Abstract
RNAs are exported from the nucleus to the cytoplasm, where they undergo translation and produce proteins needed for the cellular life cycle. Some mRNAs are targeted by different RNA decay mechanisms and thereby undergo degradation. The 5′→3′ degradation machinery localizes to cytoplasmic complexes termed P bodies (PBs). They function in RNA turnover, translational repression, RNA-mediated silencing, and RNA storage. A quantitative live-cell imaging approach to study the dynamic aspects of PB trafficking in the cytoplasm revealed that PB movements are rather confined and dependent on an existing microtubule network. Microtubule depolymerization led to a drastic decrease in PB mobility, as well as a release of regulation on PB assembly and a dramatic increase in PB numbers. The different aspects of PB trafficking and encounters with mRNA molecules in the cytoplasm are discussed.
Key words: P body, live-cell imaging, microtubules, RNA degradation
The gene expression pathway begins in the nucleus with the activation of genes, that in turn produce messenger RNAs (mRNAs). mRNAs are processed in the nucleus, then to be packaged as an mRNA-protein complex, termed the mRNP, destined for the cytoplasm. Following mRNA export, these molecules may face a number of cytoplasmic fates. Typically, an mRNA will be translated into a functional protein by the ribosomal translation machinery, while in some cases mRNAs might be maintained in a translationally repressed state. In parallel, mRNAs undergo systematic check-up to ensure that they contain the correct protein coding sequence and that they were not aberrantly formed during nuclear transcription. Several such quality control mechanisms exist, thereby regulating the accuracy of the gene expression process. mRNAs that have reached the end of their term in the life cycle of a cell or aberrant mRNAs are subjected to degradation, referred to as mRNA decay. mRNAs are linear molecules of nucleic acids and therefore several mechanisms that tackle the ends of mRNA molecules exist to guarantee the removal of mRNAs, and are orchestrated by a number of nucleic-acid degrading enzymes called exonucleases. mRNA degradation can commence from the 5′ cap end, the 3′ polyA tail, or even from within the transcript. In eukaryotes, mRNA 3′ deadenylation usually initiates the degradation process, and then either of two major pathways is utilized. A large protein complex, the exosome, degrades mRNAs from the 3′→5′ direction, whereas other proteins are involved in the 5′→3′ degradation pathway. In this case the action of a decapping enzyme (Dcp2) is first required, followed by an exonuclease termed Xrn1.1 Cap removal irreversibly marks the mRNA for degradation.
Most cytoplasmic organelles and structures have been known and well studied for years and therefore the rather recent discovery that the proteins of the 5′→3′ degradation machinery localize in previously unidentified cytoplasmic bodies, was intriguing. First it was shown by fluorescence microscopy that the Xrn1 exonuclease localizes in discrete cytoplasmic foci within mammalian cells.2 Thereafter, the Dcp2 decapping enzyme was identified in the same bodies.3–5 These cytoplasmic complexes are a topic of much interest and research. They are referred to as P bodies (PBs), processing bodies, Dcp-bodies, mRNA-decay foci and GW182 bodies. The protein inventory of P bodies now is quite large exceeding ∼25 different factors. P bodies exist naturally in the cytoplasm of yeast cells were they were first functionally recognized6 and in mammalian cells.7 They are thought to be cytoplasmic centers in which mRNA decapping and 5′→3′ degradation can occur.8 Aside of these functions, further evidence associates PBs with additional regulatory processes such as translational repression, RNA-mediated silencing and mRNP storage.9–11
P bodies are protein-RNA complexes that form in the cytoplasm. They are not defined as an organelle since they are not membrane bound.12 Electron microscopy studies showed P bodies to be composed of aggregates of electron dense fibrils (8–10 nm in diameter) 100–300 nms in diameter.12 In mammalian cells, most cells contain between 3–9 distinct PBs, although these numbers can differ between cells. Smaller PBs probably also exist as seen from immunofluorescent staining performed with antibodies to different PB components. Yet, the significance of these size differences remains unknown. The variation in PB numbers and sizes between cells are dependent on the stage of the cell cycle, and correlate with the proliferation ratio of the cell, and the availability of nutrients.12 A variety of treatments can cause either the disassembly or assembly, or an increase in the size of PBs. In yeast (S. cerevisiae), deprivation of glucose leads to a stress response that includes the assembly of PBs. On the other hand, the inhibition of translation elongation dissociates PBs.6,13,14 In mammalian cells, the shutdown of mRNA transcription or translation culminates in the disassembly of PBs.7,15,16 These observations indicate that PBs are dynamic structures whose structural integrity responds to a variety of metabolic cues. For instance during certain stress responses, cells can assemble another set of cytoplasmic bodies, termed stress granules (SGs). These large structures accumulate stalled translational pre-initiation mRNA-ribosome complexes17 and are spatially associated with PBs. It is proposed that mRNAs can cycle between ribosomal polysomes, PBs and SGs, depending on the integrity of the mRNA and the translational status of the cell.9
The dynamic nature of PB integrity raises the issue of PB trafficking within the cytoplasm. The questions at hand are whether PBs freely roam the cytoplasm, or are PBs physically interacting with other cytoplasmic structures? If some mRNAs are able to enter PBs, how and where do the PB and mRNP complexes meet? Does the cell expend energy in a cytoplasmic scanning mechanism for mRNAs destined for degradation, or are such encounters left to their own devices?
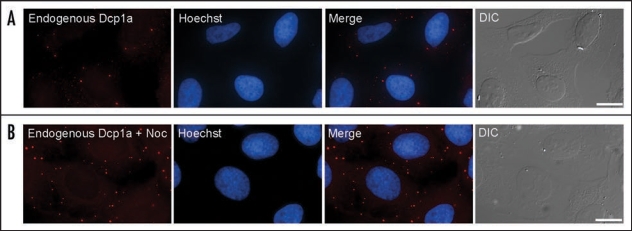
The study of the dynamic properties of PBs must take place in a living cell by use of time-lapse microscopy. Using fluorescently tagged proteins that are components of PBs it is possible to detect these structures in living cells and to follow their kinetics. Typically, green fluorescent protein (GFP) or red fluorescent protein (RFP) fusions are used. In our study, we wished to follow the dynamic aspects of PBs in human cells.18 We therefore tagged three PB components, Dcp1a, Dcp1b and Dcp2. The Dcp proteins are conserved PB components in yeast, C. elegans, Drosophila and mammalian cells.16,19–21 As aforementioned, Dcp2 is the decapping enzyme that removes the 5′ cap from the mRNA destined for degradation. Dcp1a and Dcp1b proteins share 30% homology and are encoded by two different genes.5 These proteins are thought to have a stimulatory effect on Dcp2 decapping activity. We made stable cell lines harboring each of these proteins fused to an XFP protein (Fig. 1A) and then followed the trafficking of PBs in single cells using live-cell imaging techniques performed on a fluorescent microscope. Live-cell conditions can allow for long imaging periods of many hours or days, pending the use of low levels of light exposure to avoid cytotoxic effects.
Figure 1.
Endogenous PBs were stained with an anti-Dcp1a antibody (red). The nucleus was counterstained with Hoescht (blue) and cells were also imaged in DIC (grey). (A) Normal distribution of PB in human cells. (B) Disruption of the microtubule network using nocodazole (noc) led to an increase in PB numbers. (Bar, 20 µm).
While it might be expected that PBs roam the cytoplasm in pursuit of RNAs destined for degradation, the contrary is shown by following the paths of PBs in living cells. PBs were identified in time-lapse movies, and single PBs were tracked frame by frame. PB movements were rather spatially confined, suggesting that their targets must find their own way to these sites. More specifically, rapid imaging showed erratic PB movement within small sized regions, and imaging for long periods during interphase demonstrated that most PBs tended to remain within the same cytoplasmic region throughout the cell cycle. Quantification of such movements showed that PB mobility portrayed a restricted diffusional pattern. Diffusion coefficients, which are values that describe the relative speed at which a particle moves within a defined area, were in a range that is similar to other cytoplasmic organelles (10−3 to 10−2 µm2/sec). For instance: mitochondria (5 × 10−4 µm2/sec),22 neutrophilic vesicles (2.5 × 10−2 µm2/sec),23 secretory vesicles (3.9 × 10−4 to 7.4 × 10−3 µm2/sec),24 chromaffin granules (3 × 10−3 µm2/sec).25 Slow diffusion and restricted movements of cytoplasmic organelles are known phenomena and are attributed to the non-homogeneity of the cytoplasm and to the effects that crowding, obstruction and exclusion have on cytoplasmic transport.26 On a cellular level this implies that long-range delivery of cargo within the cytoplasm must depend on mechanisms of active transport and require energy investment by the cell.
Confined motion was the major type of PB dynamics observed. Yet, few PBs were completely stationary while few others moved in a rapid and directional manner. Specifically, the latter were also observed moving along the outer rim of the nuclear membrane. This could imply a mechanism of patrolling of the nuclear periphery for association with exported mRNAs, as it has been suggested that aberrant mRNAs destined to degradation are dealt with as they emerge in the cytoplasm. In other cases, PBs traveled on a straight track for up to ten microns, and could be seen translocating rapidly from one region of the cell to the other (measured speeds of 0.5–1.1 µm/sec). All the above suggested that PBs are anchored to a cytoplasmic structure, namely the cytoskeleton. In order to visualize the cytoskeleton in living cells, GFP fusions of β-actin or tubulin proteins that integrate into the cytoskeleton were used. When a green cytoskeleton was imaged with red PBs, the cytoskeletal association was clearly demonstrated. The dynamic but confined movements of PBs were seen in association with microtubules, whereas stationary PBs were associated with actin bundles. PB mobility was in fact a result of the type of fiber it was attached to. Since actin bundles are rigid structures, no movement of PBs was seen on them. However, microtubules exhibit continual swaying motion and this is the reason for the confined erratic movement of the PBs. As for directional movement, microtubule tracks marked with GFP-tubulin showed that some PBs could translocate on microtubules, reach microtubule interscetions and change tracks. Together with the measured speeds of directional PBs it stands to reason that some PBs can be associated with a motor protein that provides translocation services to PBs. Another possibility is that PBs hitchhike on other moving organelles that are known to commute on the microtubule highways in the cytoplasm.
The microtubule and actin networks are also used for RNA trafficking in cells. A classic example is the process of RNA localization,27,28 specifically the motor driven movement of β-actin mRNA to the leading edge of moving cells.27 It is therefore plausible that mRNAs and PB encounters occur during movement on microtubules. In neuronal cells that contain long cell extremities, the microtubule system is utilized for the trafficking of mRNAs in the form of large mRNP granules. A recent study has examined whether PBs and mRNPs are transported together.29 Using a neuronal mRNP marker called Staufen (Stau), mRNPs were tagged in vivo (using GFP-Stau) together with PBs (RFP-Dcp1). Live-cell microscopy of neurons showed that mRNPs and PBs moved independently of each other. Half of the PBs moved unidirectionally both in the anterograde and retrograde direction, while the other half moved in a bidirectional manner, and showed rapid switches in direction. Interestingly, in about 50% of the cases PBs were found adjacent to mRNPs as if the two entities were docked to each other. In this study29 and in our study,18 PBs were not found associated with other organelles such as mitochondria, endoplasmatic reticulum and peroxisomes, but did associate with stress granules that are also RNA-containing structures. Chemical synaptic stimulation of neurons led to a substantial decrease in dendritic PBs, which returned to normal levels after a 30 minute period. This study implies that dendritically localized mRNPs might be stored in PBs to be released after synaptic activation.
The above study suggests that mRNPs and PBs can meet on the microtubule network of neurons. In non-neuronal cells we found PB association with microtubules, with marginal directional movements. Although mRNP-PB encounters might take place by way of use of the microtubule network, a more random mechanism might be occurring. Cytoplasmic mRNPs diffuse a 100 fold faster than PBs and other cytoplasmic vesicles (0.1 µm2/sec)30 and are at least 1,000 times more abundant,31 thereby providing the probability of interactions between RNAs and PBs based on the random movement of both. This might explain why the 5′→3′ RNA decay machinery is not membrane bound, in order to facilitate rapid RNA-PB interactions. RNA fluorescent in situ hybridization (FISH) has shown RNA encircling the P body, indicating that interactions are occurring at the PB periphery.32 Yeast and mammalian studies have shown that PBs have a role in the storage of mRNAs in an untranslated state.9,14,33 Thus, a non-membranous structure allows the robust exchange of mRNPs in and out of the PBs in response to cellular cues.
It is of interest that the 5′→3′ degradation pathway resides within PBs, while the 3′µ5′ exosomal proteins portray a diffuse cytoplasmic distribution. Such spatial compartmentalization of the two processes might be a means for obtaining different levels of regulation of RNA degradation.11 Indeed, when microtubule depolymerizing agents were used an increase in PB numbers was observed (Fig. 1B), and a similar effect has been seen in yeast cells.34 This means that a diffuse cytoplasmic pool of PB components exits. However, these two pools are constantly exchanging as shown by kinetic quantification of PB component dynamics performed by photobleaching experiments. Fluorescence recovery after photobleaching (FRAP) has shown that most PB components are constantly exchanging between PBs and the cytoplasm,16,17,35 except for the decapping enzyme Dcp2 that did not exchange.18 This suggests that Dcp2 is a core component of PBs. However, when there is no microtubule network, this control is overrun and most of the Dcp proteins can now assemble into large PBs. It is possible that the microtubules themselves act in the restriction of PB size under normal conditions.34
On the other hand, when no microtubule network was available—PB diffusion was reduced. We suggest that this slow diffusion ensures that when PBs detach from microtubules, they remain in the vicinity to re-attach, while anchoring to microtubules increases the probability of colliding with RNAs since the latter move on and in the volumes between microtubules that are not occupied by other cytoplasmic organelles. We therefore conclude that the relative mobility of the microtubule network is necessary for increasing the volumes that PBs probe and the targets that they encounter.
Future studies will reveal the kinetic aspects of RNA interactions with PBs, and will help in understanding the real function underlying PB structures.36 We still do not understand the cellular decision-making in targeting mRNAs to PBs or SGs for either RNA decay or storage. The significance of the microscopic PBs we detect also requires further investigation, namely why are there different sized PBs and do these differences reflect levels of functionality. Since the disassembly of large PBs during different treatments probably does not inhibit mRNA decay, then it remains to be seen if PB function can take place diffusely throughout the cytoplasm or whether small-sized PBs are the sites of PB action. Or in other words, does the 5′→3′ decay machinery require a large protein structure for its catalytic action, or can these particular biochemical reactions occur anywhere in the cytoplasm. A combination of live-cell microscopy and biochemical studies will shed light on these types of questions that involve PB structure and function.
Acknowledgements
Y.S.T. thanks the following agencies for their support: the Israel Science Foundation, the German-Israeli Foundation, DIP, the USA-Israel Binational Science Foundation, the Israel Cancer Research Fund, the Ministries of Health & Science, and the Israel Cancer Association. Y.S.T. thanks the Israel Science Foundation for the equipment grant for the fluorescence live-cell imaging microscope. Y.S.T. is the Jane Stern Lebell Family Fellow in Life Sciences at Bar-Ilan University.
Abbreviations
- PB
P body
- SG
stress granule
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/7773
Supplementary Material
References
- 1.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 2.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 5.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 10.Jakymiw A, Pauley KM, Li S, Ikeda K, Lian S, Eystathioy T, et al. The role of GW/P-bodies in RNA processing and silencing. J Cell Sci. 2007;120:1317–1323. doi: 10.1242/jcs.03429. [DOI] [PubMed] [Google Scholar]
- 11.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: At the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 12.Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 13.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly and disassembly in vivo. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lall S, Piano F, Davis RE. Caenorhabditis elegans decapping proteins: Localization and functional analysis of Dcp1, Dcp2 and DcpS during embryogenesis. Mol Biol Cell. 2005;16:5880–5890. doi: 10.1091/mbc.E05-07-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, Lyons GE, et al. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell. 2006;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmeen I, Zacmanidis P, Jesion G, Feldkamp LA. Motion of mitochondria in cultured cells quantified by analysis of digitized images. Biophys J. 1985;48:681–686. doi: 10.1016/S0006-3495(85)83825-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felder S, Kam Z. Human neutrophil motility: Time-dependent three-dimensional shape and granule diffusion. Cell Motil Cytoskeleton. 1994;28:285–302. doi: 10.1002/cm.970280403. [DOI] [PubMed] [Google Scholar]
- 24.Burke NV, Han W, Li D, Takimoto K, Watkins SC, Levitan ES. Neuronal peptide release is limited by secretory granule mobility. Neuron. 1997;19:1095–1102. doi: 10.1016/s0896-6273(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 25.Steyer JA, Horstmann H, Almers W. Transport, docking and exocytosis of single secretory granules in live chromaffin cells. Nature. 1997;388:474–478. doi: 10.1038/41329. [DOI] [PubMed] [Google Scholar]
- 26.Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: Volume, viscosity, diffusion, intracellular surface area. Int Rev Cytol. 2000;192:189–221. doi: 10.1016/s0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- 27.Shav-Tal Y, Singer RH. RNA localization. J Cell Sci. 2005;118:4077–4081. doi: 10.1242/jcs.02543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez de Heredia M, Jansen RP. mRNA localization and the cytoskeleton. Curr Opin Cell Biol. 2004;16:80–85. doi: 10.1016/j.ceb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, et al. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci. 2008;28:7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 32.Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, et al. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178:1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Sweet TJ, Boyer B, Hu W, Baker KE, Coller J. Microtubule disruption stimulates P-body formation. RNA. 2007;13:493–502. doi: 10.1261/rna.355807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.