease. Am J Clin Nutr. 1999;69:179–197. doi: 10.1093/ajcn/69.2.179. [DOI] [PubMed] [Google Scholar]
- 58.Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat Res. 2006;600:46–57. doi: 10.1016/j.mrfmmm.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 59.Devaskar SU, Thamotharan M. Metabolic programming in the pathogenesis of insulin resistance. Rev Endocr Metab Disord. 2007;8:105–113. doi: 10.1007/s11154-007-9050-4. [DOI] [PubMed] [Google Scholar]
- 60.Simmons RA. Developmental origins of beta-cell failure in type 2 diabetes: the role of epigenetic mechanisms. Pediatr Res. 2007;61:64R–67R. doi: 10.1203/pdr.0b013e3180457623. [DOI] [PubMed] [Google Scholar]
- 61.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 62.Ling C, Del Guerra S, Lupi R, et al. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–622. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kloc A, Zaratiegui M, Nora E, et al. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Wiggins JE, Goyal M, Sanden SK, et al. Podocyte hypertrophy, “adaptation,” and “decompensation” associated with glomerular enlargement and glomerulosclerosis in the aging rat: prevention by calorie restriction. J Am Soc Nephrol. 2005;16:2953–2966. doi: 10.1681/ASN.2005050488. [DOI] [PubMed] [Google Scholar]
- 66.Patel SR, Kim D, Levitan I, et al. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bouchard M, Souabni A, Mandler M, et al. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002;16:2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ting AH, Suzuki H, Cope L, et al. A requirement for DICER to maintain full promoter CpG island hypermethylation in human cancer cells. Cancer Res. 2008;68:2570–2575. doi: 10.1158/0008-5472.CAN-07-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 70.Plaisance V, Abderrahmani A, Perret-Menoud V, et al. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 71.Gomi H, Mizutani S, Kasai K, et al. Granuphilin molecularly docks insulin granules to the fusion machinery. J Cell Biol. 2005;171:99–109. doi: 10.1083/jcb.200505179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dolan V, Murphy M, Sadlier D, et al. Expression of gremlin, a bone morphogenetic protein antagonist, in human diabetic nephropathy. Am J Kidney Dis. 2005;45:1034–1039. doi: 10.1053/j.ajkd.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 73.Walsh DW, Roxburgh SA, McGettigan P, et al. Co-regulation of Gremlin and Notch signalling in diabetic nephropathy. Biochim Biophys Acta. 2008;1782:10–21. doi: 10.1016/j.bbadis.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 74.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martello G, Zacchigna L, Inui M, et al. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 76.Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–2539. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- 77.Patari-Sampo A, Ihalmo P, Holthofer H. Molecular basis of the glomerular filtration: nephrin and the emerging protein complex at the podocyte slit diaphragm. Ann Med. 2006;38:483–492. doi: 10.1080/07853890600978149. [DOI] [PubMed] [Google Scholar]
- 78.Rodewald R, Karnovsky MJ. Porous substructure of the glomerular slit diaphragm in the rat and mouse. J Cell Biol. 1974;60:423–433. doi: 10.1083/jcb.60.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi S, Yu L, Chiu C, et al. Podocyte-selective deletion of Dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol. 2008;19:2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harvey SJ, Jarad G, Cunningham J, et al. Podocyte-specific deletion of Dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol. 2008;19:2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ho J, Ng KH, Rosen S, et al. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Holzman LB, St John PL, Kovari IA, et al. Nephrin localizes to the slit pore of the glomerular epithelial cell. Kidney Int. 1999;56:1481–1491. doi: 10.1046/j.1523-1755.1999.00719.x. [DOI] [PubMed] [Google Scholar]
- 83.Roselli S, Gribouval O, Boute N, et al. Podocin localizes in the kidney to the slit diaphragm area. Am J Pathol. 2002;160:131–139. doi: 10.1016/S0002-9440(10)64357-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen L, Al-Awqati Q. Segmental expression of Notch and Hairy genes in nephrogenesis. Am J Physiol Renal Physiol. 2005;288:F939–F952. doi: 10.1152/ajprenal.00369.2004. [DOI] [PubMed] [Google Scholar]
- 85.Huang TH, Laux DE, Hamlin BC, et al. Identification of DNA methylation markers for human breast carcinomas using the methylation-sensitive restriction fingerprinting technique. Cancer Res. 1997;57:1030–1034. [PubMed] [Google Scholar]
- 86.Hayashizaki Y, Hirotsune S, Okazaki Y, et al. Restriction landmark genomic scanning method and its various applications. Electrophoresis. 1993;14:251–258. doi: 10.1002/elps.1150140145. [DOI] [PubMed] [Google Scholar]
- 87.Toyota M, Ho C, Ahuja N, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 88.Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 89.Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gitan RS, Shi H, Chen CM, et al. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12:158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cross SH, Charlton JA, Nan X, et al. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 92.Oberley MJ, Tsao J, Yau P, et al. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 2004;376:315–334. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
- 93.Tost J, Schatz P, Schuster M, et al. Analysis and accurate quantification of CpG methylation by MALDI mass spectrometry. Nucleic Acids Res. 2003;31:e50. doi: 10.1093/nar/gng050. [DOI] [PMC free article] [PubMed] [Google Scholar]