Abstract
Insect silks are secreted from diverse gland types; this chapter deals with the silks produced by labial glands of Holometabola (insects with pupa in their life cycle). Labial silk glands are composed of a few tens or hundreds of large polyploid cells that secrete polymerizing proteins which are stored in the gland lumen as a semi-liquid gel. Polymerization is based on weak molecular interactions between repetitive amino acid motifs present in one or more silk proteins; cross-linking by disulfide bonds may be important in the silks spun under water. The mechanism of long-term storage of the silk dope inside the glands and its conversion into the silk fiber during spinning is not fully understood. The conversion occurs within seconds at ambient temperature and pressure, under minimal drawing force and in some cases under water. The silk filament is largely built of proteins called fibroins and in Lepidoptera and Trichoptera coated by glue-type proteins known as sericins. Silks often contain small amounts of additional proteins of poorly known function. The silk components controlling dope storage and filament formation seem to be conserved at the level of orders, while the nature of polymerizing motifs in the fibroins, which determine the physical properties of silk, differ at the level of family and even genus. Most silks are based on fibroin β-sheets interrupted with other structures such as α-helices but the silk proteins of certain sawflies have predominantly a collagen-like or polyglycine II arrangement and the silks of social Hymenoptera are formed from proteins in a coiled coil arrangement.
Key words: silk, proteinaceous polymers, α-helices, β-sheets, polyglycines, coiled coils, collagen
Introduction
Silks are (1) ectodermal secretions that (2) are stored as a hydrated jelly within cells or, more often, in multicellular cavities; (3) the gels polymerise into water-insoluble filaments during passage to the external environment. Materials that meet this definition occur in all terrestrial arthropod subphyla: Chelicerata (in three orders of Arachnida, i.e., spidermites, pseudoscorpions and spiders), Myriapoda (in Symphyla, Pauropoda and primitive millipedes, absent in the centipedes) and Hexapoda (Fig. 1). Silk is always secreted by specialised ectodermal cells that make up a silk gland.1 Rudall and Kenchington distinguished eight structural silk categories based on X-ray diffraction data and suggested that all silk proteins are phylogenetically homologous.2 This view is no longer tenable. The diversity of silk-producing taxa, their silk glands and the silk composition suggest that silk has evolved independently a number of times as a product of the cuticle-secreting cells. The cuticle is composed of chitin and a blend of proteins, many of which contain repeats of short amino acid motifs;3 several of such proteins could be the precursors of silks.
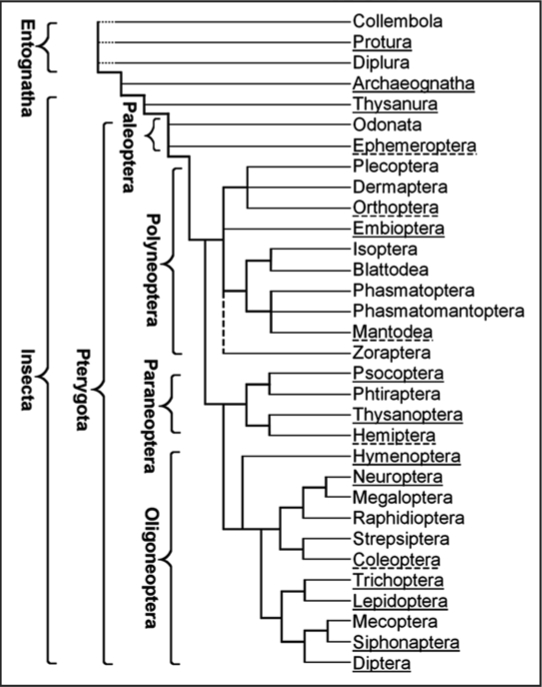
Figure 1.
Approximate evolutionary relationships and the distribution of silk production in the orders of Hexapoda. Silk production is common in orders underlined with a solid line and exceptional (limited to a small number of species) in those underlined with a dashed line.
Insect silks are produced by a variety of dermal glands, Malpighian tubuli, colleterial sex glands, the gut and the labial glands (Table 1). The latter typically produce saliva but half of each gland in the Psocoptera and the entire glands in the larvae of many Holometabola, are specialized for the production of silk. Labial silk glands develop during embryogenesis as ectodermal tubular invaginations composed of several tens to a few hundred cells. They grow by polyploidization, extend into the abdomen and often reach a large size, second only to the gut and the fat body. Most information on insect silks concerns the secretions of labial glands that are the subject of this review; little is known on the other silk types.1,4 Holometabola are monophyletic (Fig. 1) but the over-all composition of the silky secretions from their labial glands has greatly diversified (Table 2). The mechanism of silk secretion, silk composition and the silk structure has been examined most systematically in Lepidoptera, being stimulated by the interests of commercial sericulture. Since lepidopteran silk is often used as a prototype of the other silks, it is appropriate to begin this chapter with a description of the silk production in caterpillars.
Table 1.
Silk production in different hexapod orders (developmental stage, type of glands and silk function)
| Protura | ?? | Adults (Larvae?) | Abdominal glands | ?? |
| Archaeognatha | All species? | Adults | Dermal glands (cerci) | Sperm support |
| Thysanura | All species? | Adults | Dermal glands (cerci) | Sperm support |
| Ephemeroptera | One family | Larvae | Malpighian tubuli | ?? |
| Odonata | One species | Adults | unknown | Egg coverings |
| Orthoptera69 | One family | Adults and larvae | maxillary glands | Nests and burrows |
| Embioptera | All species | Larvae and adults | Dermal glands (tarsi) | Tunnels and egg coverings |
| Mantodea | All species | Adult females | Colleterial glands | Ootheca |
| Psocoptera | Some species | Adults | Labial glands | Egg covering and retreats |
| Thysanoptera70 | Some species | Adults and Larvae | Anal glands in adults | Domiciles or cocoons |
| Hemiptera71 | One species | Unknown | unknown | Shelter |
| Hymenoptera | Many species | Grown larvae | Labial glands | Cocoon |
| Neuroptera | Some species | Grown larvae | Malpighian tubuli | Cocoon |
| All species | Adult females | Colleterial glands | Egg stalk or cover | |
| Coleoptera | Some families | Grown larvae | Malpighian tubuli or Peritrophic membrane | Cocoon, underwater tunnels |
| Few hydrophilidae | Adult females | Colleterial glands | Floating rafts for eggs | |
| Trichoptera | All species | Larvae | Labial glands | Diverse |
| Lepidoptera | Most species | Larvae | Labial glands | Diverse |
| Siphonaptera | All species? | Grown larvae | Labial glands | Nests? and cocoon |
| Diptera | One family | Larvae | Labial glands | Prey capture |
| Few families | Larvae | Labial glands | Tunnels | |
| One family | Tarsal glands | Dermal glands (tarsi) | Prey package |
Table 2.
Dominant amino acids in the major silk proteins produced from the labial glands of holometabolous insects
| Hymenoptera | Diptera-Nematocera | Trichoptera | Lepidoptera | ||||||||
| Cg | Vs | Am | Cte BR1 | Cth 220 | Ha | Ld | Ye | Gm | Bm | Ap | |
| Gly | 21.7% | 24.6% | 27.7% | 31.6% | 45.9% | 27.3% | |||||
| Ala | 34.6% | 32.7% | 26.3% | 23.8% | 30.2% | 43.1% | |||||
| Ser | 32.6% | 22.7% | 21.4% | 17.8% | 17.3% | 17.8% | 18.1% | ||||
| Lle, Leu, Val | 15.1% | 20.5% | 21.5% | 16.0% | |||||||
| Arg, His, Lys | 39.0% | 16.7% | |||||||||
| Asn | 38.4% | ||||||||||
| Cys | 17.5% |
Analysis is based on 254 residues from Cotesia glomerata (Cg),61 1652 (four full length proteins) from Vespa simillima (Vs)67 and 1243 (four full length proteins) from Apis mellifera (Ap)66 silk proteins. Silk proteins BR1 (210 residues)72 and sp220 (full sequence, 1704 residues)73 were analysed in Chironomus tentans (Cte) and Ch. thummi (Cth), respectively. The composition of H-fibroins is presented in the caddisflies Hydropsyche angustipennis (Ha, 1042 residues) and Limnephilus decipiens (Ld, 516 residues),44 and in the moths Yponomeuta evonymella (Ye, 955 residues),26 Galleria mellonella (Gm, 1277 residues),38 Bombyx mori (full sequence of 5263 residues),35 and Antheraea pernyi (Ap, full sequence of 2639 residues).28
The Silks of Caterpillars (Larvae of Lepidoptera)
Spinning from the labial glands of larvae probably evolved in the ancestor of the Lepidoptera and the Trichoptera, more than 250 million years ago. Caterpillars of some species spin virtually continuously and live in silky tubes or domiciles; other species produce just a small pad or a girdle that provides support during moulting. Many species spin durable cocoons in which the larvae pupate. Silk composition has been analysed in some detail in the suprafamilies Yponomeutoidea, Pyraloidea and Bombycoidea. The conclusions are probably valid for the entire clade Ditrysia that harbours 98% of about 160,000 described lepidopteran species.5
The silk fiber of Ditrysia is a highly organised structure composed of several proteins that are derived from the posterior and middle silk gland sections (PSG and MSG, respectively). The fiber consists of two filaments (one from each gland) that are polymers of a large protein called heavy chain fibroin (H-fibroin, 350–500 kDa in different species). It is produced in the PSG along with two other proteins, light chain fibroin (L-fibroin, cca 25 kDa) and P25 glycoprotein (also called fibrohexamerin) that occurs in two forms due to various levels of glycosylation (27–31 kDa). These three proteins are assembled into elementary units in the endoplasmic reticulum,6 form microfibers in the Golgi vacuoles and are released as secretory granules into the gland lumen,7 where they accumulate as a highly concentrated gel. The gel moves into the MSG and becomes enveloped by several layers of sericins. Major sericins of the domestic silkworm, Bombyx mori, include a 150 kDa protein produced in the most distal part of MSG, a 400 kDa protein from the central and a 250 kDa protein from the proximal MSG region.8 Silk proteins polymerise into a fiber during spinning when they pass through the anterior silk gland section and the spinneret. Filament polymerisation is based on cross-linking of aligned and closely packed H-fibroin molecules. This process apparently requires the withdrawal of water and application of a shearing force. Sericins polymerise with a delay: inner sericins (from the distal MSG) seal the filament doublet into a single fiber and outer sericins glue the fibers to substrates or to one another during cocoon construction.
Commercial silks are products of the domestic silkworm Bombyx mori, several silkmoth species (most of them from the genus Antheraea) and a few other moths whose larvae spin large and closed cocoons. To release the fiber, cocoons are soaked in hot and slightly alkaline water that dissolves the outer sericin layer. Several fibers with a sticky surface are reeled together into a raw silk thread. Measurements of the physical properties and structural silk analyses by X-ray diffraction and other techniques are typically done on fibers liberated from the cocoons in hot water bath; it is assumed that the established values characterise the filaments, i.e., primarily the H-chain fibroin. Sericins and additional,9,10 minor silk components are not considered.
At least six and as many as 15 sericin-type proteins were extracted from the silk of B. mori.11,12 Some of them may be based on identical peptides but differ in the degree of glycosylation. B. mori may harbour up to five sericin genes13 of which ser1 and ser2 have been identified. The ser1 gene of nine exons is expressed in the distal and central MSG and yields 4–5 mRNAs by differential splicing.14,15 A large central exon encodes 60 copies of a 38 amino acid repeat.16 Recombinant proteins based on a common version of this repeat (SRT SGG TST YGY SSS HRG GSV SST GSS SNT DSS TKN AG) were showed to self-assemble into β-sheets and crystallites.17 Based on this observation, filament sealing into a fiber may be attributed to hydrogen bonding between serine residues of the H-fibroin and serine residues of the inner-layer sericins. Recent unpublished analysis of ser2, which is spliced into two mRNAs,18,19 disclosed 11 exons, two of which encode multiple copies of the repeat ALTEKAKPNDRSPSHKDT. The secondary structure and the manner of presumed Ser2 polymerisation are unknown. It is not excluded that the Ser2 proteins actually hinder polymerisation and hold the cocoon fibers together by electrostatic forces that are destroyed by cooking in the alkaline environment. The ser3 gene of three exons incodes repeats composed of an 86- and an 8-amino acids motif, respectively.75 Genes similar to ser1 and ser2 in the overall structure and the splicing patterns, but different in repetitive sequences, were found in the wax moth Galleria mellonella of Pyraloidea.20 Sericins have not been sufficiently examined in any other species.
The investigations on B. mori showed that H-fibroin, L-fibroin and P25 assemble in the endoplasmic reticulum in a ratio of 6:6:1 into elementary secretory units.21 H-fibroin interacts with the two other proteins by its nonrepetitive N- and C-termini. These terminal sequences are conserved across Lepidoptera and Trichoptera (Fig. 2). The N-terminus harbours an amphiphilic proline-flanked region of 35 residues and the C-terminal 26–27 residues include three Cys (only one in the Saturniidae family) in conserved positions. The redox state of the endoplasmic reticulum22 favours formation of disulfide bonds: four internal in P25, one within L-fibroin, one within H-fibroin and one linking Cys170 of the L-fibroin to Cys−22 in the H-fibroin C-terminus.23 Disulfide linkage between L-fibroin and H-fibroin is indispensable for the secretion of both proteins.24 Non-covalent interaction of P25 with the H-fibroin N-terminus facilitates transport and secretion of the highly insoluble H-fibroin/L-fibroin heterodimers.25 At the same time, L-fibroin plays a protective role in P25 glycosylation—full deglycosylation causes disintegration of the elementary secretory unit.21 Only a portion of the 30 kDa P25 and none of the less glycosylated 27 kDa form of P25 is present in the elementary secretory units,6 but both forms occur in the spun-out silk. It is therefore not excluded that filament formation is associated with a partial deglycosylation or other changes in P25.
Figure 2.
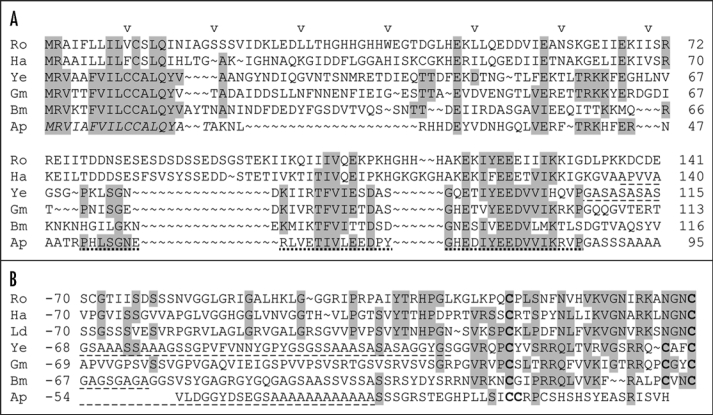
H-fibroin nonrepetitive C-terminus (A) numbered from the translation-initiating Met (the signal sequence is shown in italics in Ap and N-terminus (B) numbered in reverse from the last amino acid residue in the caddisflies) Rhyacophila obliterata (Ro, unpublished), Hydropsyche angustipennis (Ha)44 and Limnephilus decipiens (Ld)44 and in the moths Yponomeuta evonymella (Ye),26 Galleria mellonella (Gm),38 Bombyx mori (Bm)35 and Antheraea pernyi (Ap).28 Small parts of the repetitive region are included (dash-underlined). Residues conserved at least in three species are highlighted grey, Cys residues in the secreted protein are printed boldface. A region conserved in the N-terminus of lepidopteran H-fibroins and in a very degenerate form in the trichopteran H-fibroins, is dot-underlined.
The conservation of L-fibroin, P25 and the nonrepetitive H-fibroin ends (Fig. 2) in the basal superfamily Yponomeutoidea,26 suggests strongly that their interaction represents a plesiomorphic mechanism of the silk secretion and gel/filament conversion in Ditrysia. It was proposed that silk filament construction from the three protein types had been conserved because the hydrophobic nature of H-fibroin requires association with the L-fibroin and P25.27 However, the H-fibroin of Yponomeuta evonymella is amphiphilic.26 A different mechanism of the gel/filament conversion evolved in the family Saturniidae (Bombycoidea) that possesses relatively small H-fibroins (200–250 kDa) with alternating hydrophilic and hydrophobic motifs in their repeats.28,29 This amphiphilicity probably allows gel formation and the gel/filament transition without the L-fibroin and the P25 that have been lost in this family.30,31
Filament polymerisation rests on weak molecular interactions between repetitive motifs that occupy about 95% of the H-fibroin length (Fig. 3). X-ray diffraction studies of lepidopteran silks revealed the presence of β-sheets whereby the carbonyl oxygen and amide nitrogen of adjacent peptide chains are linked by hydrogen bridges. Pleated β-sheets are stacked into crystallites held by interactions between amino acid side chains. From the crystallite dimensions it was predicted that they are largely based on Gly and Ala in B. mori32 and on Ala in Antheraea pernyi.33 This was later confirmed when the protein sequences became available. Most of the B. mori H-fibroin is built from the motif GAGAGS.34 Strings of GAGAGS hexamers (S is occasionally replaced with Y or V) followed by GAAS or a similar tetrapeptide form a second tier repeat and two to six such repeats followed by a highly conserved sequence of 43 residues consisting of a chain of GA dipeptides followed by charged residues (Fig. 3) make up a third order iteration.35 The 43 residue sequence functions as spacers breaking the repetitive region into 12 large modules. The repetitive H-fibroin region in A. pernyi also contains 12 modules, each consisting of 5–8 repeats. The repeats fall into four categories (Fig. 3) that are all composed of a relatively hydrophilic part (11–22 residues) and a hydrophobic string of 12–14 (exceptionally 5 and 15) alanines.28 We wish to emphasize that the number or repeats per module varies; the same is true for the number of motifs concatenated in the repeats of B. mori. Such heterogeneity, occasional residue replacements and the intercalation of spacers separating the repeats probably prevent excessive crystallization that would occur with long strings of the precisely registered motifs.36
Figure 3.
H-fibroin repeats in the moths Y. evonymella,26 G. mellonella,38 B. mori35 and A. pernyi28 and the caddisflies H. angustipennis and L. decipiens.44 Several motifs are recognised: Ser and/or Ala rich, may alternate with Gly (dash-underlined), bulky hydrophobic central residues flanked with charged residues (highlighted grey), GlyGlyXxx triplets (dot-underlined) and motifs containing Pro (for example GPGLV) or Trp surrounded by charged residues (solid-underlined).
Bombyx and Antheraea represent families Bombycidae and Saturniidae that are characterised by low silk production in early larval instars and high production for the construction of extensive cocoons after the termination of feeding. During cocoon construction, up to 20% of the body biomass is converted to silk. Since sufficient endurance and strength are the major functional demands put on the cocoon silk, the use of simple crystalline motifs GAGAGS or poly(Ala) in the fibroin sequence is ideal. It is likely that these motifs are the products of selection for a high rate of silk synthesis at limited nutrient supply. Different selection forces operated in Pyraloidea and Yponomeutoidea that spin elastic hides during most of their larval life and eventually also construct cocoons for pupation. Their silk must be multifunctional and this is probably why their H-fibroins contain a variety of motifs. This variety presumably provides required combination of physical silk properties. Some motifs contain metabolically expensive amino acids. They are obtained from the food (most spinning is done during the feeding period) and from the old silk that is partly consumed and thereby being recycled.
Crystallites formed by stacked β-sheets were detected in the silks of all examined caterpillars37 but differences in periodicity indicated that they are made by different amino acids. Analysis of H-fibroins in the representatives of Pyraloidea38,39 and Yponomeutoidea26 revealed that their repeats are formed by arrays of diverse motifs; motif concatenations are short and rare (Fig. 3). The Ala-rich motifs, which often consist of the strings of Ala or Ser, are likely to form crystallites; the replacement of Ala by Ser does not disturb crystallinity.40 The formation of β-sheets and crystallites is also likely for the motifs in which Gly alternates with Ser or another residue, i.e., GXGX or GXXG. On the other hand, the GGX motif probably yields a spiral conformation.41 Little is known about the conformation of the motifs that occur in the H-fibroin repeats of Pyraloidea and Yponomeutoidea (Fig. 3). These “amphiphilic” motifs consist of three hydrophobic amino acid residues flanked on each side by 2–4 polar residues. Motifs of this kind also occur in the nonrepetitive H-fibroin N-terminus in all Lepidoptera and in a more degenerate form in Trichoptera (Fig. 2). When present in the repeats, the motif is imbedded in a highly crystalline region and probably restrains the formation and size of the crystallites, thereby affecting fiber stiffness. Frequent association of this motif with Pro contributes to the idea that they function to disturb β-sheet formation.
The H-fibroins of Yponomeutoidea and Pyraloidea contain several repeat types (Fig. 3). Repeat composition from simple motifs as well as repeat arrangement in higher order modules is species specific. For example, G. mellonella H-fibroin includes highly conserved repeats of 63 amino acid residues (A), 43 residues (B1) and 18 residues (B2).38 The repeat assembly AB1AB1AB1AB2AB2(AB2) makes up a module and 12 such modules constitute the major part of the H-fibroin. High repeat homogeneity is probably essential for precise alignment of specific motifs that occupy precise but widely distributed regions of the repeats. The homogeneity is extreme in silks of high functional demands, such as in G. mellonella. Larvae of this species live in beehives and their survival depends on continuous spinning of tight tubes in which they hide. The tubes are elastic to allow the larva to turn around. On the other hand, stiffness and close fiber packing into a protective wall are needed in the cocoons that are spun at the end of larval development. The silk of G. mellonella probably meets all these requirements due to a proper blend of motifs and their precise alignment in the highly conserved repeats. Related species that are less dependent on the silk performance produce H-fibroins with less conserved repeats. In the Mediterranean flour moths, Ephestia kuehniella, the regular repeat arrangement is disturbed primarily in the central part of the repetitive region and in the Indian meal moth, Plodia interpunctella, the repeats seem to be erratic throughout the region.39
Caddisflies (Trichoptera)
The silk gland morphology and the silk composition in Trichoptera are similar to those of Lepidoptera. Histological evidence42,43 and our unpublished cDNA sequences demonstrated the presence of sericin-like materials but none have yet been characterized. PSG proteins of the silk filament have been analysed in all three trichopteran suborders and found to be similar, in spite of differences in the silk use. The larvae of Hydropsyche angustipennis (suborder Annulipalpia, family Hydropsychidae) spin catching nets and hiding tubes and, at the end of larval development, construct small domes from the sand grains. The larvae of Limnephilus decipiens (Integripalpia, Limnephilidae) use silk to stick together pieces of plants and other materials (depending on the species) into protective cases; at the end of larval development they close the cases and pad them with silk lining. The larvae of Rhyacophila obliterata (Spicipalpia, Rhyacophilidae) use silk only before pupation to spin parchment-like cocoons. The silk of all three species contains homologues of lepidopteran H-fibroin and L-fibroin but not of P25.44 Terminal regions of the H-fibroins and the entire L-fibroin sequences contain conserved spacing of residues with specific properties (hydrophobicity, charge), including the cysteines that link H-fibroin and L-fibroin in the silkworm silk. The structures of both the H-fibroin and L-fibroin genes are also similar to those of Lepidoptera, suggesting that the H-fibroin/L-fibroin interaction evolved in the ancestor of Trichoptera and Lepidoptera. P25 does not occur in Trichoptera but it is uncertain whether it has been secondarily lost in this order or if it represents an innovation specific to Lepidoptera-Ditrysia.
H-fibroin repeats in all analysed trichopteran H-fibroins are blends of several motifs that are rarely reiterated in string-like fashion (Fig. 3). Motif diversification is less pronounced than in Lepidoptera; the patterns of motif arrangement in the repeats, rather than differences in the motifs characterize caddisfly suborders. Existence of higher-order modules remains to be proven. The Ser-rich and Gly-rich motifs and an “amphiphilic” motif (2–3 hydrophobic residues flanked by two hydrophilic ones or a proline) resemble certain lepidopteran H-fibroins. The strings of SX dipeptides apparently form β-sheets and crystallites based on hydrogen bonding via polar zipper interactions,45 whereas some of the Gly-rich strings, such as GGLGGLGH are more likely to have a spiral conformation. The GPGXX motif, which probably makes a β-spiral and confers filament elasticity,46 occurs in the H-fibroin of H. angustipennis whereas species from other suborders contain Pro in other arrangements. Caddisfly H-fibroins do not contain the typical lepidopteran motifs GAGA and poly(Ala) and their Ala content is strikingly low with Ala virtually absent in the repeats of L. decipiens. Unique to caddis-flies is a highly symmetrical motif with a central Trp (R. obliterata: GPGGRRGWGRRGPG). Its occurrence in all caddisfly suborders suggests functional importance. All examined caddisflies further contain repeats with an amphiphilic region that begins and ends with a (Ser-X)4–6 string and includes 30–35 amino acid residues.
Since the silks of caddisflies are produced and persist in water, one would expect H-fibroins with hydrophobic repeats but those identified so far are amphiphilic.43 The high content of hydrophilic residues, both neutral and charged, probably facilitates the secretion and storage of the covalently linked L-fibroin/H-fibroin dimer in the absence of P25. Mechanism stabilising the spun-out silk in water remains to be discovered. It is not excluded that covalent cross-linking by disulfide bridges plays a role. The L-fibroins of caddisflies contain 2–5 additional Cys in comparison with the lepidopteran L-fibroins (Fig. 2). More importantly, several cDNAs from the silk gland-specific libraries of the caddisflies we studied encoded Cys-rich repeats (unpublished). A 37 kDa silk protein composed of five repeats, each 63–65 amino acids long and including 4–5 Cys residues, was discovered in another Hydropsyche species.47
Fleas (Siphonaptera)
Larvae of Siphonaptera (fleas) produce silk from the labial glands to construct nests, pupation cases and individual and group cocoons. There is no data on the silk composition, except for an over-all analysis of the representation of individual amino acid residues. The silk proteins have predominantly an α-helical conformation and the silk composition is characterised by low levels of glycine and a high proportion of glutamic acid residues.2,37,48
Midges, Glow Worms and Flies (Diptera)
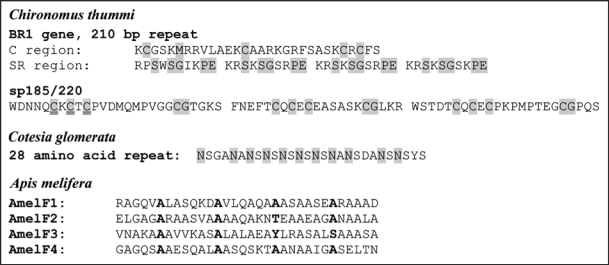
Silk has evolved in the larvae of some nematoceran Diptera, which produce silk from the labial glands and independently in adult male hilarine flies (Diptera-Brachycera: Empididae). The male hilarine flies secrete class III silk from the dermal glands located in the fore leg basitarsi and use it to wrap gifts that are offered to females during courtship.49 The silk contains a 220 kDa protein with unique repeats.50 The silks of Nematocera have been described in the glow worm (Mycetophilidae; Arachnocampa luminosa) and analysed in detail in the water midges (Chironomidae). Glow worm larvae spin silken nests on the roof of caves or overhangs and suspend silken snares from this nest to trap prey. The silk proteins have not yet been described but X-ray diffraction data indicates that they are in a cross-beta structure.51 Chironomid larvae spin protective tunnels throughout their development. Silk composition changes in the last larval instar when a pupation tube is made. The glands of Chironomus tentans include 38–40 cells producing about 15 secretory proteins that fall into three size groups: about 1,000 kDa, 100–200 kDa and less than 100 kDa.52 The largest ones (spIa, spIb, spIc and spId), are encoded by four closely related genes BR1, BR2.1, BR2.2 and BR6, respectively; they are called BR to emphasize their localisation in the puffs (Balbiani rings) that are visible on the polytene chromosomes. These genes have unique 5′ and 3′ termini and a large central region consisting of uninterrupted blocks of 125–150 nearly identical repeats, each with a constant (C) and a slightly variable subrepeat (SR) region. The C region encodes about 30 amino acid residues with four Cys, one Met and one Phe in conserved spacing. The SR region codes for 30–60 residues that are arranged in shorter (3–11 residue) tandem repeats, rich in Pro, often flanked by basic and acidic amino acids (Fig. 4). Repeats encoded by BR6 are extremely charged.
Figure 4.
Some of the repeats identified in the silk proteins in the chironomid larvae and in the larvae of Hymenoptera. Repeats encoded by the BR genes of Chironomus contain a constant region C and the SR region composed of 4 subrepeats.74 Smaller silk proteins, such as sp185/220 are composed of degenerate repeats.75 The Cotesia silk protein contains the dipeptide repeat Asn (sometimes Gly, Glu or Tyr): Ser/Ala in a conserved 28 amino acid repeat.62 Residues conserved in all repeats of the respective protein are highlighted grey. The primary sequence of the coiled coil silk proteins (examples from Apis melifera are shown) have heptad repeats characteristic of coiled coil proteins in the seqeunce HPPHPPP where H = generally hydrophobic residues and P = generally polar or charged residues.66 The initial residue of the heptad repeats is shown in boldface.
Seven silk proteins under 200 kDa are built from sequences related to the C and/or SR regions. For example, silk protein 115/140 includes 65–68 copies of an SR region, sp17 contains a single C/SR repeat and sp12 is a 7.6 kDa protein that may represent a diverged SR region.53 The data suggest that 11 silk genes have evolved after sequence duplication and recombination of a single ancestral gene. The conservation of the C and SR sequences indicates the functional importance of their combination. Extreme deviation was found in the gene BR3 that encodes one of the major silk proteins (designated sp185) in 38 exons (Fig. 4). The protein contains 72 copies of comparatively variable repeats with a C region. It has been proposed that splitting of the repetitive region into variable sized exons prevents the homogenization of the repeats observed in other BR silk genes. Structural studies on synthetic peptides representing the C or SR region indicated that the C region is primarily α-helical and the SR-region forms an extended poly(Gly)II-type helix.54 It is assumed that the extended helical spI proteins assemble into a silk backbone by electrostatic interaction between the SR regions and covalent disulfide bridges between the C regions (there are >400 Cys per molecule). The propensity of spI silk proteins to form disulfide bridges was confirmed after a recombinant 83-mer protein containing typical C and SR regions was expressed in Escherichia coli.55
The silk glands of Chironomus thummi and related species have a lobe composed of four cells distinguishable from the others. These cells express a 2.5 kb transcript that encodes a putative 77 kDa protein with a comparatively nonrepetitive sequence unrelated to other chironomid silk proteins.56 The sequence of 756 amino acid residues contains 24 Cys, 22 Met and strings of Ser and Thr. A secreted protein of 160 kDa is associated with both N- and O-linked glycan moieties. Some of these characteristics resemble lepidopteran sericins.
Sawflies, Ants and Bees (Hymenoptera)
Cocoon silks produced from the labial glands of Hymenoptera are based on a surprising array of protein structures. Below is a description of the phylogenetic distribution of the molecular structure of cocoon silk in six of the 21 hymenopteran superfamilies with the number of species from which the data was obtained indicated in brackets. The distribution of silk types between and among the two hymenopteran suborders Symphyta and Apocrita suggests that β-sheet structured silks are plesiomorphic, while coiled coil structured silks, polyglycine silks and collagen-like silks represent derived conditions.
Among Symphyta, production of silky cocoons is common. Silks of the superfamily Megalodontoidea (two species analysed) is β-sheet structured, similar to that of families Argidae (3 species), Cimbicidae (two species), Diprionidae (three species) and Pergidae (two species) from the Tenthredionoidea.2,37,57 Species from the family Tenthredinidae produce silk based on either β-sheet structured proteins, collagen-like proteins or polyglycine II structured proteins (4, 6 and 7 species, respectively);2,58 with more than one of these protein structures detected in some of the species. Collagen is a structure where three left-handed polypeptide helices twist together to form a right-handed triple helix that is stabilized by numerous hydrogen bonds.59 The interior of the triple helix cannot accommodate any side chains larger than hydrogen and this places a restriction on the primary sequence so that every third residue is required to be glycine. The collagen triple helix is typically stabilised by high levels of proline or 4-hydroxyproline (produced by post translational enzymatic hydroxylation of proline) in the nonglycine positions. As expected, the collagen from the Tenthredinidae cocoons contains around one third glycine but no hydroxyproline; instead it contains substantial amounts of hydroxylysine.57 Polyglycine II is a structure of parallel poly-glycine chains, each with a three-fold screw axis and each hydrogen-bonded to six neighbors.60 Consistent with the structural analysis, the polyglycine II silks of the sawflies contain high levels of glycine (66%).2
In the Apocrita, species from the parasitic wasp superfamilies Ichneumonoidea (five species)2,37,57 and Chrysoidea (2 species)61 produce silk that is primarily based on β-sheet structured proteins. A partial gene sequence (785 bp) encoding a fragment of a very large silk protein (>500 kDa) has been described from the parasitic wasp Cotesia glomerata (Ichneumonoidea, Braconidae).62 The partial sequence encodes a product that contains repeats of 84 amino acid residues (Fig. 4), including strings of the dipeptide (N-S)14 (where Asn is substituted on one occasion for Gly, Asp or Tyr and Ser at four positions for Ala). Dipeptide repeats are characteristic of the β-sheet silk proteins but the presence of Asn as one of the residues is unusual. A comparison of 23 other Microgastrinae cocoon silks revealed that 19 had comparably high Asn levels (25–51%), whereas silks of other Braconidae was composed of predominantly Ala, Ser and Gly.63 Where investigated, X-ray diffraction patterns indicate that the Braconidae silks are predominantly β-sheet structured.2,37,57
Larvae from the social superfamilies Apoidea and Vespoidea of the Apocrita produce silk from proteins in a tetrameric coiled coil structure.57,64 Coiled coils are formed when multiple α-helical polypeptides wind around each other. The sequence of coiled coil proteins is characterized by a seven-residue periodicity where large, nonpolar residues generally occupy the first and fourth position of each heptad.65 The coiled coil silk proteins have been described from five species: Apis mellifera, Bombus terrestris (Apoidea); Oecophylla smargdina; Myrmecia forficata; Vespa simillima (Vespoidea). In all species, four homologous genes were found.61,66,67 They encoded small (30–50 kDa) proteins that consisted of a continuous predicted coiled coil region of around 210-residues (Fig. 4), flanked by shorter (20–180 residue) N- and C-termini. In the species from Vespoidea, the N- and C-terminals were rich in Ser. Despite the similarity in architecture, the proteins had diverged to less than 45% sequence identity between Apoidea and Vespoidea. This level of divergence was not unexpected as the character of the residues in the seven-residue periodicity of coiled coils is important rather than the amino acid identity. However, the composition of the seven-residue periodicity of the silks was unusual in comparison to classic coiled coils, containing very high levels of Ala in all positions and high levels of small polar residues (Ser and Thr) and low levels of large hydrophobic residues (Leu, Ile and Met) in the positions located within the core of the coiled coil.
All four of the above described silk protein structures are produced from the insects labial glands for the construction of cocoons that protect the metamorphosing juvenile. Are they possibly based on four different ancestral proteins the use of which was promoted in some and suppressed in the other taxa? Analysis of minor silk components may provide an answer to this question. Or did the genes encoding collagen, polyglycine or coiled coil silk proteins evolve from genes encoding β-sheet silk proteins? In all of the structures, space between and within the polypeptide chains is effectively filled generating structures with maximal van der Waals interactions and mechanically effective protein fibers. In order to pack effectively, there are constraints on the architecture and composition of the proteins that make up these filaments. Maximal van der Waals interactions will form in β-sheets when every second residue is consistent in size and bulkiness. Similarly, a protein is only able to form a polyglycine II lattice in the absence of nonglycine amino acids as there is no room for side chains larger than the hydrogen within the lattice. The structure of collagen is restricted to glycine in every third position in order for the requisite inter-chain hydrogen bonds to form. In comparison to the above structures, coiled coils are able to accommodate a wide range of structures, although glycine and proline are not favoured.
The co-incidence in the phylogenetic distribution of beta-sheet, polyglycine II, and collagen-like structured silks in the Hymenoptera coupled with their common usage and method of synthesis, suggests an evolutionary link. However, it is difficult to envisage a gradual evolution of one protein structure to the other without intermediates with compromised mechanical integrity. Although the mechanical properties of most of the hymenopteran silks have not been described, it is known that in many cases the mechanical strength of silks appears to exceed that required for the silk function.68 Therefore it is possible that the extraordinary mechanical properties of insect silks are incidental consequence of the silks tightly packed molecular structure. If the final structure offered other selective advantages then less mechanically efficient silk fiber intermediates may not be selected against. The lack of experimental comparison of the silks from each of these structures does not allow us to ascertain the selective advantage of either collagen-like or polyglycine II structured silk.
Although it is similarly difficult to trace an evolutionary path from the large, highly repetitive beta-sheet silk depicted by the Cotesia silk gene to the small nonrepetitive silks genes of the ants and bees, there is more information on the characteristics of the different silks that suggests why the coiled coil silks became so established. The coiled coil silks evolved in a common ancestor predating the emergence of sociality.61 The coiled coil silks are more resistant to denaturation than the β-sheet silks and evolution of silks with enhanced stability would allow the construction of longer lasting domiciles.61,66 Sociality is intrinsically linked to the ability to build durable nests and therefore evolution of sociality in the Apoidea and Vespoidea might be linked to the acquisition of this more stable silk.
Acknowledgements
Writing this chapter was in part supported by grant ME 907 from the Ministry of Education, Youth, and Sports of the Czech Republic.
Note
This manuscript has been previously published: Sehnal F, Sutherland T. Silks produced by insect labial glands. In: Scheibel T, editor. Fibrous Proteins. Austin: Landes Bioscience; 2008. pp. 106–116.
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/7489
References
- 1.Sehnal F, Akai H. Insect silk glands: Their types, development and function and effects of environmental factors and morphogenetic hormones on them. Int J Insect Morphol Embryol. 1990;19:79–132. [Google Scholar]
- 2.Rudall KM, Kenchington W. Arthropod silks: The problem of fibrous proteins in animal tissues. Annu Rev Entomol. 1971;16:73–96. [Google Scholar]
- 3.Willis JH, Iconomidou VA, Smith RF, et al. Cuticular proteins. In: Gilbert LI, Iatrou K, Gill S, editors. Comprehensive insect science. Vol. 4. Oxford: Elsevier; 2005. pp. 79–109. [Google Scholar]
- 4.Craig CL. Evolution of arthropod silks. Annu Rev Entomol. 1997;42:231–267. doi: 10.1146/annurev.ento.42.1.231. [DOI] [PubMed] [Google Scholar]
- 5.Kristensen NP, Scoble MJ, Karsholt O. Lepidoptera phylogeny and systematics: The state of inventorying moth and butterfly diversity. Zootaxa. 2007;1668:699–747. [Google Scholar]
- 6.Inoue S, Tanaka K, Tanaka H, Ohtomo K, Kanda T, Imamura M, et al. Assembly of the silk fibroin elementary unit in endoplasmic reticulum and a role of L-chain for protection of α1,2-mannose residues in N-linked oligosaccharide chains of fibrohexamerin/P25. Eur J Biochem. 2004;271:356–366. doi: 10.1046/j.1432-1033.2003.03934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akai H, Nagashima T, Aoyagi S. Ultrastructure of posterior silk gland cells and liquid silk in Indian tasar silkworm, Antherae mylitta Drury (Lepidoptera • Saturniidae) Int J Insect Morphol Embryol. 1993;22:497–506. [Google Scholar]
- 8.Takasu Y, Yamada H, Tsubouchi K. Isolation of three main sericin components from the cocoon of the silkworm, Bombyx mori. Biosci Biotech Biochem. 2002;66:2715–2718. doi: 10.1271/bbb.66.2715. [DOI] [PubMed] [Google Scholar]
- 9.Žurovec M, Yang C, Kodrík D, Sehnal F. Identification of a novel type of silk protein and regulation of its expression. J Biol Chem. 1998;273:15423–15428. doi: 10.1074/jbc.273.25.15423. [DOI] [PubMed] [Google Scholar]
- 10.Nirmala X, Mita K, Vanisree V, Zurovec M, Sehnal F. Identification of four small molecular mass proteins in the silk of Bombyx mori. Insect Mol Biol. 2001;10:437–445. doi: 10.1046/j.0962-1075.2001.00282.x. [DOI] [PubMed] [Google Scholar]
- 11.Gamo T, Inokuchi T, Laufer H. Polypeptides of fibroin and sericin secreted from the different sections of the silk gland in Bombyx mori. Insect Biochem. 1977;7:285–295. [Google Scholar]
- 12.Sprague K. The Bombyx mori silk proteins: characterization of large polypeptides. Biochemistry. 1975;14:925–931. doi: 10.1021/bi00676a008. [DOI] [PubMed] [Google Scholar]
- 13.Grzelak K. Control of expression of silk protein genes. Comp Biochem Physiol. 1995;110:671–681. doi: 10.1016/0305-0491(94)00215-g. [DOI] [PubMed] [Google Scholar]
- 14.Hamada Y, Yamashita O, Suzuki Y. Haemolymph control of sericin gene expression studied by organ transplation. Cell Differ. 1987;20:65–76. doi: 10.1016/0045-6039(87)90466-0. [DOI] [PubMed] [Google Scholar]
- 15.Michaille JJ, Garel A, Prudhomme JC. The expression of five middle silk gland specific genes is territorially regulated during the larval development of Bombyx mori. Insect Biochem. 1989;19:19–27. [Google Scholar]
- 16.Garel A, Deleage G, Prudhomme JC. Structure and organization of the Bombyx mori Sericin 1 gene and of the Sericin 1 deduced from the sequence of the Ser 1B cDNA. Insect Biochem Mol Biol. 1997;27:469–477. doi: 10.1016/s0965-1748(97)00022-2. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Valluzzi R, Bini E, Vernaglia B, Kaplan DL. Cloning, expression and assembly of sericin-like protein. J Biol Chem. 2003;278:46117–46123. doi: 10.1074/jbc.M307792200. [DOI] [PubMed] [Google Scholar]
- 18.Michaille JJ, Garel A, Prudhomme JC. Cloning and characterization of the highly polymorphic Ser2 gene of Bombyx-mori. Gene. 1990;86:177–184. doi: 10.1016/0378-1119(90)90277-x. [DOI] [PubMed] [Google Scholar]
- 19.Michaille JJ, Garel A, Prudhomme JC. Expression of Ser1 and Ser2 genes in the middle silk gland of Bombyx mori during the fifth instar. Sericologia. 1990;30:49–60. [Google Scholar]
- 20.Žurovec M, Sehnal F, Scheller K, Krishna KA. Silk gland specific cDNAs from Galleria mellonella L. Insect Biochem Mol Biol. 1992;22:55–67. [Google Scholar]
- 21.Inoue S, Tanaka K, Arisaka F, Kimura S, Ohtomo K, Mizuno S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain and P25, with a 6:6:1 molar ratio. J Biol Chem. 2000;275:40517–40528. doi: 10.1074/jbc.M006897200. [DOI] [PubMed] [Google Scholar]
- 22.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka K, Kajiyama N, Ishikura K, Waga S, Kikuchi A, Ohtomo K, et al. Determination of the site of disulfide linkage between heavy and light chains of silk fibroin produced by Bombyx mori. Biochim Biophys Acta. 1999;1432:92–103. doi: 10.1016/s0167-4838(99)00088-6. [DOI] [PubMed] [Google Scholar]
- 24.Takei F, Kikuchi Y, Kikuchi A, Mizuno S, Shimura K. Further evidence for importance of the subunit combination of silk fibroin in its efficient secretion from the posterior silk gland cells. J Cell Biol. 1987;105:175–180. doi: 10.1083/jcb.105.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka K, Inoue S, Mizuno S. Hydrophobic interaction of P25, containing Asn-linked oligosaccharide chains, with the H-L complex of silk fibroin produced by Bombyx mori. Insect Biochem Mol Biol. 1999;29:269–276. doi: 10.1016/s0965-1748(98)00135-0. [DOI] [PubMed] [Google Scholar]
- 26.Yonemura N, Sehnal F. The design of silk fiber composition in moths has been conserved for more than 150 million years. J Mol Evol. 2006;63:42–53. doi: 10.1007/s00239-005-0119-y. [DOI] [PubMed] [Google Scholar]
- 27.Sehnal F, Žurovec M. Construction of silk fiber core in Lepidoptera. Biomacromolecules. 2004;5:666–674. doi: 10.1021/bm0344046. [DOI] [PubMed] [Google Scholar]
- 28.Sezutsu H, Yukuhiro K. Dynamic rearrangement within the Antheraea pernyi silk fibroin gene is associated with four types of repetitive units. J Mol Evol. 2000;51:329–338. doi: 10.1007/s002390010095. [DOI] [PubMed] [Google Scholar]
- 29.Hwang JS, Lee JS, Goo TW, Yun EY, Lee KS, Kim YS, et al. Cloning of the fibroin gene from the oak silkworm, Antheraea yamamai and its complete sequence. Biotech Lett. 2001;23:1321–1326. [Google Scholar]
- 30.Tamura T, Inoue H, Suzuki Y. The fibroin genes of the Antheraea yamamai and Bombyx mori are different in the core regions but reveal a striking sequence similarity in their 5′-ends and 5′-flanking regions. Mol Gen Genet. 1987;206:189–195. [Google Scholar]
- 31.Tanaka K, Mizuno S. Homologues of fibroin L-chain and P25 of Bombyx mori are present in Dendrolimus spectabilis and Papilio xuthus but not detectable in Antheraea yamamai. Insect Biochem Mol Biol. 2001;31:665–677. doi: 10.1016/s0965-1748(00)00173-9. [DOI] [PubMed] [Google Scholar]
- 32.Marsh RE, Corey RB, Pauling L. An investigation of the structure silk fibroin. Biochem Biophys Acta. 1955;16:1–34. doi: 10.1016/0006-3002(55)90178-5. [DOI] [PubMed] [Google Scholar]
- 33.Marsh RE, Corey RB, Pauling L. Structure of tussah silk fibroin. Acta Cryst. 1955;8:710–715. doi: 10.1016/0006-3002(55)90178-5. [DOI] [PubMed] [Google Scholar]
- 34.Mita K, Ichimura S, James TC. Highly repetitive structure and organization of the silk fibroin gene. J Mol Evol. 1994;38:583–592. doi: 10.1007/BF00175878. [DOI] [PubMed] [Google Scholar]
- 35.Zhou CZ, Confalonieri F, Jacquet M, Perasso R, Li ZG, Janin J. Silk fibroin: Structural implications of a remarkable amino acid sequence. Proteins Struct Func Genet. 2001;448:119–122. doi: 10.1002/prot.1078. [DOI] [PubMed] [Google Scholar]
- 36.Fedic R, Žurovec M, Sehnal F. The silk of Lepidoptera. J Insect Biotech Sericol. 2002;71:1–15. [Google Scholar]
- 37.Warwicker JO. Comparative studies of fibroins II. The crystal structures of various fibroins. J Mol Biol. 1960;2:350–362. doi: 10.1016/s0022-2836(60)80046-0. [DOI] [PubMed] [Google Scholar]
- 38.Žurovec M, Sehnal F. Unique molecular architecture of silk fibroin in the waxmoth, Galleria mellonella. J Biol Chem. 2002;277:22639–22647. doi: 10.1074/jbc.M201622200. [DOI] [PubMed] [Google Scholar]
- 39.Fedic R, Žurovec M, Sehnal F. Correlation between fibroin amino acid sequence and physical silk properties. J Biol Chem. 2003;278:35255–35264. doi: 10.1074/jbc.M305304200. [DOI] [PubMed] [Google Scholar]
- 40.Simmons A, Ray E, Jelinski L. Solid-state C-13 NMR of Nephila-clavipes dragline silk establishes structure and identity of crystalline regions. Macromolecules. 1994;27:5235–5237. [Google Scholar]
- 41.Ashida J, Ohgo K, Komatsu K, Kubota A, Asakura T. Determination of the torsion angles of alanine and glycine residues of model compounds of spider silk (AGG)10 using solid state NMR methods. J Biomol NMR. 2003;25:91–103. doi: 10.1023/a:1022220428948. [DOI] [PubMed] [Google Scholar]
- 42.Zaretschnaya SN. Glands of caddisworm III. Spinning glands. Proc Acad Sci USSR. 1965;12:293–303. [Google Scholar]
- 43.Engster M. Studies on silk secretion in the Trichoptera (f. Limnephilidae) I. Histology, histochemistry and ultrastructure of the silk glands. J Morphol. 1976;150:183–211. doi: 10.1002/jmor.1051500109. [DOI] [PubMed] [Google Scholar]
- 44.Yonemura N, Sehnal F, Mita K, Tamura T. Protein composition of silk filaments spun under water by caddisfly larvae. Biomacromolecules. 2006;7:3370–3378. doi: 10.1021/bm060663u. [DOI] [PubMed] [Google Scholar]
- 45.Bini E, Knight DP, Kaplan DL. Mapping domain structures in silks from insects and spiders related to protein assembly. J Mol Biol. 2004;335:27–40. doi: 10.1016/j.jmb.2003.10.043. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi CY, Lewis RV. Evidence from flagelliform silk cDNA for the structural basis of elasticity and modular nature of spider silks. J Mol Biol. 1998;275:773–784. doi: 10.1006/jmbi.1997.1478. [DOI] [PubMed] [Google Scholar]
- 47.EEum JH, Yoe SM, Seo YR, Kang SW, Han SS. Characterization of a novel repetitive secretory protein specifically expressed in the modified salivary gland of Hydropsyche sp (Trichoptera; Hydropsychidae) Insect Biochem Mol Biol. 2005;35:435–441. doi: 10.1016/j.ibmb.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Lucas F, Shaw JTB, Smith SG. Comparative studies of fibroins I. The amino acid composition of various fibroins and its significance in relation to their crystal structure and taxonomy. J Mol Biol. 1960;2:339–349. doi: 10.1016/s0022-2836(60)80045-9. [DOI] [PubMed] [Google Scholar]
- 49.Yang JH, Merritt DJ. The ultrastructure of the silk-producing basitarsus in the Hilarini (Diptera: Empidinae) Arthrop Struct Dev. 2003;32:157–165. doi: 10.1016/S1467-8039(03)00006-9. [DOI] [PubMed] [Google Scholar]
- 50.Sutherland TD, Young JH, Sriskantha A, Weisman S, Okada S, Haritos VS. An independently evolved Dipteran silk with features common to Lepidopteran silks. Insect Biochem Mol Biol. 2007;37:1036–1043. doi: 10.1016/j.ibmb.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 51.Rudall KM. Silk and other cocoon proteins. In: Florkin M, Mason HS, editors. Comparative Biochemistry. Vol. 4. New York: Academic Press; 1962. pp. 397–443. [Google Scholar]
- 52.Wieslander L. The Balbiani ring multigene family—coding repetitive sequences and evolution of a tissue-specific cell-function. Progr Nucl Acid Res Mol Biol. 1994;48:275–313. doi: 10.1016/s0079-6603(08)60858-2. [DOI] [PubMed] [Google Scholar]
- 53.Galli J, Wieslander L. Structure of the smallest salivary-gland secretory protein gene. Chironomus-tentans. J Mol Evol. 1994;38:482–488. doi: 10.1007/BF00178848. [DOI] [PubMed] [Google Scholar]
- 54.Wellman SE, Hamodrakas SJ, Kamitsos EI, Case ST. Secondary structure of synthetic peptides derived from the repeating unit of a giant secretory protein from Chironomus tentans. Biochim Biophys Acta. 1992;1121:279–285. doi: 10.1016/0167-4838(92)90157-9. [DOI] [PubMed] [Google Scholar]
- 55.Smith SV, Correia JJ, Case ST. Disulfide bonds in a recombinant protein modeled after a core repeat in an aquatic insects silk protein. Protein Sci. 1995;4:945–954. doi: 10.1002/pro.5560040514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffman RT, Schmidt ER, Case ST. A cell-specific glycosylated silk protein from Chironomus thummi salivary glands—Cloning, chromosomal localization and characterization of cDNA. J Biol Chem. 1996;271:9809–9815. doi: 10.1074/jbc.271.16.9809. [DOI] [PubMed] [Google Scholar]
- 57.Lucas F, Rudall KM. Extracellular fibrous proteins: The silks. In: Florkin M, Stotz EH, editors. Comprehensive Biochemistry. Vol. 26. Amsterdam: Elsevier; 1968. pp. 475–558. [Google Scholar]
- 58.Lucas F, Rudall KM. Symposium on Fibrous Proteins. Canberra: Butterworths; 1967. Variety in composition and structure of silk fibroins: Some new types of silk from the Hymenoptera; pp. 45–55. [Google Scholar]
- 59.Ramachandran GN, Kartha G. Structure of collagen. Nature. 1955;176:593–595. doi: 10.1038/176593a0. [DOI] [PubMed] [Google Scholar]
- 60.Crick FHC, Rich A. Structure of polyglycine II. Nature. 1955;176:780–781. doi: 10.1038/176780a0. [DOI] [PubMed] [Google Scholar]
- 61.Sutherland TD, Weisman S, Trueman HE, Sriskantha A, Trueman JW, Haritos VS. Conservation of essential design features in coiled coil silks. Mol Biol Evol. 2007;24:2424–2432. doi: 10.1093/molbev/msm171. [DOI] [PubMed] [Google Scholar]
- 62.Yamada H, Shigasada K, Igarashi Y, Takasu Y, Tsubouchi K, Kato Y. A novel asparagine-rich fibrous proteins (Xenofibron) from the cocoons of the parasitic wasp Cotesia (=Apanteles) glomerata. Int J Wild Silkmoth and Silk. 2004;9:61–66. [Google Scholar]
- 63.Quicke D, Shaw M, Takahashi M, Yanechin B. Cocoon silk chemistry of noncyclostome braconidae, with remarks on phylogenetic relationships within the Microgastrinae (Hymenoptera: Braconidae) J Natural History. 2004;38:2167–2181. [Google Scholar]
- 64.Atkins EDT. A four-strand coiled coil model for some insect fibrous proteins. J Mol Biol. 1967;24:139–141. [Google Scholar]
- 65.Woolfson DN. The design of coiled coil structures and assemblies. In: Parry DAD, Squire JM, editors. Fibrous Proteins: Coiled-coils, Collagen and Elastomers. San Diego: Elsevier; 2005. pp. 79–112. [DOI] [PubMed] [Google Scholar]
- 66.Sutherland TD, Campbell PM, Weisman S, Trueman HE, Sriskantha A, Wanjura WJ, et al. A highly divergent gene cluster in honeybees encodes a novel silk family. Genome Res. 2006;16:1414–1421. doi: 10.1101/gr.5052606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sezutsu H, Kajiwara H, Kojima K, Mita K, Tamura T, Tamada Y, et al. Identification of four major hornet silk genes with a complex of alanine-rich and serine-rich sequences in Vespa simillima xanthoptera Cameron. Biosci Biotechnol Biochem. 2007;71:2725–2734. doi: 10.1271/bbb.70326. [DOI] [PubMed] [Google Scholar]
- 68.Denny MW. Silks—Their properties and functions. In: Vincent JFV, Currey JD, editors. The Mechanical Properties of Biological Materials. Vol. 34. Cambridge: University press Cambridge; 1980. pp. 247–272. [Google Scholar]
- 69.Rentz DCF. The world's most unusual gryllacridid (Orthoptera: Gryllacrididae) J Orthoptera Res. 1997;6:57–68. [Google Scholar]
- 70.Morris DC, Schwarz MP, Cooper SJ, Mound LA. Phylogenetics of Australian Acacia thrips: The evolution of behaviour and ecology. Mol Phylogenet Evol. 2002;25:278–292. doi: 10.1016/s1055-7903(02)00258-0. [DOI] [PubMed] [Google Scholar]
- 71.Fletcher MJ, Kent DS. Feeding by kahaono leafhoppers in silken shelters (Hemiptera: Cicadellidae: Typhlocybinae: Dikraneurini) Austral Entomol. 2002;29:115–118. [Google Scholar]
- 72.Paulsson G, Höög C, Bernholm K, Wieslander L. Balbiani ring 1 gene in Chironomus ten-tans: Sequence organization and dynamics of a coding minisatellite. J Mol Biol. 1992;225:349–361. doi: 10.1016/0022-2836(92)90926-b. [DOI] [PubMed] [Google Scholar]
- 73.Case ST, Cox C, Bell WC, Hoffman RT, Martin J, Hamilton R. Extraordinary conservation of cysteines among homologous Chironomus silk proteins sp185 and sp220. J Mol Evol. 1997;44:452–462. doi: 10.1007/pl00006165. [DOI] [PubMed] [Google Scholar]
- 74.Höög C, Wieslander L. Different evolutionary behavior of structurally related, repetitive sequences occurring in the same Balbiani ring gene in Chironomus tentans. Proc Natl Acad Sci USA. 1984;81:5165–5169. doi: 10.1073/pnas.81.16.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takasu Y, Yamada H, Tamura T, Sezutsu H, Mita K, Tsubouchi K. Identification and characterization of a novel sericin gene expressed in the anterior middle silk gland of the silkworm Bombyx mori. Insect Bioch Mol Biol. 2007;37:1234–1240. doi: 10.1016/j.ibmb.2007.07.009. [DOI] [PubMed] [Google Scholar]