Abstract
Biomaterials, having evolved over millions of years, often exceed man-made materials in their properties. Spider silk is one outstanding fibrous biomaterial which consists almost entirely of large proteins. Silk fibers have tensile strengths comparable to steel and some silks are nearly as elastic as rubber on a weight to weight basis. In combining these two properties, silks reveal a toughness that is two to three times that of synthetic fibers like Nylon or Kevlar. Spider silk is also antimicrobial, hypoallergenic and completely biodegradable.
This article focuses on the structure-function relationship of the characterized highly repetitive spider silk spidroins and their conformational conversion from solution into fibers. Such knowedge is of crucial importance to understanding the intrinsic properties of spider silk and to get insight into the sophisticated assembly processes of silk proteins. This review further outlines recent progress in recombinant production of spider silk proteins and their assembly into distinct polymer materials as a basis for novel products.
Key words: biomimetics, biotechnology, protein folding, protein assembly, spinning
Preface
Spider silk has attracted human interest for thousands of years,1–3 mostly due to its toughness and ductility, but also because spider silk seems not to cause inflammation and allergic reactions. Therefore spider silk has been employed for hunting and fishing as well as for bandages.1
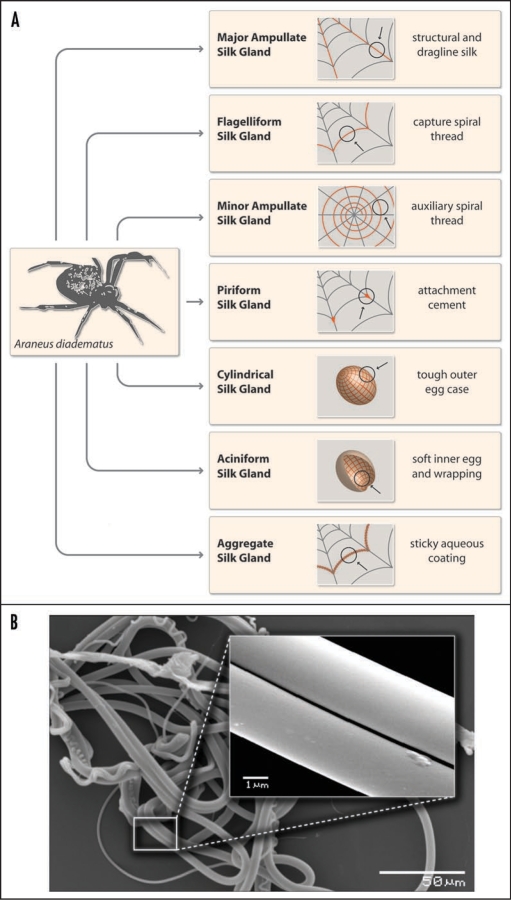
In nature, spiders use their silk for several applications such as for webs, wrapping of prey, protection of their offspring and as a lifeline which ensures their safe escape from predators (Fig. 1A). The variety of uses for silk among spiders is much larger than among insects like silkworms (see chapter 8), which use their silk often for making cocoons.
Figure 1.
(A) Schematic overview of different types of spider silk as produced by female orb weaving spiders such as the European garden spider Araneus diadematus. (B) Electronmicrographs of natural spider silk taken from an orb web of Araneus diadematus. The various morphologies of the distinct silk types can be easily depicted. The inset shows the smooth surface of MA silk.
Catching Prey in Mid-Flight
Currently, more than 34,000 different spider species are known and roughly 50% thereof use webs to catch prey. Further, more than 130 different shapes of spider webs are known. Among the most studied webs are the so-called orb webs which consist of several different types of silk as shown for the European garden spider Araneus diadematus (Fig. 1A and B).4
Frame and radii of orb webs are made of strong and rather rigid silk. The underlying proteins (typically two different types) are produced in the major ampullate glands and therefore silk fibers made of these proteins are named MA silk. This particular silk is further used by spiders as a lifeline (or roping thread) which has to be ready at hand to escape predators—it is therefore always dragged, hence the nickname “dragline silk.” The capture spiral of an orb web comprises fibers of only one type of protein which is produced in the flagelliform (Flag) gland of spiders. Flag silk is highly elastic (up to 300%) and perfectly dissipates the impact energy of prey. For example, a typical honey bee with a body weight of 120 mg and a maximum flight velocity of about 3.1 m/s crashes into a spider's web with a kinetic energy of approx. 0.55 mJ.5 Flag silk with diameters of only 1–5 µm can sufficiently withstand that (on this scale) massive impact. The enormous resilience of these threads is crucial for catching and holding prey which is sometimes even bigger than the spider itself.
In addition to MA and Flag silk, orb weaving spiders utilize two further silks for constructing webs. Silk fibers made of proteins produced in the minor ampullate gland are used to build an auxiliary spiral. This temporary spiral stabilizes the body of the web and provides a template for the capture spiral.6 Connections between Flag and MA silk (the web scaffolding connection joints) and attachment of the web frame to the substratum (trees, undergrowth, masonry…) are made of a sophisticated silk cement consisting of proteins produced in the piriform gland.7
Importantly, Flag silk is not sticky. To retain prey in the web, additional glue is applied around the capture threads.8 Evolutionarily, the first adhesive was a special silk fiber which was hackled by comb-like devices, cribella, on the spider's hind legs in order to achieve a maximum surface area. The adhesive properties of cribellate silk are mainly based on a large sum of van-der-Waals forces provided by huge surface areas similar to the adhesion principle of gecko feet, where an extremely large surface area is provided by nano hairs on the toes.9,10 Although an excellent adhesive, hackling of silk consumes a huge amount of time and energy. Thus, another type of spiders, ecribellate spiders, developed a different strategy of gluing. Specific silk threads are covered with a sticky aqueous layer containing organic molecules, salts, fatty acids and small glycoproteins which are produced in the aggregate gland of spiders.11,12 Recently, it was proposed that the wet glue also contains small peptides which are thought to function as metal chelators.8 The presence of metal ions might contribute to the inhibition of microbial growth on silk threads which is crucial to protecting the web or preventing damage to developing eggs. Wet glue is more energy- and time-efficient (compared to the cribellate system) and is therefore used by the majority of today's spiders.
The Structure of Spider Silk
Primary structure.
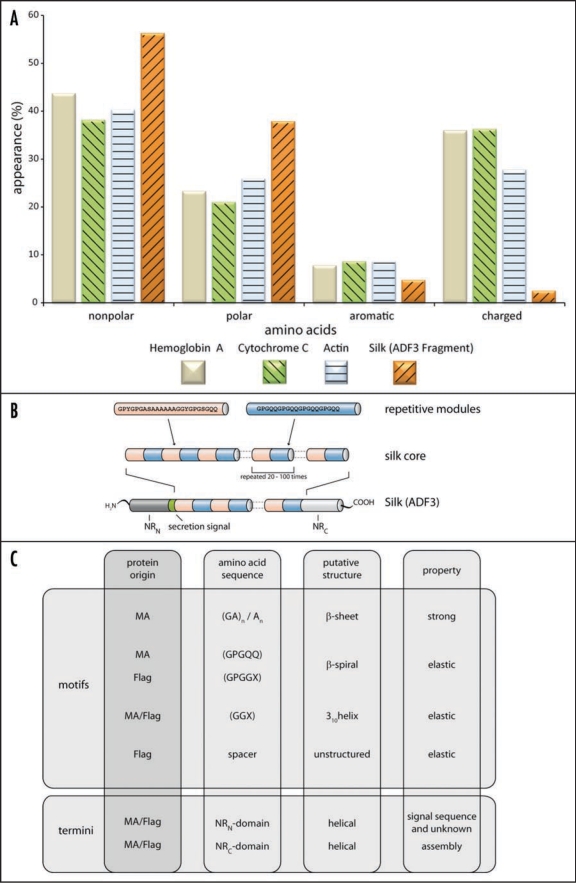
Spider silk primarily consists of proteins that possess large quantities of nonpolar and hydrophobic amino acids like glycine or alanine, but for example, no or only very little tryptophan.4,13,14 In comparison to common cellular enzymes, it is evident that silk proteins exhibit a quite aberrant amino acid composition (Fig. 2A). Furthermore, spider silk proteins contain highly repetitive amino acid sequences, especially in their large core domain (Fig. 2B).
Figure 2.
(A) Amino acid composition of three common proteins in comparison to spider silk. (B) Model of the hierarchical structure of a MA silk protein. For example, Araneus diadematus Fibroin-3 (ADF3) has a highly repetitive core domain flanked by two nonrepetitive domains (aminoterminal domain: NRN; carboxyterminal domain: NRc). The aminoterminal domain also comprises a secretion signal sequence to allow protein export. (C) Amino acid motifs of silk proteins. The different amino acid motifs present in MA and Flag silk are correlated with their putative structure and their impact on the final properties of the thread. Although the function of the nonrepetitive parts is not fully understood, it is thought that these parts play a role in triggering the assembly of spider silks.
The repetitive sequences often account for more than 90% of the whole spider silk protein and are composed of short polypeptide stretches of about 10–50 amino acids. These motifs can be repeated more than a hundred times within one individual protein. Each polypeptide repeat therefore has distinct functional features resulting in the outstanding mechanical properties of spider silk threads.15 MA and Flag silks contain up to four typical oligopeptide motifs which are repeated several times: [I] (GA)n/(A)n, [II] GPGGX/GPGQQ, [III] GGX (X = A, S or Y) and [IV] “spacer” sequences which contain charged amino acids (Fig. 2C).16,17 Structural analysis revealed that oligopeptides with the sequence (GA)n/(A)n tend to form α-helices in solution and β-sheet structures in assembled fibers.18,19 The structures acquired by oligopeptides with the sequences GPGGX/GPGQQ and GGX have not yet been identified. Several studies describe these regions to adopt amorphous rubber-like structures,20,21 whereas others suggest formation of a 31-helical structure.18 Flagelliform silk, typically rich in GPGGX and GGX motifs, preferably folds into β-turn structures resulting in a right-handed β-spiral helix upon stacking (13, 14, 30).16,22
Apart from the repetitive core domain, nonrepetitive regions are located at the protein's termini.23 These nonrepetitive terminal domains of the proteins are crucial for the assembly of spider silk proteins into fibers. The regions comprise approx. 100–200 amino acids and show—in contrast to the repetitive core—well defined secondary and tertiary structures in solution.24,25 Due to conserved cysteine residues, these domains can establish intermolecular disulfide bonds and are thus able to stabilize dimers and multimers under oxidizing conditions. Therefore, these domains are thought to initiate and specify assembly of silk proteins.26–28 Several carboxy-terminal nonrepetitive sequences of different silks and spiders have been identified, revealing a high sequence homology amongst these domains.24,29,30
However, only one full length sequence of a Flag silk protein from Nephila clavipes covering both termini has been identified so far. Further, only recently the first full length sequence of MA silk from the black widow spider has been reported.31
In contrast to the Flag silk gene, both analyzed MA silk genes lack introns consisting instead of unusually large exons (>9,000 bp of coding sequence). The large exons might be a result of gene duplication processes during evolution. Additionally, genes with shorter introns generally show a higher expression rate than genes with large introns32—and spider silks are highly expressed throughout the lifetime of the spider.
The primary structure of spider silk proteins shows a specific hydrophobicity pattern with alternating hydrophilic and hydrophobic blocks in their core domains. Such amphiphilic composition is reminiscent of surfactants or biological membranes and, in the case of spider silks, is thought to be crucial for phase separation during the spinning process (see below).33–35 Additionally, the unusual amphiphilic pattern might be responsible for formation of micelles postulated as intermediate structures during thread assembly (see below).36
Quaternary structure and protein stability.
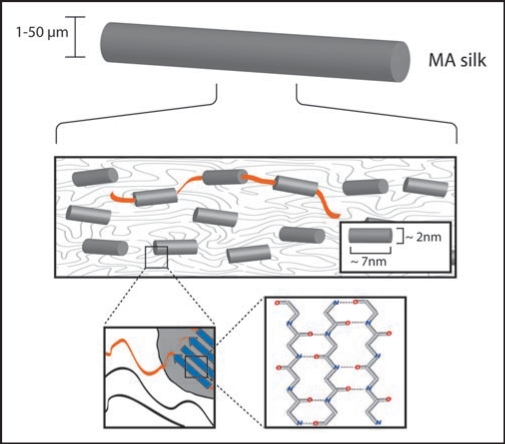
After secretion from the silk glands, silk proteins are in aqueous solution and lack considerable secondary or tertiary structure.37 Particularly in their repetitive core domains, however, the long repetitive sequences permit weak but numerous intra- and intermolecular interactions between neighboring domains and proteins upon passage through the spinning duct. These interactions result in the formation of secondary, tertiary and quaternary structure. Roentgen diffraction analysis of the final structure of MA silk threads led to the identification of areas of high electron density embedded in areas with low electron density (Fig. 3).2,3,38 In a postulated model of this structure, the high electron density regions comprise crystalline sub-structures with high β-sheet content.39 These sub-structures are thought to be responsible for the mechanical strength of the silk thread. The elasticity of silk is based on the areas with low electron density, which are characterized by amorphous structures with few defined elements of secondary or supersecondary structure.40,41 Such arrangement closely resembles that of protein hydrogels.42 Upon tensile loading, the hydrogel-like areas can partially deform, contributing to the elasticity and flexibility of the thread.
Figure 3.
Schematic structure of spider MA silk. The thread is composed of small crystalline b-sheet rich subunits (see close-ups) which are embedded into an amorphous structure. The crystalline and noncrystalline parts are covalently connected, ensuring the coexistence of strength and ductility. Diameters of MA threads depend on species, but also on age, weight and state of health of a specific individual.
Different types of silk reveal different structural distributions (e.g., different compositions of crystalline-and hydrogel-parts). MA silk which is used for constructing the frame of the web contains a high amount of crystalline (β-sheet) structures. In contrast, the much more flexible Flag silk consists almost exclusively of amorphous hydrogel-like regions. Thus, the correlation between the structure and function of individual spider silk proteins becomes evident. However, in the future more detailed analysis is necessary to characterize the structure-function relationship of individual spider silk proteins.
The Assembly of Spider Silk
Starting with a highly concentrated spinning dope…
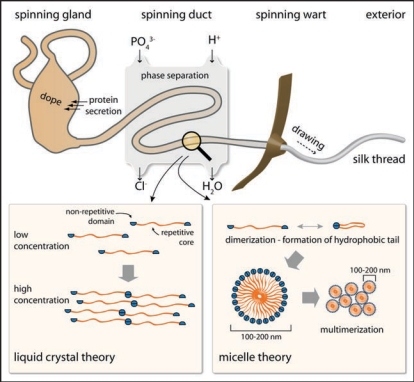
As described above, assembly does not commence with globular folded protein monomers, but with mainly intrinsically unfolded proteins at extremely high concentrations.43 Several mechanisms are thought to be necessary to gain and maintain the high silk protein concentrations (up to 50% w/v)44 in the gland, including lyotropic liquid crystallinity, glycosylation of the outer surface of the folded silk proteins and phase separation induced by a polyol or by a phospholipid surfactant.45 Starting with an almost entire random coil structure in the gland, silk proteins rapidly assemble upon passage through the spinning duct and the silk structure becomes water-insoluble (Fig. 4).46 The tightly controlled assembly behavior requires bi-stable folding of the involved protein and precise control of the environmental conditions in the spinning duct (e.g., pH, ionic concentration, water content).
Figure 4.
Model of the silk spinning process. The highly concentrated spider silk protein solution is secreted and stored inside the spinning gland. Upon initiation of thread formation, the solution is directed through a narrow ion exchange channel, in which a phase separation process takes place. Mechanical drawing of the silk thread (hind legs/gravity) results in rapid assembly of the silk fiber. Two theories exist concerning the molecular process inside the spinning duct. The first theory supports a liquid-crystalline behavior of the silk proteins, whereas the other theory favors a micellar organization of the proteins, before elongation (through laminar force) finally leads to thread assembly.
Phase transition in the spinning duct: two theories.
Successful silk assembly is based on extending, aligning and packaging of individual silk proteins in the laminar flow inside the spinning duct. Hydrophobic residues (typically polyalanines of the repetitive core region) align upon multimerization initiated by the terminal domains and additionally by shear forces in the spinning duct. These polyalanine segments thereby expose an increasingly hydrophobic surface which triggers the formation of β-pleated structures with numerous intra- and interchain hydrogen bonds.46
Two theories on the mechanism of silk fiber assembly have been proposed.36,37,47 One is based on the crystalline alignment of the underlying proteins in the laminar flow inside the spinning duct (Fig. 4). Monomers or disulfide-linked multimers pass the spinning duct at very high concentrations. The alignment in one direction together with the high concentration results in a liquid-crystalline like behavior of the spinning dope. The proposed liquid crystalline state is the basis for the formation of intermolecular interactions like van-der-Waals forces and hydrogen bonds between neighboring molecules.37 Upon further loss of solvent the conformational conversion is finalized and a silk fiber can be drawn out of the spinning duct.
In the second model, silk proteins first assemble into small micelles with a diameter of approx. 100–200 nm due to their amphiphilic properties inside the spinning dope.36 A multitude of these micelles form globules with diameters in the micrometer range (Fig. 4). Shear forces, which arise during passage through the spinning duct, force these globules into an elongated shape finally leading to fiber formation.
Mechanical properties of spider silk.
The most outstanding property of spider silk is its maximal resilience. Distinct spider silk threads are able to absorb three-times more energy than for example Kevlar, one of the sturdiest materials on a weight-to-weight basis (Table 1).39 It is interesting to note that synthetic materials typically show a higher stiffness and strength compared to natural fibers, whereas natural fibers tend to be more elastic. Synthetic carbon fibers, for example, have a yield point at approx 4 GPa. This is more than five times higher than the best insect silk. The elasticity of carbon fibers, on the other hand, is only marginal. As soon as a carbon fiber is elongated more than 1 percent or bent to a certain degree it will instantly break. Spider silk shows a well-balanced combination of strength and elasticity and therefore mechanically outperforms other natural fibers as well as synthetic threads under certain circumstances.48
Table 1.
| Material | Density [g cm−3] | Strength [Gpa] | Elasticity [%] | Toughness [MJ m−3] |
| MA silk* | 1.3 | 1.1 | 27 | 180 |
| Flag silk* | 1.3 | 0.5 | 270 | 150 |
| Insect silk# | 1.3 | 0.6 | 18 | 70 |
| Nylon 6.6 | 1.1 | 0.95 | 18 | 80 |
| Kevlar 49 | 1.4 | 3.6 | 2.7 | 50 |
| Carbon fiber | 1.8 | 4 | 1.3 | 25 |
| Steel | 7.8 | 1.5 | 0.8 | 6 |
European garden spider Araneus diadematus.
Silkworm Bombyx mori.
In addition to its outstanding resilience, MA silk shows a torsional shape memory that prevents the spider from twisting and turning during its descent on a MA silk thread.49,50 Interestingly, MA silk needs no extra stimulus for total recovery after being turned from its initial position. Instead, it scarcely oscillates after twisting because of its high damping coefficient. Spider silk also shows a high supercontraction rate.51,52 Absorption of water leads to shrinkage and tightens the thread. This process is important to ensure the rigidity of the spider's web during its lifetime and is thought to be caused by the organization and arrangement of individual silk proteins.53,54
Spider silk vs insect silk.
Spider silk is often compared to insect silk, preferably taken from the silkworm Bombyx mori. The commercially available silkworm silk is reeled from cocoons of caterpillar pupae. This process has only been slightly optimized over centuries and is highly cost-efficient. MA spider silk can be obtained by manually drawing the silk thread out of the spinning wart of immobilized spiders. However, this process is only suitable for MA silk (and not for the other spider silks), it is time consuming and highly expensive, especially since most spiders are cannibals, rendering farming costly.
The differences between insect and spider silks are evident on all levels, from the molecules involved to the structural arrangement of the proteins to the mechanical properties of the thread. On a molecular level, insect silk comprises a large amount of sericin-proteins, which are absent in spider silk. The proteins which are responsible for the fibrillar structure (so-called fibroins in insect silk) are, in contrast to spider silk spidroins, composed of light and heavy chain counterparts. Mechanically, silkworm silk is much weaker and less extensible as compared to for example MA silk of spiders.55,56 Interestingly, depending on spinning conditions, silkworm silk is either strong or elastic, whereas spider silk combines both properties.57
Although the mechanical properties of both types of silk crucially depend on spinning conditions, it is primarily the proteins involved that make the real difference. Therefore, techniques have long been sought to recombinantly produce and engineer natural spider silk proteins.
Mimicking Nature
Recombinant spider silk.
Several biotechnological methods for recombinantly producing spider silk proteins have been analyzed during the last decades, since recombinant production of sufficient amounts of silk proteins is essential for understanding their structure and their assembly behavior.48 Due to the highly repetitive character of individual spider silk molecules it is quite complicated to determine the complete cDNA sequence of a silk gene. As mentioned above only limited information about complete silk genes is available. Sequencing of repetitive proteins often gives incorrect or unreliable results, providing little information on gene size or the number of repeat units in particular.
The first attempt to obtain recombinant spider silk proteins was the direct transformation of original or fragmented silk genes into bacterial hosts. Bacteria, however, were not suitable for this task due to the large size of the genes.58 Recombinant production of spider silk proteins in bacteria has been further complicated by the different codon usage of spiders compared to bacteria.14 Thus, eukaryotic expression systems like the yeast Pichia pastoris were tested as expression hosts.59 Here, problems occurred during protein purification rather than during the production process. Similar problems were encountered upon attempts to use plants (such as potato or tobacco) as transgenic expression host, however, such systems are still under consideration since they appear to be attractive for larger scale productions.60
Other expression systems for the direct transformation of spider silk genes/fragments have also been used during the last decades. The most prominent one is the expression of spider silk in the milk glands of transgenic goats by the Canadian company Nexia Biotechnologies. Although this technique initially produced promising results, the concentration of soluble protein in the milk was found to be low and the proteins could not be efficiently purified for thorough analysis.61 Tests were also performed in mammalian cell lines with comparable results.62
Finally, insect cells which are phylogeneticaly closely related to that of spiders were employed using the baculovirus expression system.63,64 This particular system benefits from the fact that the easily modifiable baculovirus selectively infects insect cells. Partial cDNAs comprising the known carboxyterminal repetitive and nonrepetitive sequences of the two MA proteins of Araneus diadematus, ADF3 and ADF4, were cloned into the baculoviral genome. The recombinant virus was then used to infect insect cell lines (e.g., sf9 cells of the fall armyworm Spodoptera frugiperda or high five cells of the cabbage looper Trichopulsia ni). During cytoplasmic production, spider silk either remained soluble or assembled into solid fibers—depending on the type of MA silk protein.48,64 A great advantage of the baculovirus expression system, in comparison to expression in mammalian cells, is the relatively easy culturing process and the potential to easily modify the expression conditions.48 For large scale production, however, the Baculovirus system might be too inefficient and too expensive.
The difficulties encountered made it evident that a direct transformation of original silk genes and silk fragments is not the method of choice for recombinant spider silk protein production. To achieve stable production with efficient yields, i.e., by bacterial hosts, the spider genes must be adapted to the bacterial codon usage. To this end, genes coding for spider silk-like proteins were generated using a cloning strategy which is based on a combination of synthetic DNA modules and PCR-amplified authentic gene sequences.28 This approach includes the carboxyterminal nonrepetitive regions present in all MA silks and the codon-optimized repetitive regions. Using a seamless cloning technique, a controlled combination of engineered modules as well as of authentic gene fragments without generation of unwanted gaps or artifacts inside the coding sequence is possible.65 Employing this strategy, it was possible to dramatically increase the expression yields.66 The modular system further enabled the production of special engineered proteins with tailor-made properties for experimental analysis.
Artificial spinning of spider silk.
Due to the availability of recombinant spider silk proteins, scientists will be able to analyze the assembly of spider silk threads in a functional in vitro spinning process in the near future. This process has to ensure that the generated silk fiber resembles natural silk in its microstructure, chemical composition and mechanical properties (Fig. 4).46
To adapt the sophisticated spinning machinery of spiders, several aspects have to be taken into account. Importantly, besides the protein composition of the spinning dope and the phase separation process in the spinning duct, several mechanical parameters play critical roles in silk assembly. In nature, spiders draw the thread with the hind legs (or by the force of gravity in case of roping) out of the spinning wart.15,36,37 This drawing process has been copied in the laboratory by forced silking of captive spiders. Analysis of the silking process revealed that spinning speeds range from 0.1 to 400 mm per second. Interestingly, large differences in resilience, ductility and thread diameters were reported depending on spinning speed and temperature.55 It was shown that silks produced at higher reeling speeds have a slightly higher yield point, but are less extensible and weaker than silks spun at lower speeds.
Several attempts have been made to spin recombinant spider silk proteins for scientific purposes. At first wet-spinning processes were employed with promising results.67,68 Using silicon micro-spinnerets several meters of insect or spider silk fibers could be produced. Although the wet-spun silks visually resembled natural ones, their diameter was up to ten times bigger than naturally occurring silks leading to worse mechanical properties than found in natural silk fibers. Other attempts using special postspinning techniques yielded silks with even larger diameters.62,69 Until now, even the best mechanical properties obtained by artificial spinning techniques are much lower than that of natural dragline silks.68
Exceeding nature—proteins as polymer materials.
Spiders mainly employ silk as linear thread. However, spider silk proteins have much more potential, because they can assemble in all three dimensions to build distinct macroscopic structures (Fig. 5). Nowadays, several attempts are underway to employ silk proteins as biopolymer for novel materials.
Figure 5.
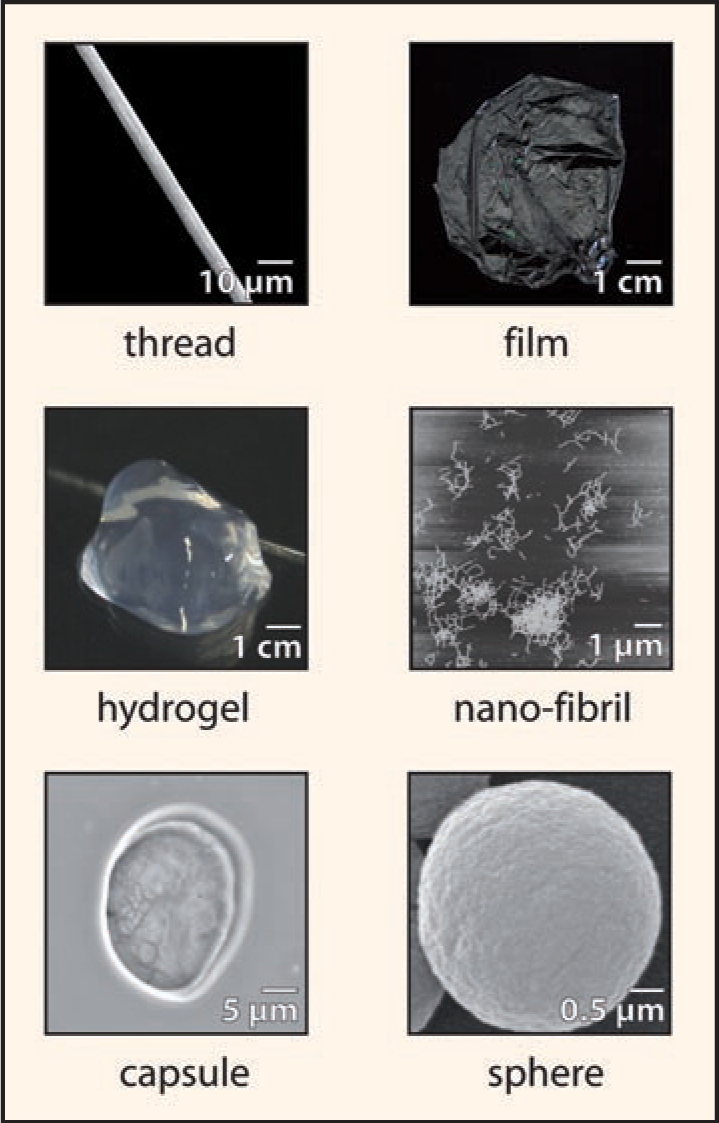
Possible shapes of spider silk. In nature, spider silk proteins are exclusively converted into silk threads. However, in vitro it is possible to transform silk proteins in other two- or three-dimensional shapes. The figure shows images of a capsule, a sphere, a thread and nano-fibrils (all electron microscopy), as well as a hydrogel and a film (photograph) made by recombinantly produced engineered spider silk protein.35,42,48,71
For example, silk films can be made from a diluted spider silk solution.70,71 A silk protein solution is poured onto a surface and the solvent (e.g., water) is allowed to evaporate. During the evaporation process, the silk proteins assemble on the surface and form a sturdy, transparent film. The films can be manufactured with thickness from few nanometers to several micrometers sharing different chemical and mechanical characteristics, depending on the choice of solvent and environmental conditions. Although silk films themselves cannot fulfill the role of model systems for the assembly behavior of all spider silk proteins, they give valuable insights into secondary and tertiary structure formation of these proteins under laboratory conditions. Analysis confirmed that MA proteins are intrinsically unfolded while in aqueous solution. Upon film formation, the proteins rapidly switch to a helical structure. Post treatment of the films with organic solvents like methanol resulted in further rearrangements of protein structure, increasing the β-sheet content dramatically.71–73
In vitro spider silk is further able to self-assemble into small nanofibrils upon incubation in potassium phosphate buffer for several days at room temperature.74,75 The resulting fibrils are structurally comparable to amyloid fibrils.
Taken together, analyzing the structure-function relationship of spider silk proteins in the near future will not only unravel the “secrets” behind the extreme toughness of silk threads, but will also help to engineer and design novel polymeric materials which are inspired by nature. The direct control of silk assembly will enable scientists to manufacture tailor-made spider silk materials with outstanding properties whose diversity cannot be achieved by conventional materials.
Acknowledgements
We thank John Hardy and Eileen Lintz for critical comments on the manuscript. This work was supported by the DFG (SCHE 603/4-3).
This manuscript has been previously published: Römer L, Scheibel T. In: Fibrous, Proteins. Scheibel T, editor. Austin: Landes Bioscience; 2008. The elaborate structure of spider silk; Structure and function of a natural high performance fiber; pp. 121–129.
Footnotes
Previously published online as a Prion E-publication: http://www.landesbioscience.com/journals/prion/article/7490
References
- 1.Gerritsen VB. An airbus could tiptoe on spider silk. Protein Spotlight. 2000;24:1–2. [Google Scholar]
- 2.Kaplan D, Adams WW, Farmer B, Viney C. Silk polymers: material science and biotechnology. 1st ed. Washington DC: ACS Symposium Series; 1993. [Google Scholar]
- 3.Fraser RD, MacRae TP. Conformation in Fibrous Proteins. 1st ed. New York: Academic Press; 1973. [Google Scholar]
- 4.Vollrath F. Strength and structure of spiders' silks. J Biotechnol. 2000;74:67–83. doi: 10.1016/s1389-0352(00)00006-4. [DOI] [PubMed] [Google Scholar]
- 5.Nentwig W. Why do only certain insects escape from a spider's web? Oecologica. 1982;53:412–417. doi: 10.1007/BF00389023. [DOI] [PubMed] [Google Scholar]
- 6.Zschokke S. The influence of the auxiliary spiral on the capture spiral in Araneus diadematus Clerck (Araneidae) Bull Br Arachnol Soc. 1993;9:167–173. [Google Scholar]
- 7.Gosline JM, DeMont EM, Denny MW. The structure and properties of spider silk. Endeavour. 1986;10:37–43. [Google Scholar]
- 8.Hu X, Yuan J, Wang X, Vasanthavada K, Falick AM, Jones PR, et al. Analysis of aqueous glue coating proteins on the silk fibers of the cob weaver, Latrodectus hesperus. Biochemistry. 2007;46:3294–3303. doi: 10.1021/bi602507e. [DOI] [PubMed] [Google Scholar]
- 9.Gao H, Yao H. Shape insensitive optimal adhesion of nanoscale fibrillar structures. Proc Natl Acad Sci USA. 2004;101:7851–7856. doi: 10.1073/pnas.0400757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arzt E, Gorb S, Spolenak R. From micro to nano contacts in biological attachment devices. Proc Natl Acad Sci USA. 2003;100:10603–10606. doi: 10.1073/pnas.1534701100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Townley MA, Tillinghast EK, Neefus CD. Changes in composition of spider orb web sticky droplets with starvation and web removal and synthesis of sticky droplet compounds. J Exp Biol. 2006;209:1463–1486. doi: 10.1242/jeb.02147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vollrath F, Tillinghast EK. Glycoprotein glue beneath a spider web's aqueous coat. Naturwissenschaften. 2005;78:557–559. [Google Scholar]
- 13.Rising A, Nimmervoll H, Grip S, Fernandez-Arias A, Storckenfeldt E, Knight DP, et al. Spider silk proteins—mechanical property and gene sequence. Zoolog Sci. 2005;22:273–281. doi: 10.2108/zsj.22.273. [DOI] [PubMed] [Google Scholar]
- 14.Xu M, Lewis RV. Structure of a protein superfiber: spider dragline silk. Proc Natl Acad Sci USA. 1990;87:7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis RV. Spider silk: Ancient ideas for new biomaterials. Chem Rev. 2006;106:3762–3774. doi: 10.1021/cr010194g. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi CY, Lewis RV. Evidence from flagelliform silk cDNA for the structural basis of elasticity and modular nature of spider silks. J Mol Biol. 1998;275:773–784. doi: 10.1006/jmbi.1997.1478. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi CY, Lewis RV. Spider flagelliform silk: Lessons in protein design, gene structure and molecular evolution. Bioessays. 2001;23:750–756. doi: 10.1002/bies.1105. [DOI] [PubMed] [Google Scholar]
- 18.van Beek JD, Hess S, Vollrath F, Meier BH. The molecular structure of spider dragline silk: folding and orientation of the protein backbone. Proc Natl Acad Sci USA. 2002;99:10266–10271. doi: 10.1073/pnas.152162299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmons AH, Michal CA, Jelinski LW. Molecular orientation and two-component nature of the crystalline fraction of spider dragline silk. Science. 1996;271:84–87. doi: 10.1126/science.271.5245.84. [DOI] [PubMed] [Google Scholar]
- 20.Gosline JM, Denny MW, DeMont EM. Spider silk as rubber. Nature. 1994;309:551–552. [Google Scholar]
- 21.Termonia Y. Monte Carlo diffusion model of polymer coagulation. Phys Rev Lett. 1994;72:3678–3681. doi: 10.1103/PhysRevLett.72.3678. [DOI] [PubMed] [Google Scholar]
- 22.Becker N, Oroudjev E, Mutz S, Cleveland JP, Hansma PK, Hayashi CY, et al. Molecular nanosprings in spider capture-silk threads. Nat Mater. 2003;2:278–283. doi: 10.1038/nmat858. [DOI] [PubMed] [Google Scholar]
- 23.Scheibel T. Protein fibers as performance proteins: new technologies and applications. Curr Opin Biotechnol. 2005;16:427–433. doi: 10.1016/j.copbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Rising A, Hjälm G, Engström W, Johansson J. N-terminal nonrepetitive domain common to dragline, flagelliform and cylindriform spider silk proteins. Biomacromolecules. 2006;7:3120–3124. doi: 10.1021/bm060693x. [DOI] [PubMed] [Google Scholar]
- 25.Ittah S, Michaeli A, Goldblum A, Gat U. A model for the structure of the C-terminal domain of dragline spider silk and the role of its conserved cysteine. Biomacromolecules. 2007;8:2768–2773. doi: 10.1021/bm7004559. [DOI] [PubMed] [Google Scholar]
- 26.Sponner A, Vater W, Rommerskirch W, Vollrath F, Unger E, Grosse F, et al. The conserved C-termini contribute to the properties of spider silk fibroins. Biochem Biophys Res Commun. 2005;338:897–902. doi: 10.1016/j.bbrc.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 27.Sponner A, Unger E, Grosse F, Weisshart K. Conserved C-termini of Spidroins are secreted by the major ampullate glands and retained in the silk thread. Biomacromolecules. 2004;5:840–845. doi: 10.1021/bm034378b. [DOI] [PubMed] [Google Scholar]
- 28.Huemmerich D, Helsen CW, Quedzuweit S, Oschmann J, Rudolph R, Scheibel T. Primary structure elements of spider dragline silks and their contribution to protein solubility. Biochemistry. 2004;43:13604–13612. doi: 10.1021/bi048983q. [DOI] [PubMed] [Google Scholar]
- 29.Hu X, Vasanthavada K, Kohler K, McNary S, Moore AM, Vierra CA. Molecular mechanisms of spider silk. Cell Mol Life Sci. 2006;63:1986–1999. doi: 10.1007/s00018-006-6090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motriuk-Smith D, Smith A, Hayashi CY, Lewis RV. Analysis of the conserved N-terminal domains in major ampullate spider silk proteins. Biomacromolecules. 2005;6:3152–3159. doi: 10.1021/bm050472b. [DOI] [PubMed] [Google Scholar]
- 31.Ayoub NA, Garb JE, Tinghitella RM, Collin MA, Hayashi CY. Blueprint for a high-performance biomaterial: full-length spider dragline silk genes. PLoS ONE. 2007;2:514. doi: 10.1371/journal.pone.0000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nat Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- 33.Exler JH, Hummerich D, Scheibel T. The amphiphilic properties of spider silks are important for spinning. Angew Chem Int Ed Engl. 2007;46:3559–3562. doi: 10.1002/anie.200604718. [DOI] [PubMed] [Google Scholar]
- 34.Hermanson KD, Huemmerich D, Scheibel T, Bausch AR. Engineered Microcapsules Fabricated from Reconstituted Spider Silk. Advanced Materials. 2007;19:1810–1815. [Google Scholar]
- 35.Jin HJ, Kaplan DL. Mechanism of silk processing in insects and spiders. Nature. 2003;424:1057–1061. doi: 10.1038/nature01809. [DOI] [PubMed] [Google Scholar]
- 36.Vollrath F, Knight DP. Liquid crystalline spinning of spider silk. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 37.Riekel C, Bränden C, Craig C, Ferrero C, Heidelbach F, Müller M. Aspects of X-ray diffraction on single spider fibers. Int J Biol Macromol. 1999;24:179–186. doi: 10.1016/s0141-8130(98)00084-1. [DOI] [PubMed] [Google Scholar]
- 38.Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The mechanical design of spider silks: From fibroin sequence to mechanical function. J Exp Biol. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 39.Kubik S. High-performance fibers from spider silk. Angew Chem Int Ed Engl. 2002;41:2721–2723. doi: 10.1002/1521-3773(20020802)41:15<2721::AID-ANIE2721>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure and mechanical properties of spider silk proteins. Int J Biol Macromol. 1999;24:271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 42.Rammensee S, Huemmerich D, Hermanson KD, Scheibel T, Bausch A. Rheological characterisation of recombinant spider silk nanofiber networks. Appl Phys A. 2006;82:261–264. [Google Scholar]
- 43.Dicko C, Kenney JM, Knight D, Vollrath F. Transition to a beta-sheet-rich structure in spidroin in vitro: The effects of pH and cations. Biochemistry. 2004;43:14080–14087. doi: 10.1021/bi0483413. [DOI] [PubMed] [Google Scholar]
- 44.Hijirida DH, Do KG, Michal C, Wong S, Zax D, Jelinski LW. 13C NMR of Nephila clavipes major ampullate silk gland. Biophys J. 1996;71:3442–3447. doi: 10.1016/S0006-3495(96)79539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.SenGupta S, Scheibel T. Folding, self-assembly and conformational switches of proteins. In: Zbilut JP, Scheibel T, editors. Protein folding and misfolding. New York: Nova Publishers; 2007. pp. 1–34. [Google Scholar]
- 46.Scheibel T. Spider silks: Recombinant synthesis, assembly, spinning and engineering of synthetic proteins. Microb Cell Fact. 2004;3:14. doi: 10.1186/1475-2859-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ko FK, Jovicic J. Modeling of mechanical properties and structural design of spider web. Biomacromolecules. 2004;5:780–785. doi: 10.1021/bm0345099. [DOI] [PubMed] [Google Scholar]
- 48.Vendrely C, Scheibel T. Biotechnological production of spider-silk proteins enables new applications. Macromol Biosci. 2007;7:401–409. doi: 10.1002/mabi.200600255. [DOI] [PubMed] [Google Scholar]
- 49.Emile O, Le Floch A, Vollrath F. Biopolymers: Shape memory in spider draglines. Nature. 2006;440:621. doi: 10.1038/440621a. [DOI] [PubMed] [Google Scholar]
- 50.Emile O, Le Floch A, Vollrath F. Time-resolved torsional relaxation of spider draglines by an optical technique. Phys Rev Lett. 2007;98:167402. doi: 10.1103/PhysRevLett.98.167402. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Shao Z, Vollrath F. Relationships between supercontraction and mechanical properties of spider silk. Nat Mater. 2005;4:901–905. doi: 10.1038/nmat1534. [DOI] [PubMed] [Google Scholar]
- 52.Perez Rigueiro J, Elices M, Guinea GV. Controled supercontraction tailors the tensile behaviour of spider silk. Polymer. 2003;44:3733–3736. [Google Scholar]
- 53.Shao Z, Vollrath F, Sirichaisit J, Young RJ. Analysis of spider silk in native and supercontracted states using raman spectroscopy. Polymer. 1999;40:2493–2500. [Google Scholar]
- 54.Yang Z. Supercontraction and backbone dynamics in spider silk: C-12 and H-2 NMR studies. J Am Chem Soc. 2000;122:9019–9025. [Google Scholar]
- 55.Vollrath F, Madsen B, Shao Z. The effect of spinning conditions on the mechanics of a spider's dragline silk. Proc Biol Sci. 2001;268:2339–2346. doi: 10.1098/rspb.2001.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilding MA, Hearle J. Fiber structure. In: Salamone JC, editor. Polymeric materials encyclopedia. Vol. 11. Bota Raton: CRC; 1996. pp. 8307–8322. [Google Scholar]
- 57.Shao Z, Vollrath F. Surprising strength of silkworm silk. Nature. 2002;418:741. doi: 10.1038/418741a. [DOI] [PubMed] [Google Scholar]
- 58.Arcidiacono S, Mello C, Kaplan D, Cheley S, Bayley H. Purification and characterization of recombinant spider silk expressed in Escherichia coli. Appl Microbiol Biotechnol. 1998;49:31–38. doi: 10.1007/s002530051133. [DOI] [PubMed] [Google Scholar]
- 59.Fahnestock SR, Bedzyk LA. Production of synthetic spider dragline silk protein in Pichia pastoris. Appl Microbiol Biotechnol. 1997;47:33–39. doi: 10.1007/s002530050884. [DOI] [PubMed] [Google Scholar]
- 60.Scheller J, Gührs KH, Grosse F, Conrad U. Production of spider silk proteins in tobacco and potato. Nat Biotechnol. 2001;19:573–577. doi: 10.1038/89335. [DOI] [PubMed] [Google Scholar]
- 61.Trivedi BP. Lab spins artificial spider silk, paving the way to new materials. 2002. http://news.nationalgeographic.com/news/2002/01/0117_020117TVspidermammals.html.
- 62.Lazaris A, Arcidiacono S, Huang Y, Zhou JF, Duguay F, Chretien N, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 63.Miao Y, Zhang Y, Nakagaki K, Zhao T, Zhao A, Meng Y, et al. Expression of spider flagelliform silk protein in Bombyx mori cell line by a novel Bac-to-Bac/BmNPV baculovirus expression system. Appl Microbiol Biotechnol. 2006;71:192–199. doi: 10.1007/s00253-005-0127-2. [DOI] [PubMed] [Google Scholar]
- 64.Huemmerich D, Scheibel T, Vollrath F, Cohen S, Gat U, Ittah S. Novel assembly properties of recombinant spider dragline silk proteins. Curr Biol. 2004;14:2070–2074. doi: 10.1016/j.cub.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 65.Padgett KA, Sorge JA. Creating seamless junctions independent of restriction sites in PCR cloning. Gene. 1996;168:31–35. doi: 10.1016/0378-1119(95)00731-8. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt M, Römer L, Strehle M, Scheibel T. Conquering isoleucine auxotrophy of Escherichia coli BLR(DE3) to recombinantly produce spider silk proteins in minimal media. Biotechnol Lett. 2007;29:1741–1744. doi: 10.1007/s10529-007-9461-z. [DOI] [PubMed] [Google Scholar]
- 67.Jelinski LW, Blye A, Liivak O, Michal C, LaVerde G, Seidel A, et al. Orientation, structure, wet-spinning and molecular basis for supercontraction of spider dragline silk. Int J Biol Macromol. 1999;24:197–201. doi: 10.1016/s0141-8130(98)00085-3. [DOI] [PubMed] [Google Scholar]
- 68.Seidel A, Liivak O, Jelinski LW. Artificial spinning of spider silk. Macromolecules. 1998;31:6733–6736. [Google Scholar]
- 69.Arcidiacono S, Mello CM, Butler M, Welsh E, Soares JW, Allen A. Aqueous processing and fiber spinning of recombinant silks. Macromolecules. 2002;35:1262–1266. [Google Scholar]
- 70.Vendrely C, Ackerschott C, Römer L, Scheibel T. Modular design of performance proteins with repetitive sequences: recombinant flagelliform spider silk as basis for biomaterials. Methods in Molecular Biology. 2008;474:3–14. doi: 10.1007/978-1-59745-480-3_1. [DOI] [PubMed] [Google Scholar]
- 71.Slotta U, Tammer M, Kremer F, Koelsch P, Scheibel T. Structural analysis of spider silk films. Supramol Chem. 2006;18:465–471. [Google Scholar]
- 72.Metwalli E, Slotta U, Darko C, Roth SV, Scheibel T, Papadakis CM. Structural changes of thin films from recombinant spider silk proteins upon post treatment. Appl Phys A. 2007;89:655–661. [Google Scholar]
- 73.Huemmerich D, Slotta U, Scheibel T. Films from recombinant spider silk proteins. Appl Phys A. 2006;82:219–222. [Google Scholar]
- 74.Kenney JM, Knight D, Wise MJ, Vollrath F. Amyloidogenic nature of spider silk. Eur J Biochem. 2002;269:4159–4163. doi: 10.1046/j.1432-1033.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 75.Slotta U, Hess S, Spiess K, Stromer T, Serpell L, Scheibel T. Spider silk and amyloid fibrils: A structural comparison. Macromol Biosci. 2007;7:183–188. doi: 10.1002/mabi.200600201. [DOI] [PubMed] [Google Scholar]