Abstract
Infectious amyloid forms of the release factor, Sup35, comprise the yeast prion [PSI+]. This protein-based unit of inheritance is an evolutionary capacitor able to release cryptic genetic variation during environmental stress and generate potentially beneficial phenotypes. Genetic data have uncovered a sophisticated proteostasis network that tightly regulates [PSI+] formation, propagation and elimination. Central to this network, is the AAA+ ATPase and protein disaggregase, Hsp104. Shifting the balance of the cytosolic Hsp70:Hsp40 chaperone machinery and associated nucleotide exchange factors also influences the [PSI+] prion cycle. Yet, a precise understanding of how these systems co-operate to directly modulate the protein folding events required for sustainable Sup35 prionogenesis has remained elusive. Here, we spotlight recent advances that begin to clarify this issue. We suggest that the Hsp70:Hsp40 chaperone machinery functions collectively as a rheostat that adjusts Hsp104's basic prion-remodeling activities.
Key words: Sup35, prion, Hsp104, Hsp70, Hsp40, chaperone
The yeast prion [PSI+] arises when Sup35, a release factor, populates self-templating amyloid forms that transmit heritable increases in nonsense suppression.1 This requires N-terminal regions of Sup35's prion domain to switch from an intrinsically disordered state to the cross-β form of amyloid fibers.1 The modest reductions in translation termination fidelity conferred by [PSI+] alter mRNA stability, gene expression and protein function.1–4 Consequently, complex multigenic traits develop that are often (but not always) advantageous.1,3,4 [PSI+] induction increases with increasing environmental stress,5 which has reinforced proposals that [PSI+] is a transient ‘bet-hedging’ adaptation that enhances survival in variable environments by promoting evolvability.1,3–6
A sophisticated proteostasis7 network orchestrates [PSI+] formation, propagation and elimination.8 Central to this network is the hexameric AAA+ ATPase and protein-disaggregase, Hsp104.9 Either too much or too little Hsp104 can eliminate [PSI+].10 To understand this dosage relationship, pure components have been used to define the direct effects of Hsp104 on Sup35 prionogenesis.11,12 The prion nature of various Hsp104-remodeled Sup35 conformers was stringently tested by infecting [psi−] cells.12 At low concentrations, Hsp104 promotes Sup35 prion nucleation and severs Sup35 prions to expose new surfaces for prion growth11–13 (Fig. 1A). At higher concentrations, Hsp104 converts Sup35 prions to SDS-soluble monomeric species and non-infectious amyloid-like material11–15 (Fig. 1A). This ‘amyloid-like’ material is a non-fibrillar aggregated species, that retains SDS-resistance and the ability to bind Thioflavin-T, but has diminished capacity to template the prion conformation.11,12 Similar SDS-soluble and SDS-resistant Sup35 conformers appear to accumulate in [PSI+] cells expressing high levels of Hsp104.16 Moreover, SDS-resistant Sup35 polymers with reduced seeding activity can, in some circumstances, be isolated from [psi−] cells.17 The basic prion-remodeling activities of Hsp104 reconstituted in vitro would seem to explain how Hsp104 dictates [PSI+] inheritance patterns.11,12 Surprisingly, however, these activities did not require Hsp70 and Hsp40,11,12 which assist Hsp104 in renaturing chemically or thermally denatured aggregates.18 Furthermore, several aspects of the cellular milieu are not recreated in this minimal system and Hsp104 is unlikely to operate in isolation in vivo. Genetic data suggest that the Hsp70:Hsp40 chaperone machinery also affect [PSI+] induction, propagation and elimination.8 However, these data might reflect complex pleiotropic effects rather than direct effects on Sup35 folding. Thus, how Hsp104, Hsp70 and Hsp40 collaborate to directly affect the conformational states of Sup35 has remained unclear. Recent advances that combine pure protein biochemistry with prion infection studies have shed light on this issue.19
Figure 1.
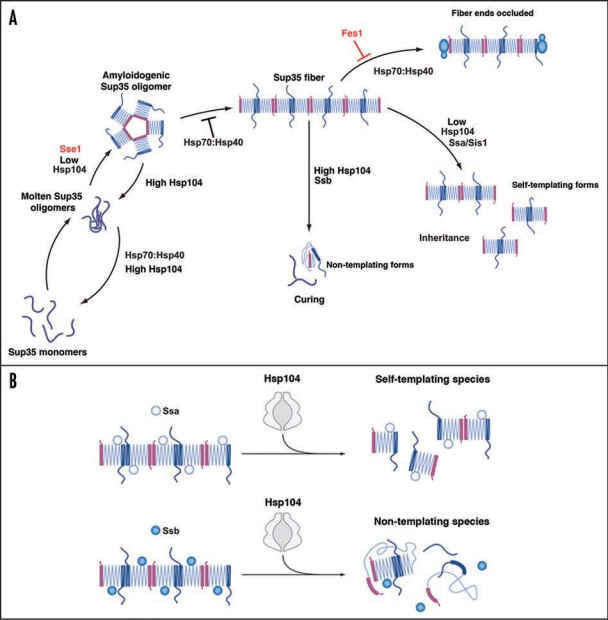
The role of Hsp104, Hsp70 and Hsp40 in Sup35 prionogenesis. (A) Sup35 prions assemble after a lag phase during which a dynamic ensemble of monomeric and molten oligomeric species form. The intermolecular contacts that nucleate prion assembly are likely established within amyloidogenic oligomers. Sup35 fibers are held together by an alternating sequence of Head-to-Head (pink) or Tail-to-Tail (dark blue) intermolecular contacts that are separated by a central core region (light blue) comprised of intramolecular contacts. Once formed, fibers stimulate their own assembly by recruiting and converting monomers at their ends. Various steps are promoted or antagonized by Hsp104, Hsp70, Hsp40 and the NEFs Fes1 and Sse1 as indicated. At high concentrations, Hsp70 and Hsp40 can bind Sup35 prions and occlude prion recognition elements. The C-terminal domain of Sup35 is not depicted for clarity. (B) Incorporation of Ssa1 and Ssb1 into Sup35 prions has distinct effects on subsequent Hsp104 remodeling. Ssa1 incorporation protects fibers against remodeling by Hsp104 to non-infectious conformations, perhaps by maintaining active fiber ends. Ssb1 incorporation promotes the remodeling of fibers to non-infectious conformations, perhaps by promoting inactivation of fiber ends. The majority of Ssb1 is associated with the ribosome.29 However, a considerable fraction is found throughout the cytoplasm,29 which is likely to be able to interact with Sup35 prions.
Reconstructing these events using pure components19 poses a challenge due to the complexity of the cytosolic Hsp70:Hsp40 chaperone network. Two cytosolic Hsp70 subfamilies affect [PSI+]: Ssa and Ssb. Ssa can promote20–24 or antagonize21,25,26 [PSI+], whereas Ssb is a consistent [PSI+] antagonist.22,25,27 These functional differences are conferred by the distinct substrate binding domains of Ssa and Ssb.22 The four members of the Ssa subfamily are found throughout the cytoplasm.28 By contrast, the two Ssb proteins are mostly associated with the ribosome, although a considerable portion can be found throughout the cytoplasm.29 Hsp70s couple their ATPase cycle to rounds of substrate binding and release.30 Substrates are typically bound via extended polypeptide segments that become exposed in non-native forms.30 In the ATP-bound state, substrate binding is dynamic, while in the ADP state substrate is stably bound.30 Yet, basal Hsp70 ATPase activity is too slow to drive chaperone activity.30 Therefore, Hsp70s function with their obligate Hsp40 cochaperones, which via their J-domain accelerate Hsp70 ATPase activity and stabilize substrate binding.30 For Ssa1 two key Hsp40s are Ydj1 and Sis1.31 By contrast, Ssb1 requires a heterodimer of Zuo1, an Hsp40 and Ssz1, an atypical Hsp70.32 Nucleotide exchange factors (NEFs) facilitate exchange of ADP for ATP and promote substrate release.30 Fes1 and Sse1 serve as NEFs for both Ssa1 and Ssb1.33–36 Recently, it became clear that Ssa1/2 are major components of ex vivo Sup35 prion polymers, with ∼one Ssa molecule per two Sup35 molecules. Ssb1, Ydj1 and Sis1 are also found to be physically associated, although in smaller amounts.37
First, the direct effects of the Hsp70:Hsp40 chaperone system on Sup35 prionogenesis were determined.19 Sup35 prions form after a lag phase during which a dynamic ensemble of monomers and molten oligomers form38 (Fig. 1A). Molten oligomers mature into amyloidogenic oligomers that nucleate prion assembly11,12,38 (Fig. 1A). During this process, prion recognition elements within the N-terminal prion domain termed the ‘Head’ and ‘Tail’ are proposed to make homotypic intermolecular contacts. Thus, fibers are held together by an alternating sequence of ‘Head-to-Head’ and ‘Tail-to-Tail’ interactions, which are separated by a central core comprised of intramolecular contacts39–41 (Fig. 1A). Once formed, fibers seed their own assembly by recruiting and converting monomers at their growing ends.38 Hsp70:Hsp40 pairs were potent antagonists of de novo Sup35 prion assembly, particularly the ribosome-associated Ssb1:Zuo1:Ssz1 complex.19 Ydj1 and Sis1 selectively bound Sup35 oligomers and fibers, but not monomers, and facilitated Ssa1 and Ssb1 binding.19 Hsp70:Hsp40 pairs blocked prion nucleation by disassembling molten oligomers and by binding mature oligomers19 (Fig. 1A). Although able to disassemble molten oligomers, Hsp70:Hsp40 could not disassemble preformed Sup35 prions.19 Rather, recruitment of Ssa1 or Ssb1 to Sup35 fibers occluded prion recognition elements and prevented seeded assembly19 (Fig. 1A). These biochemical data help explain several genetic correlates, including why overexpression of Hsp70:Hsp40 can cure certain [PSI+] variants,25,26 and why deletion of Ssb, the most potent antagonist (in combination with Zuo1 and Ssz1) of synthetic Sup35 prionogenesis, increases [PSI+] induction ∼10-fold.27
How do NEFs affect these Hsp70 activities? Curiously, Sse1 directly stimulated nucleation of the prion domain of Sup3524 (Fig. 1A). Indeed, Sse1 facilitates [PSI+] induction even in the absence of [PIN+].24 By contrast, Fes1 exerted no direct effects on Sup35 prionogenesis,19 allowing direct assessment of how nucleotide exchange activity affected Hsp70 activities. Fes1 partially antagonized the inhibitory activities of Hsp70:Hsp40 (Fig. 1A), which provides a direct explanation for why Fes1 deletion destabilizes [PSI+] and Fes1 overexpression promotes [PSI+].21 Fes1 NEF activity promotes substrate release by Ssa1 and Ssb1, which may remove Hsp70 from Sup35 prions and allow conformational replication to proceed. This is consistent with the transient and iterative interactions between Hsp70 and polyglutamine aggregates observed in vivo, where Hsp70 rapidly cycles on and off the aggregate surface.42
In general, the Hsp70:Hsp40 chaperone machinery directly inhibits de novo and templated Sup35 prion assembly. Yet, how does this machinery affect prion remodeling by Hsp104? At low concentrations, Hsp104 accelerates Sup35 prionogenesis.12 This activity was able to override the inhibitory activities of Ssa1:Sis1 or Ssb1:Sis1, but not Ssa1:Ydj1 and Ssb1:Ydj1.19 Thus, opposing Hsp40-dependent chaperone machineries balance the promotion (Sis1) or inhibition (Ydj1) of Sup35 prionogenesis.
Incorporation of Hsp70 and Hsp40 into nascent Sup35 prions made them better substrates for remodeling by high concentrations of Hsp104.19 This supports genetic data that Hsp70 assists prion fragmentation by Hsp104.20,21 Importantly, the consequences of Hsp104-remodeling depended upon the incorporated Hsp70. Ssb1 promoted the elimination of Sup35 prions, whereas Ssa1 inhibited it (Fig. 1A and B). Thus, Ssb1 likely increases the Hsp104 activity that converts Sup35 prions to non-replicating amyloid-like species, perhaps by ensuring inactivation of fiber ends (Fig. 1B). This might involve destabilization of fiber end structure or by sequestration of fiber ends to make them inaccessible (Fig. 1B). By contrast, Ssa1 likely maintains active fiber ends despite remodeling by Hsp104 (Fig. 1B). These differences in Ssa1 and Ssb1 activity likely reflect differences in their interactions with Sup35 prions conferred by their distinct substrate binding domains.22 Furthermore, these biochemical observations help explain why high levels of Ssa1 inhibit curing by overexpression of Hsp104, whereas high levels of Ssb1 promote it.23,27 The protection of self-replicating activity by Ssa1 despite remodeling by Hsp104 helps explain several genetic correlates that Ssa1 can promote [PSI+].23,24,27 The presence of Ssa1/2 as a major component of Sup35 prions in vivo37 might help ensure long-term prion stability. Taken together, these observations suggest that the Hsp70:Hsp40 chaperone machinery functions collectively as a rheostat that finely tunes Hsp104's basic prion-remodeling activities.
Hsp70 and Hsp40 likely help, but are not absolutely required, to present specific polypeptide loops of Sup35 fibers to Hsp104 to promote prion remodeling. Such a role is consistent with a function for Hsp70 and Hsp40 early in protein disaggregation.43 Accumulating evidence suggests that Hsp104 drives protein disaggregation and prion propagation by translocating substrates across its large central cavity after they are fed into its N-terminal entrance.15,43–49 Recent genetic data reinforce the facilitatory role of the Hsp70:Hsp40 machinery in Sup35 prion severing by Hsp104. Depletion of Sis1 leads to gradual increases in polymer size and loss of [PSI+] over ∼60 generations,46,50 suggesting that severing of Sup35 prions by Hsp104 is partially impaired by the absence of Sis1. Similarly, incorporation of Sis1 into Sup35 prions increases their susceptibility to Hsp104-driven remodeling in vitro.19 Intriguingly, depletion of Sis1 causes a rapid loss of [RNQ+] and [URE3] indicating a greater importance of the Hsp70:Hsp40 chaperone machinery for the propagation of these prions.50 Little is known about the direct effects of Hsp104, Hsp70 or Hsp40 on synthetic Rnq1 prionogenesis. However, Hsp104 is able to effectively fragment Ure2 prions in vitro, but is not able to eliminate their self-templating activity.12 Therefore, in vivo, additional factors associated with Ure2 prions may make the Hsp70:Hsp40 system more essential for Hsp104 activity. Unfortunately, in contrast to Sup35 and Rnq1,37,51 little is known about the factors that associate with Ure2 prions in vivo.
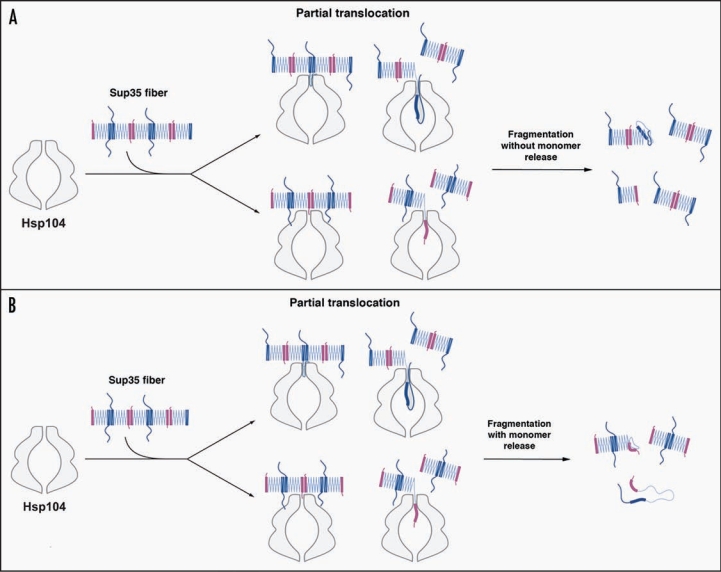
Within the framework of the ‘Head-to-Head’ and ‘Tail-to-Tail’ model of Sup35 prion structure39–41 (Fig. 2A), Hsp104 need only break the ‘Head-to-Head’ or ‘Tail-to-Tail’ intermolecular contact to fragment prions. This might only require partial translocation across the central channel of Hsp104 hexamers. For example, Hsp104 might pull on the extreme N-terminal region of Sup35, which is partially accessible in Sup35 prions,39,52 to melt only the Head-to-Head contact and then release (Fig. 2A). Alternatively, Hsp104 might translocate polypeptide loops just C-terminal to the Tail to forcibly separate only the Tail-to-Tail contact and then release (Fig. 2A). In this way, fragmentation of Sup35 prions at low Hsp104 concentrations may not involve the liberation of Sup35 monomers (Fig. 2A). In either case, the released portion of Sup35 may occasionally fail to fold back into the appropriate β-sheet conformation leading to inactivation of the fiber end (Fig. 2A). This event may be promoted or inhibited by Hsp70 (Fig. 1B). At higher Hsp104 concentrations, Hsp104 hexamers likely co-operate to drive Sup35 fiber disassembly,11,12 which facilitates the release of Sup35 monomers (Fig. 2B) and perhaps more frequent inactivation of fiber ends. Even with Sup35 monomer release, translocation might be partial, such that the Head-to-Head and Tail-to-Tail contacts of individual Sup35 molecules are broken, but the large C-terminal GTPase domain of Sup35 is not translocated across the channel (Fig. 2B). The large central cavity of Hsp104 might facilitate such an activity.15 Indeed, this mode of partial threading is sufficient for Hsp104 to promote the disaggregation of denatured aggregates of model substrates.53 In this manner, Hsp104 might release Sup35 with a functional C-terminal GTPase domain and rapidly reverse the [PSI+] phenotype.
Figure 2.
Possible modes of Sup35 prion fragmentation by Hsp104. (A, B) Partial translocation might disrupt Head-to-Head (pink) or Tail-to-Tail (blue) contacts without (A) or with (B) monomer release. Partial translocation might occasionally leave some fiber ends inactivated. The C-terminal domain of Sup35 is not depicted for clarity.
Our model outlined above emphasizes the importance of the large central cavity of Hsp104, which has been revealed by cryo-EM reconstructions.15 Several interesting Hsp104 mutants (L462R, P557L and D704N) have been described that confer defects in [PSI+] propagation, but not thermotolerance.54 These mutations were all suggested to be close to the ‘lateral channels’ of Hsp104.54 However, these assignments were based on cryo-EM reconstructions and domain fitting of tClpB hexamers,55 which appear unlikely to accurately reflect Hsp104 hexamer structure.15 Indeed, in the cryo-EM reconstruction and domain fitting of Hsp104, L462 and D704 do not appear to be so close to the lateral channels.15 In this model, L462 resides on helix L2 of Hsp104's coiled-coil domain in proximity to nucleotide in nucleotide binding domain (NBD) 1 and D704 is located on NBD2 at the contact interface with the coiled-coil of the adjacent protomer.15 P557 is quite close to the lateral channels but faces the central cavity on a short linker between the NBDs.15 Hence, the importance of the lateral channels and how mutations at these positions selectively affect Hsp104 prion-remodeling activity remains unclear.
We must also note that the precise atomic structure of Sup35 prions remains unknown and several distinct models of Sup35 prion structure have been proposed.39,40,52,56–58 Regardless of the exact prion structure, fragmentation by Hsp104 requires separation of intermolecular prion contacts. However, the very nature of these intermolecular contacts can also vary as Sup35 can assemble into multiple structurally distinct amyloid forms or ‘strains’, which encode distinct [PSI+] variants.1,39,59–61 [PSI+] variants range from ‘weak’ to ‘strong’ as the extent of Sup35 aggregation and inactivation increases.1,59 In vitro, Sup35's prion domain forms fibers at 4°C that encode strong [PSI+] variants, whereas fibers formed at 25°C encode weak [PSI+] variants.39,60,61 These distinct prion conformations are distinguished by the amount of primary sequence that is sequestered in cross-β structure.39,52 More specifically, the central core is longer,39 the position of the Tail-to-Tail contact is more C-terminal39 and residues N-terminal to the Head are more structured in 25°C fibers than in 4°C fibers.39,52 This renders 4°C fibers more fragile and sensitive to detergents.60,61 Fragmentation of these distinct prion conformations likely makes distinct demands on Hsp104. For example, in 4°C fibers, the shorter stretch of cross-β structure is likely to increase the accessibility of intermolecular contacts and require less ATP hydrolysis by Hsp104 to unfold. By contrast, the intermolecular contacts of 25°C fibers are likely to be less accessible and the longer stretch of cross-β structure likely requires more ATP hydrolysis by Hsp104 to unfold. Thus, prion fragmentation by Hsp104 with or without monomer release (Fig. 2A and B) is likely to be more difficult for Sup35 prion conformations that encode weak [PSI+] and perhaps leads to more frequent fiber end inactivation. This is in keeping with weak [PSI+] variants possessing fewer, longer Sup35 polymers per cell.16,61 The sensitivity of [PSI+] variants to curing by excess Hsp104 is inversely proportional to their strength.62 That is, weak [PSI+] variants are more readily cured by Hsp104 overexpression than strong [PSI+] variants.62 We suspect that excess Hsp104 remodels prion conformers that encode weak [PSI+] in a manner that more rapidly generates various non-templating species (Fig. 1A and B). Future biochemical studies will address the foregoing possibilities, as well as how the Hsp70:Hsp40 machinery affects different Sup35 prion strains.
It is also important to consider that the protein-remodeling activities of Hsp104 might have downstream consequences for prions in vivo that are not reconstituted in vitro. Several recent studies emphasize the importance of spatial quality control of protein aggregates in yeast.63–66 For example, Hsp104 appears to be intimately involved in the retention of carbonylated, aggregated proteins in the mother cell, which are associated with aging.63 Moreover, yeast lacking Hsp104 have reduced longevity and fail to retain carbonylated proteins in the mother.63 Overexpressed Sup35, Ure2 and Rnq1 form aggregates that are found throughout the cytoplasm, but a subset are partitioned to a discrete perivacuolar compartment.64,65 This partitioning of prion aggregates may play an important and currently unappreciated role in yeast prion protein regulation.67 For example, the cell may be unable to recover Sup35 from non-replicating amyloid-like aggregates generated by excessive remodeling by Hsp104. These terminally aggregated conformers might then accumulate at a perivacuolar site65 and be targeted for retention and possibly autophagic degradation.
Finally, Hsp104 rapidly remodels amyloid and preamyloid oligomers.11,12 Curiously, there is no known orthologue or analogue of Hsp104 in metazoa.9 Therefore, reintroduction of Hsp104 might have therapeutic utility for several protein-misfolding disorders.9 Indeed, expression of Hsp104 has ameliorative effects in rodent models of Huntington68,69 and Parkinson disease.70 Moreover, Hsp104 directly disassembles α-synuclein fibers and oligomers connected with Parkinson disease.70,71 Hsp104 also directly antagonizes Aβ42 amyloidogenesis, which is connected with Alzheimer's disease.72 An accurate understanding of Hsp104 structure and function might enable enhancements that potentiate its activity against the specific protein-misfolding events that distinguish several fatal neurodegenerative disorders.9
Acknowledgements
We thank Mary Leonard for expert graphical assistance. This work was supported by an NIH Director's New Innovator Award (DP2OD002177); an American Heart Association Scientist Development Grant; and University of Pennsylvania Institute on Aging and Alzheimer's Disease Core Center pilots (J.S.).
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/7952
References
- 1.Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Gen. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 2.Namy O, Galopier A, Martini C, Matsufuji S, Fabret C, Rousset JP. Epigenetic control of polyamines by the prion [PSI+] Nat Cell Biol. 2008;10:1069–1075. doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- 3.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 4.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 5.Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biology. 2008;6:2605–2613. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King OD, Masel J. The evolution of bet-hedging adaptations to rare scenarios. Theor Popul Biol. 2007;72:560–575. doi: 10.1016/j.tpb.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 8.Rikhvanov EG, Romanova NV, Chernoff YO. Chaperone effects on prion and nonprion aggregates. Prion. 2007;1:217–222. doi: 10.4161/pri.1.4.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shorter J. Hsp104: A weapon to combat diverse neurodegenerative disorders. Neurosignals. 2008;16:63–74. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 11.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 12.Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narayanan S, Walter S, Reif B. Yeast prion-protein, sup35, fibril formation proceeds by addition and substraction of oligomers. Chembiochem. 2006;7:757–765. doi: 10.1002/cbic.200500382. [DOI] [PubMed] [Google Scholar]
- 15.Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, Saibil HR. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 17.Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, Ter-Avanesyan MD. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–8812. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- 18.Glover JR, Lindquist S. Hsp104, Hsp70 and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 19.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung G, Jones G, Wegrzyn RD, Masison DC. A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics. 2000;156:559–570. doi: 10.1093/genetics/156.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones G, Song Y, Chung S, Masison DC. Propagation of Saccharomyces cerevisiae [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol Cell Biol. 2004;24:3928–3937. doi: 10.1128/MCB.24.9.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, et al. Hsp70 chaperones as modulators of prion life cycle: Novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newnam GP, Wegrzyn RD, Lindquist SL, Chernoff YO. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadlish H, Rampelt H, Shorter J, Wegrzyn RD, Andreasson C, Lindquist S, Bukau B. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS ONE. 2008;3:1763. doi: 10.1371/journal.pone.0001763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 26.Kryndushkin DS, Smirnov VN, Ter-Avanesyan MD, Kushnirov VV. Increased expression of Hsp40 chaperones, transcriptional factors and ribosomal protein Rpp0 can cure yeast prions. J Biol Chem. 2002;277:23702–23708. doi: 10.1074/jbc.M111547200. [DOI] [PubMed] [Google Scholar]
- 27.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone ssb in formation, stability and toxicity of the [PSI+] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 29.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 30.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 31.Lu Z, Cyr DM. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem. 1998;273:27824–27830. doi: 10.1074/jbc.273.43.27824. [DOI] [PubMed] [Google Scholar]
- 32.Huang P, Gautschi M, Walter W, Rospert S, Craig EA. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- 33.Dragovic Z, Shomura Y, Tzvetkov N, Hartl FU, Bracher A. Fes1p acts as a nucleotide exchange factor for the ribosome-associated molecular chaperone Ssb1p. Biol Chem. 2006;387:1593–1600. doi: 10.1515/BC.2006.198. [DOI] [PubMed] [Google Scholar]
- 34.Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol Cell Biol. 2002;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] Infection Is Transmitted by Sup35 Polymers within [PSI+] Aggregates with Heterogeneous Protein Composition. Mol Biol Cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 39.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447:556–561. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Duennwald ML, Roberts BE, Rozeboom LM, Zhang YL, Steele AD, et al. Direct and selective elimination of specific prions and amyloids by 4,5-dianilinophthalimide and analogs. Proc Natl Acad Sci USA. 2008;105:7159–7164. doi: 10.1073/pnas.0801934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S, Nollen EA, Kitagawa K, Bindokas VP, Morimoto RI. Polyglutamine protein aggregates are dynamic. Nat Cell Biol. 2002;4:826–831. doi: 10.1038/ncb863. [DOI] [PubMed] [Google Scholar]
- 43.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 44.Lum R, Niggemann M, Glover JR. Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J Biol Chem. 2008;283:30139–30150. doi: 10.1074/jbc.M804849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lum R, Tkach JM, Vierling E, Glover JR. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem. 2004;279:29139–29146. doi: 10.1074/jbc.M403777200. [DOI] [PubMed] [Google Scholar]
- 46.Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shorter J, Lindquist S. Navigating the ClpB channel to solution. Nat Struct Mol Biol. 2005;12:4–6. doi: 10.1038/nsmb0105-4. [DOI] [PubMed] [Google Scholar]
- 49.Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 50.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci USA. 2008;105:16596–16601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 53.Haslberger T, Zdanowicz A, Brand I, Kirstein J, Turgay K, Mogk A, et al. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat Struct Mol Biol. 2008;15:641–650. doi: 10.1038/nsmb.1425. [DOI] [PubMed] [Google Scholar]
- 54.Kurahashi H, Nakamura Y. Channel mutations in Hsp104 hexamer distinctively affect thermotolerance and prion-specific propagation. Mol Microbiol. 2007;63:1669–1683. doi: 10.1111/j.1365-2958.2007.05629.x. [DOI] [PubMed] [Google Scholar]
- 55.Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, et al. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 56.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 58.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI+] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 62.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ganusova EE, Ozolins LN, Bhagat S, Newnam GP, Wegrzyn RD, Sherman MY, et al. Modulation of prion formation, aggregation and toxicity by the actin cytoskeleton in yeast. Mol Cell Biol. 2006;26:617–629. doi: 10.1128/MCB.26.2.617-629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Meriin AB, Zaarur N, Romanova NV, Chernoff YO, Costello CE, et al. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J. 2008 doi: 10.1096/fj.08-117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cox BS, Byrne LJ, Tuite MF. Prion stability. Prion. 2007;1:170–178. doi: 10.4161/pri.1.3.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vacher C, Garcia-Oroz L, Rubinsztein DC. Overexpression of yeast hsp104 reduces polyglutamine aggregation and prolongs survival of a transgenic mouse model of Huntington's disease. Hum Mol Gen. 2005;14:3425–3433. doi: 10.1093/hmg/ddi372. [DOI] [PubMed] [Google Scholar]
- 69.Perrin V, Regulier E, Abbas-Terki T, Hassig R, Brouillet E, Aebischer P, et al. Neuroprotection by Hsp104 and Hsp27 in lentiviral-based rat models of Huntington's disease. Mol Ther. 2007;15:903–911. doi: 10.1038/mt.sj.6300141. [DOI] [PubMed] [Google Scholar]
- 70.Lo Bianco C, Shorter J, Regulier E, Lashuel H, Iwatsubo T, Lindquist S, et al. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 2008;118:3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kong B, Chae YK, Lee K. Regulation of in vitro fibril formation of synuclein mutant proteins by Hsp104p. Protein Pept Lett. 2003;10:491–495. doi: 10.2174/0929866033478717. [DOI] [PubMed] [Google Scholar]
- 72.Arimon M, Grimminger V, Sanz F, Lashuel HA. Hsp104 targets multiple intermediates on the amyloid pathway and suppresses the seeding capacity of Abeta fibrils and protofibrils. J Mol Biol. 2008;384:1157–1173. doi: 10.1016/j.jmb.2008.09.063. [DOI] [PubMed] [Google Scholar]