Abstract
Recent years have seen a huge increase in the generation of genomic and proteomic data. This has been due to improvements in current biological methodologies, the development of new experimental techniques and the use of computers as support tools. All these raw data are useless if they cannot be properly analysed, annotated, stored and displayed. Consequently, a vast number of resources have been created to present the data to the wider community. Annotation tools and databases provide the means to disseminate these data and to comprehend their biological importance. This review examines the various aspects of annotation: type, methodology and availability. Moreover, it puts a special interest on novel annotation fields, such as that of phenotypes, and highlights the recent efforts focused on the integrating annotations.
Keywords: genome annotation, proteome annotation, sequencing
1. Introduction
Over the last decade, significant developments in the biological and computer sciences have made it possible to generate large amounts of raw genomic and proteomic data. However, meaningful biological inferences can only be gained where expert organization and interpretation of these data are carried out. In 1999, the DNA sequence of chromosome 22, the first human chromosome to be fully sequenced, was published (Dunham et al. 1999) and the first draft of the human genome assembly was completed in 2001 (Lander et al. 2001). The human genome took 10 years to sequence. Today, the sequencing of entire genomes has become routine, resulting in ever increasing numbers of published genomes across the kingdoms of life (see Pop & Salzberg (2008) for the new challenges presented by this fact) and, even, the appearance of metagenomics research (Venter et al. 2004; Tringe et al. 2005). Consequently, this has led to a significant increase in the number of genes and corresponding translated proteomic sequences deposited into databases such as TrEMBL (Boeckmann et al. 2003), which now comprises over 6 million sequences. Likewise, the increased efficiency in tools for the elucidation of protein structure has helped to accelerate the number of experimentally determined structures which are released by the PDB each month. This is in part due to the advent of structural genomics initiatives (Burley 2000; Brenner 2001), which generally attempt to solve every representative structure of an interesting family or aim to cover fold space by picking targets unlike any other structures previously solved. Importantly, much of these genomic and proteomic data are experimentally uncharacterized, putting much emphasis on the need for accurate analytical tools and up-to-date specialist databases. This review discusses the various aspects of genome and proteome annotation, with particular focus on the way in which these methods have started to be integrated.
1.1 Overview of annotation process: genome to proteome
The increasing efficiency of genome sequencing has led to a significant rise in the release rate of sequenced genomes. Sequencing can be done at different levels ranging from whole-genome tiling arrays (Yazaki et al. 2007; low accuracy) to methods in which the genome is divided into contigs, each of which is sequenced separately and the data recombined (Mardis 2008). Subsequent genome annotation involves the prediction of a number of features on the DNA: coding genes; pseudogenes; promoters–regulatory regions; untranslated regions; and repeats, to name a few. Although growing attention is focused on non-coding RNA (extensively reviewed in Huttenhofer et al. (2005), Mendes Soares & Valcarcel (2006) and Amaral et al. (2008)), traditionally, most interest is in the prediction of the coding genes, since peptides are seen as potential targets for drug discovery (Ofran et al. 2005). This prediction is not a trivial process as gene structure is not common among all the organisms (Wong et al. 2001; Blencowe 2006). For example, eukaryotes have exons and introns in their genes, whereas prokaryotes have the whole coding sequence as a continuum. In addition, different species have different rates of alternative splicing (Kan et al. 2001; Modrek et al. 2001). Alternative splicing is often predicted using expressed sequence tags (ESTs)—short sequences of a transcribed spliced nucleotide sequence that have been used extensively to identify gene transcripts and have been important in gene discovery and gene sequence determination (Adams et al. 1991). Subsequent studies have shown that the alternative splicing prediction by this method in the main sequencing projects depends on the EST coverage and that, in effect, the more ESTs, the more alternative splicing is reported (Brett et al. 2002; Gupta et al. 2004). Finally, not only cis-splicing is possible, but also some examples of trans-splicing (involving more than one pre-mRNA) have been found (Caudevilla et al. 1998; Dorn et al. 2001; Horiuchi & Aigaki 2006). Looking at all these facts together, it is easy to understand the reason for not yet having an accurate number of genes and transcripts from the fully sequenced genomes (reviewed in Southan (2004) and Brent (2008)).
Once the number and location of the genes are known, the corresponding protein sequences can be generated via translation of the gene sequences. This is not always a trivial process since some genetic features reduce the accuracy of correctly translating the correct region of a given gene sequence. For example, the existence of exons that can be translated using different frames without finding any STOP codon (Clark & Thanaraj 2002) or multiple NAGNAG tandem splice acceptor sites (Hiller et al. 2004), which lead to two possible peptides having different numbers of residues. In addition, many transcripts seem to be potential targets for the non-sense-mediated decay mechanism (Lewis et al. 2003), which eliminates mRNAs containing premature ends of translation. Thus, the biological relevance of the majority of these transcripts is unclear as there is no certainty about their biological role (Neu-Yilik et al. 2004; Ravasi et al. 2006). Experimental methods can also be used to generate the amino acid sequence from an expressed protein, such as Edman degradation or mass spectrometry.
The process of making sense of protein sequence data is also complex, and a huge number of tools and specialist databases have been developed in order to characterize these protein sequences (and their three-dimensional structures; figure 1 and table 1). More information can often be elucidated about the protein by experimentally determining its three-dimensional structure by X-ray crystallography and NMR. This is supplemented by the prediction of homology models using a template (Martí-Renom et al. 2000) or physico-chemical rules (Kuhlman et al. 2003). The ultimate aim is to relate protein structure data to the corresponding functional information. Here, one of the main problems is that the existence of a gene, a transcript or a peptide does not mean that the protein has a functional role (Lewis et al. 2003; Neu-Yilik et al. 2004; Tress et al. 2007).
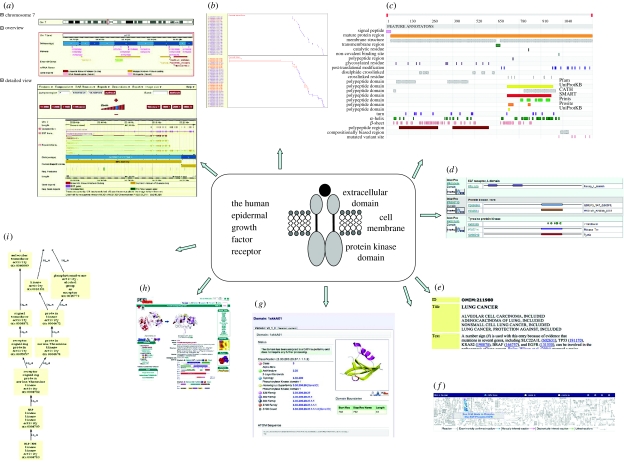
Figure 1.
A selection of resources showing data for the human epidermal growth factor receptor (EGFR). (a) Ensembl (genomic information), (b) ASAP II (alternative splicing information), (c) Dasty2 protein DAS client (protein kinase domain), (d) InterProScan (functional annotation), (e) OMIM (disease information), (f) Reactome (EGFR signalling information): signalling by EGFR (Homo sapiens), (g) CATH domain database (protein kinase domain), (h) PDBsum (structural information) and (i) GO (MAP/ERK kinase kinase activity)
Table 1.
Illustrating a selection of data resources (annotation tools, databases and repositories). (The resources are divided into those which serve genomic data, proteomic sequence and proteomic structure. An indication of ‘automatic’ or ‘manual’ annotation is associated with each method to describe how the data are generated. Those which just provide a central point for a particular set of data are annotated as a ‘repository’. This list includes a great variety of resources that the authors consider useful; however, other specialized reviews can afford a bigger coverage of tools or databases for specific problems (Casadio et al. 2008; Meinnel & Giglione 2008).)
| data resource | description | URL | type: manual automatic repository |
|---|---|---|---|
| genome | |||
| ASAP II | database of splicing variants including tissue and cancer analysis (Kim et al. 2007) | http://bioinformatics.ucla.edu/ASAP2/ | A |
| ASPicDB | database of splicing pattern of human genes (Castrignano et al. 2008) | http://t.caspur.it/ASPicDB/ | A |
| ASTD | database containing alternative transcripts generated by either alternative splicing or alternative start or end points (Stamm et al. 2006) | http://www.ebi.ac.uk/astd/ | A |
| dbSNP | a catalogue of variation from the National Center for Biotechnology Information (Smigielski et al. 2000) | http://www.ncbi.nlm.nch.gov/projects/snp | R |
| Ensembl | pipeline which includes prediction of genes, transcripts and peptides (Flicek et al. 2008) | http://www.ensembl.org | A |
| FlyBase | database of Drosophila genomes (Grumbling & Strelets 2006) | http://flybase.bio.indiana.edu/ | M |
| GenBank | database containing all publicly available DNA sequences (Benson et al. 2008) | http://www.ncbi.nlm.nih.gov/Genbank/ | R |
| GOLD | resource monitoring the worldwide genome projects (Liolios et al. 2008) | http://www.genomesonline.org/ | A |
| NCBI tools | repository of tools to perform analysis in several types of data: genes; proteins; and genomes (Wheeler et al. 2007) | http://www.ncbi.nlm.nih.gov/Tools/ | A |
| OMIM | database of human-inherited diseases and the genes causing them (Hamosh et al. 2002) | http://www.ncbi.nlm.nih.gov/omim/ | M |
| RefSeq | non-redundant database of annotated sequences (genomic DNA, transcripts and proteins; Pruitt et al. 2007) | http://www.ncbi.nlm.nih.gov/RefSeq/ | M/A |
| SNPeffect | database for the annotation of the effect of SNPs (Reumers et al. 2005) | http://snpeffect.vib.be/index.php | A |
| TAIR | database containing genetic and molecular biology data for Arabidopsis thaliana (Swarbreck et al. 2008) | http://www.arabidopsis.org/ | M/A |
| UCSC genome browser | browser for displaying genomic data (Karolchik et al. 2008) | http://genome.ucsc.edu/ | A |
| Vega | repository of manually curated data for finished vertebrate genomes (Wilming et al. 2008) | http://vega.sanger.ac.uk | M |
| WormBase | database containing genomic information for Caenorhabditis elegans and other nematodes (Rogers et al. 2008) | http://www.wormbase.org/ | M |
| proteomic/sequence | |||
| a suite of tools to analyse post-translational modifications from the CBS | predicting the attachment of chemical groups: phosphorylation (NetPhos; Blom et al. 1999; NetPhosK; Blom et al. 2004; NetPhosYeast; Ingrell et al. 2007); O-linked glycosylation (NetOGlyc; Julenius et al. 2005; YinOYang; Gupta & Brunak 2002; DictyOGlyc; Gupta et al. 1999); N-linked glycosylation (NetNGlyc); C-linked glycosylation (NetCGlyc; Julenius 2007); glycation (NetGlycate; Johansen et al. 2006); acetylation (NetAcet; Kiemer et al. 2005); sulphation; and lipid attachment (LipoP; Juncker et al. 2003); | http://www.cbs.dtu.dk/services/ | A |
| tools for the indication of peptide cleavage: signal peptides (SignalP; Bendtsen et al. 2004; LipoP; Juncker et al. 2003; TatP; Bendtsen et al. 2005a,b); propeptides (ProP; Duckert et al. 2004); transit peptides (TargetP; Emanuelsson et al. 2007; ChloroP; Emanuelsson et al. 1999); viral polyprotein processing (NetCorona; Kiemer et al. 2004; NetPicoRNA; Blom et al. 1996); caspase cleavage and also protein sorting and subcellular localization; secretion (SecretomeP; Bendtsen et al. 2005a,b); import into mitochondria and chloroplasts (ChloroP); and nuclear export (NetNES; La Cour et al. 2004) | |||
| CSA | database containing information about catalytic residues, part manually curated, part by homology (Porter et al. 2004) | http://www.ebi.ac.uk/thornton-srv/databases/CSA/ | M/A |
| FireDB/Firestar | database containing residues with functional annotation (Lopez et al. 2007a) and a tool for predicting functional residues in unannotated sequences (Lopez et al. 2007b) | http://firedb.bioinfo.cnio.es/ | A |
| Gene3D | functional annotation database which searches similarities between unannotated proteins from whichever origin and CATH domains (Yeats et al. 2008) | http://gene3d.biochem.ucl.ac.uk/Gene3D/ | A |
| Interpro | consortium database which includes annotation from different database members (Mulder et al. 2007) | http://www.ebi.ac.uk/interpro/ | M/A |
| iProClass | integrative database for protein functional features (Wu et al. 2004) | http://pir.georgetown.edu/iproclass/ | A |
| KEGG | resource containing information about genes, functions, hierarchies, pathways and ligands (Kanehisa et al. 2008) | http://www.genome.jp/kegg/ | M/A |
| MEMSAT | predicts the structure of all-helical transmembrane proteins and the location of their constituent helical elements within a membrane (Jones 2007) | http://bioinf.cs.ucl.ac.uk/memsat/ | A |
| Panther | database of functional assignments for genes and proteins (Thomas et al. 2003) | http://www.pantherdb.org/ | M/A |
| Pfam | database containing multiple alignments of protein domains and conserved regions (Finn et al. 2008) | http://www.sanger.ac.uk/Software/Pfam/ | M/A |
| PIR | databases and tools for genomic and proteomic studies (Wu et al. 2007) | http://pir.georgetown.edu/ | A |
| PMut | server aimed at the prediction of pathological mutations using neural networks (Ferrer-Costa et al. 2005a,b) | http://mmb.pcb.ub.es/PMut/ | A |
| PRIDE | repository for proteomics data, which allows users to submit, retrieve and compare experimental data (Jones & Côté 2008) | http://www.ebi.ac.uk/pride/ | R |
| Prints | database of fingerprints characterizing protein families (Attwood 2002) | http://www.bioinf.manchester.ac.uk/dbbrowser/PRINTS/ | A |
| ProDom | database of protein domain families generated using SwissProt and TrEMBL sequences (Bru et al. 2005) | http://prodom.prabi.fr/ | A |
| Prosite | database of functional domains containing protein signatures (Hulo et al. 2006) | http://www.expasy.ch/prosite/ | A |
| ProtoNet | server which clusters proteins in order to predict structure and function (Kaplan N. et al. 2005) | http://www.protonet.cs.huji.ac.il/ | A |
| PupaSuite | Web tool focused on the analysis of SNPs (Conde et al. 2006) | http://pupasuite.bioinfo.cipf.es/ | A |
| SMART | database of functional domains based on profiles obtained through hidden Markov models from homologous sequences (Schultz et al. 1998) | http://smart.embl-heidelberg.de/ | A |
| Superfamily | database of functional domain assignments (at the SCOP superfamily level) for completely sequenced organisms (Gough et al. 2001) | http://supfam.cs.bris.ac.uk/SUPERFAMILY/ | A |
| TIGRFAMs | database of protein families collated and annotated using HMMs (Haft et al. 2003) | http://www.tigr.org/TIGRFAMs/index.shtml | A |
| TMHMM | prediction of transmembrane helices in proteins (Krogh et al. 2001) | http://www.cbs.dtu.dk/services/TMHMM/ | A |
| UniprotKB/SwissProt | database containing protein information features (The UniProt Consortium 2008) | www.ebi.ac.uk/swissprot/ | M |
| UniprotKB/TrEMBL | translated version of the EMBL database (The UniProt Consortium 2008) | http://www.ebi.ac.uk/TrEMBL/ | A |
| proteomic/structure | |||
| CATH | classification of protein domain structures mainly based on structural features (secondary structure, architecture and topology) and homology clustering (Greene et al. 2007) | http://www.cathdb.info/ | M/A |
| Genomic Threading Database | proteome annotation from structure folding recognition (McGuffin et al. 2004) | http://bioinf.cs.ucl.ac.uk/GTD/ | A |
| ModBase | database of three-dimensional models built by homology modelling (Pieper et al. 2006) | http://modbase.compbio.ucsf.edu/ | A |
| MoDEL | database containing molecular dynamics trajectories and their analysis (Rueda et al. 2007) | http://mmb.pcb.ub.es/MODEL/ | A |
| MSD | collection, management and distribution of data about macromolecular structures (Tagari et al. 2006) | http://www.ebi.ac.uk/msd/ | A |
| PDBsum | structural annotation of each three-dimensional structure deposited in the protein Data Bank (Laskowski et al. 2005a) | http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/ | A |
| PISA | tool to analyse PDB structures in order to predict the macromolecular interfaces and the quaternary state (Krissinel & Henrick 2007) | http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html | A |
| Procognate | database of cognate ligands for enzyme structures (Bashton et al. 2008) | http://www.ebi.ac.uk/thornton-srv/databases/procognate/ | A |
| ProFunc | identifies the likely biochemical function of a protein from its three-dimensional structure (Laskowski et al. 2005b) | http://www.ebi.ac.uk/thornton-srv/databases/ProFunc/ | A |
| RCSB PDB | atlas of three-dimensional protein structures into the PDB (Berman et al. 2002) | http://www.rcsb.org | R |
| SwissModel | server aimed at the construction of homology models (Schwede et al. 2003) | http://swissmodel.expasy.org/SWISS-MODEL.html | A |
| SCOP | structural classification of proteins based on evolutionary information and topology (Andreeva et al. 2004) | http://scop.mrc-lmb.cam.ac.uk/scop/ | M |
| wwPDB | repository aimed at maintaining a single protein Data Bank archive of macromolecular structural data (Berman et al. 2003) | http://www.wwpdb.org/index.html | R |
| other | |||
| ArrayExpress | database containing curated expression profiles (Parkinson et al. 2007) | http://www.ebi.ac.uk/microarray-as/aew/ | R |
| Babelomics | integrated system for performing different analyses on gene function (Al-Shahrour et al. 2006) | http://babelomics.bioinfo.cipf.es/ | A |
| Brenda | database containing enzyme functional information such as Km or substrates (Barthelmes et al. 2007) | http://www.brenda-enzymes.info/ | M/A |
| ChEBI | dictionary of small chemical compounds (Degtyarenko et al. 2008) | http://www.ebi.ac.uk/chebi/ | M/A |
| GEPAS | integrated system for performing different analyses on gene expression (Montaner et al. 2006) | http://gepas.bioinfo.cipf.es/ | A |
| GSCAN | server for the scanning of SNPs and QTLs in the genome (Valdar et al. 2006) | http://gscan.well.ox.ac.uk/ | A |
| IntAct | database containing molecular interaction data (Kerrien et al. 2007a) | http://www.ebi.ac.uk/intact/ | M |
| MACiE | database of enzymatic reactions (Holliday et al. 2007) | http://www.ebi.ac.uk/thornton-srv/databases/MACiE/ | M |
Furthermore, systems biology has started to put the genome and proteome in the context of the organism. Consequently, the expression levels of each transcript, the proteins involved in the regulation of their transcription and splicing and the generated interaction networks begin to be exhaustively studied and annotated. Also of special interest is the ChIP–chip methodology, which provides high-quality information about regulatory sequences in the DNA. Finally, not only the intra-individual systems have been studied, but also the evolutionary relationship between genes (orthology, in-paralogy and out-paralogy) is being included in the most important annotation databases.
Annotation can be approached in a number of ways: from manual curation of the literature to automatic methods. The latter are very diverse; some use the transfer of information from one characterized sequence to a homologue, while, in ab initio methods, features are predicted based on a set of derived rules. Over the last two decades, tools for the annotation of genomic and proteomic sequences and their structures have been developed and made accessible for others to use. This has added to a huge availability of characterized data. The databases that store these data often specialize in curating one particular area of annotation and are often most powerful when arranged in such a way in which the data can be probed computationally. For example, CATH (Orengo et al. 1997) is a database of protein structural domains where users can obtain a broad view of a chosen protein family or a narrower view of a particular protein structure.
Often, the tools that provide annotation also form the basis of the dataset that is represented in a given database. Over the last few years, there has been a move towards the integration of the wide range of genome and proteome annotation methods and databases in order to provide an overall view of the function of these genes (for an elegant project covering some of these points, see Fleming et al. 2006).
2. Types of annotation
Comprehensive protein feature annotation is an effective way to build up a picture of the function of the protein. Such features may include: for genes, expression levels, position of regulatory elements, binding sites, splicing junctions and individual variance; and, for proteins, position of functional residues, identification of post-translational modifications, description of residues that interact with DNA, protein or ligand, elucidation or prediction of the domain partners, description of the overall biological unit and even data describing the three-dimensional structure of the protein.
The annotation of such features can be carried out in three ways: first, manual curation from experimental data in the literature. This method provides the highest accuracy datasets; however, it relies on highly trained curators, is slow and can cover only a fraction of the data to be annotated. Alternatively, information can be derived by automatically transferring the knowledge we have about one sequence to a related sequence considered to be homologous. The accuracy of these methods depends on the evolutionary distance; the greater the distance, the less confidence you can have in accurately predicting a feature (Chothia & Lesk 1986; Wilson et al. 2000). In addition, the number of shared functional domains is also critical to the annotation transference reliability (Hegyi & Gerstein 2001). Consequently, greater coverage of the transfer of information usually results in decreasing the accuracy due to inference from more distant homologues. Finally, some annotations can be predicted using ab initio methods, which use rules trained on previous annotations or the physico-chemical properties of the molecule to predict the feature. The Rosetta method (Das et al. 2007) predicts the fold of a protein by calculating the energy for different conformations of the protein structure. In general, when choosing an annotation method, one must weigh up the often competing demands of speed and accuracy. Manual curation or experimental methods have high accuracy but are time consuming, and are probably more appropriate for small datasets. Methods that produce annotations with higher speed and coverage (e.g. transfer by homology, many ab initio methods) often do so with lower accuracy, but such a trade-off may be reasonable where datasets are large.
2.1 Manual curation
Manually curated data resources are created by human eye and, as a result, data are substantiated by highly trained and knowledgeable annotators. Through this process, annotators are able to survey and critically assess all the information available. In addition, annotators are able to access information buried deep in journal publications, which is not as accessible to more automated methods. Although labour-intensive and relatively slow compared with automatic annotation methods, manually annotated datasets provide an invaluable reliable reference resource that can provide an accurate ‘gold standard’ dataset from which users can base their annotation by similarity algorithm. A number of such data resources exist (table 1); however, we illustrate with just two examples: the Vega database (Wilming et al. 2008) and the UniprotKB/SwissProt database (Boeckmann et al. 2003; see below).
As well as providing a useful gold standard dataset, in many cases, manual curation represents the only way of extracting information from the literature and putting it into a format that can be queried in bulk or by a computer. This is certainly the case for the catalytic site atlas (CSA; Torrance et al. 2005), which was created for the very reason that catalytic residues were only ever reported in the literature and it was therefore impossible to extract computationally. Traditionally, information flows from the experimental laboratory into the literature, which provides the only reference to the data. These experimental data come from laboratories dedicated to that particular protein or family of proteins. For example, functional residues in a protein structure can be determined by site-directed mutagenesis (Grollman 1990) or location of a gene product in a cell can be located by fluorescence, electronic microscopy or radioactivity (Hoque & Cole 2008; Lewis et al. 2008; Usami et al. 2008).
Manual annotation provides us with the most accurate way of providing data flow from experimental studies reported in the literature into databases. Of course, manual annotation can also be slow. In order to try and speed up manual curation, text mining methods have been developed to extract this information automatically (Rebholz-Schuhman et al. 2007; Zweigenbaum et al. 2007; Cohen & Hunter 2008; Zhou & He 2008). However, as of yet, these methods are in their infancy. Of course, one very effective way of stopping the flow of data from experiment to literature is to change the deposition procedure such that incorporation into a database in a computer-readable format is also compulsory at the time of submission. In addition, this also means that the process must be user-friendly. Ideally, input fields would be free text, allowing the experimentalist to fully explain the nature of the new information. However, it is likely that a high level of freedom would create extra work for the database curators. A more manageable task might be created if experimentalists were to input into restricted fields—however, this still leaves the tasks of defining these fields in such a way that they are not so ambiguous that they result in an inconsistent dataset across different entries of the database. One example of this is the annotation of function. Function is associated with many mutually overlapping levels: chemical; biochemical; cellular; organism mediated; developmental; and physiological. Therefore, a simple definition of function could lead to a huge array of inconsistent information across the database entries. (The use of ontologies is described below.) Another sizeable problem is that of ensuring that the added data are of a high quality. In addition, it is also important to respect the information given in the original entry, and conflicts in data need to be dealt with carefully. Both the EMBL nucleotide database (Kulikova et al. 2007) in conjunction with GenBank (Kulikova et al. 2007; Benson et al. 2008) and DDBJ (Okubo et al. 2006) have recognized this and have begun to provide a service of this nature. This is done through an online submission procedure (WEBIN; Kulikova et al. 2007). Through the help of controlled input fields and pull-down lists, unambiguous annotations are produced. The WEBIN submission process has evolved over time so that specific information is required for each field. To obtain the final database entry, the submitter works closely with an EMBL curator so that the best annotation is achieved from the data.
2.1.1 Vega
The Vertebrate Genome Annotation database (Vega; http://vega.sanger.ac.uk; Wilming et al. 2008) provides high-quality manual annotation for 20 out of the 24 human chromosomes, four whole mouse chromosomes and approximately 40 per cent of the zebrafish Danio rerio genome. Vega also displays regions of significance from other vertebrate genomes, human haplotypes and mouse strains including the finished sequence and annotation of the major histocompatability complex from different human haplotypes, and mouse non-obese diabetes strain annotation of insulin-dependent diabetes candidate regions. The annotation process is slow and these datasets have been built up over many years; however, the result can be used as a trusted, standard data resource. For example, it has been used to provide the basis for integration between a number of organizations to form the Consensus Coding Sequence (CCDS) project (http://www.ncbi.nlm.nih.gov/CCDS/). This collaboration between the RefSeq group (Pruitt et al. 2007) at the NCBI, the Havana and the Ensembl groups at the EMBL and WTSI and the Genome informatics group at UCSC aims to provide a standardized, uniform set of protein-coding gene annotations across the human genome. Their goal is to provide a comprehensive annotation of coding and non-coding variants for each human and mouse CCDS locus to create a structured basis for a comparison with RefSeq. In addition to a high-quality dataset of gene structures, which can be used to predict gene structures on low-coverage genomes from other vertebrate species, the process of manual annotation can provide better results for the identification of polyadenylation features, non-coding genes, splice variants, pseudogenes and some of the more complex gene arrangements.
2.1.2 UniProtKB/SwissProt
UniProtKB/SwissProt (Boeckmann et al. 2003) is perhaps one of the most well-known manually curated data resources providing high-quality, well-structured database entries for almost 390 000 protein sequences (release 55.5 as of 10 June 2008). Gene sequences deposited into the EMBL nucleotide database are then translated and incorporated into TrEMBL (a databank of coding nucleic acid sequences translated into protein), which stores almost 6 000 000 sequences (release 38.5 as of 10 June 2008). A subsection of these are then manually curated and added to SwissProt.
The process of annotation can be described in a number of stages. First, the sequence is captured. SwissProt is a non-redundant database—each entry groups all peptides from a single gene—and therefore sequences are compared and discrepancies between them are noted. Each sequence then undergoes literature-based curation (where information is manually extracted from literature sources and added to the entry) and rigorous sequence analysis. Currently, database entries contain information from over 1400 different journals. All information added during the curation process is verified by expert biologists and therefore considered highly reliable. Information is added to the entry in a highly structured and uniform manner, making it easier to read computationally. Each line is added using an information type identifier; for example, RA, to annotate a reference, or GN, to annotate a gene name. Much of the functional nature of the entry is recorded using three such identifiers; FT identifier gives and records a description of a defined region, using a list of feature types, the CC line can contain a free-text comment, which is marked with a clearly defined CC topic (category), and, finally, a set of predefined keywords are carefully chosen, which best represent the entry. This enables maximum flexibility on how these data are used. The highly defined format is essential for the use of this data resource, and allows for very little ambiguity between entries and also allows maximum capacity for the data to be manipulated computationally.
Such a task needs highly trained and knowledgeable curators. There are now 120 curators spread over the European Bioinformatics Institute (EBI) and the Swiss Institute of Bioinformatics. The SwissProt team has developed a number of resources to streamline the annotation process. Often curators within the SwissProt team will have specific families which they always curate, thus speeding up the process of knowing what to look for and also making it more accurate with the curator having a bank of knowledge already about that family. In addition, curators have a number of tools available to them to help the annotation process. These tools are bound together via a text editor with a powerful C-like macro language, Crisp (Boeckmann et al. 2003), which has been manipulated to provide a platform for highly formatted textual annotation and has the ability to launch a number of sequence analysis tools. During the curation process, a number of sequence analysis tools are used to help annotate such features as signal sequences, transmembrane domains, coiled coil domains and N-glycosylation sites, to name a few. Once an entry is created, it is checked with a syntax checker in order to highlight any inconsistencies in the format or mistakes in the controlled vocabulary fields of the entry. It has also become possible to provide some methods of automatic annotation on SwissProt sequences with high accuracy. The first automatic annotation project was that of high-quality automated and manual annotation of microbial proteins (HAMAP), in which information was transferred from manually annotated proteins to homologues of complete bacterial and archaeal proteomes and based on a set of manually curated rules (Gattiker et al. 2003). UniProt has begun to mine specialist knowledge from expert communities in their annotation procedure, with a pilot scheme to involve the yeast consortium in the annotation of yeast proteins. It is hoped that this scheme can be extended to include other specialist communities in the future (http://www.uniprot.org/news/2007/09/11/release).
2.2 Automatic annotation
Once high-quality information has been transferred into a database in a computationally interpretable way, it is possible, in many cases, to transfer these annotations to homologous genes in order to provide increased annotation coverage. For example, some genomes can be annotated by comparison—much of the gene structure of the chimpanzee can be elucidated by comparison with the human genome (Chimpanzee Sequencing and Analysis Consortium 2005). However, the most successful transfer can be achieved at the proteomic level once the three-dimensional protein structure is known, as protein structure is much more conserved than sequence during evolution (Chothia & Lesk 1986). Again, this is an equivocal process since phenomena including alternative splicing or single nucleotide polymorphisms (SNPs) can alter the structure and function of the proteins (Wen et al. 2004; Hiller et al. 2005; Stetefeld & Ruegg 2005). All these processes involve the most important step of identifying homologues.
Although homology is an old morphological term, which implies an evolutionary divergence from a common ancestor (e.g. mammal and fish bodily extremities), in functional annotation, this word is sometimes not properly used (Petsko 2001). Indeed, many of the so-called ‘by homology’ annotations should be renamed as ‘by similarity’ since they are not based on evolution but on resemblance, e.g. all annotations provided by tools using similarity searches to transfer information. However, as homology is an extremely complex term, beyond the goal of this review, we would recommend the reader to look at the useful explanations given by Fitch (2000), while we use the common assumption that very similar sequences or structures are homologous.
Proteins that have evolved from a common ancestor are often found to share a related structure, function and sequence. The comparison of protein structures has the capability to identify very distant relationships between protein sequences. However, we do not know the three-dimensional structure of every sequence. Instead, the relationships hidden within sequence space must usually be teased out through the use of computational sequence comparison methods. Measuring sequence similarity to infer the evolutionary distance between proteins is a fundamental tenet of structural biology and has been drawn on to organize sequence space into clusters of proteins that have diverged from a common ancestor and therefore share a common protein fold. In order to detect as many related sequences as possible, powerful sequence comparison methods have been developed, such as PSI-BLAST (Altschul et al. 1997) and hidden Markov models (HMMs; Eddy 1996), which use sequence profiles built up of groups of related sequences to identify remote protein homologues.
Other automatic methods involve the development of rules based on the analyses of previously characterized data. The most interesting thing about these methods is that they do not require the existence of annotated homologous relatives. Rosetta from the Baker laboratory is one of the most popular and successful tools using ab initio calculations for protein structure (Das et al. 2007). The method works by finding local common conformations of small residue stretches. These local conformations are then refined together on the tertiary structure by minimizing the free energy. This method has been extended to predict protein–protein interactions by modelling thousands of conformers, which are then ranked using van der Waals, solvation and H-bond energies (Gray et al. 2003). Ab initio methodologies have also been successfully used to predict other features such as gene promoters based on stiffness and helical deformation of nucleic acids (Goñi et al. 2007) and transcription factor binding sites from DNA–amino acid interaction preferences (Kaplan, T. et al. 2005). For an extensive review on ab initio and comparative genomics methods, see Jones (2006).
A number of genome annotation sources have been created, including the UCSC genome browser (Karolchik et al. 2008) and the tools provided at the NCBI (Wheeler et al. 2007). The Ensembl pipeline (Curwen et al. 2004) provides automatic annotation of eukaryotic genomes for predicting gene structures (as well as providing other annotations such as homology mapping between species and mapping to other data resources such as expression arrays). The pipeline is a suite of programs built from observing how annotators build gene structures. The pipeline uses information from known proteins, cDNA and EST sequences. First, species-specific known protein sequences are taken from UniprotKB/SwissProt, UniprotKB/TrEMBL and RefSeq. Then, in order to increase coverage, proteins from other organisms are matched using different thresholds. In parallel, to increase coverage further, full-length cDNAs (Stoesser et al. 1998; Pruitt & Maglott 2001; Okazaki et al. 2002) are aligned to the genomic sequence. Annotation of this kind provides fast and high coverage for annotations of genomes and, as such, the genes on 39 species (release 49) have been annotated. The results of these analyses can be viewed on the Ensembl Web browser as part of the Ensembl project (Flicek et al. 2008), a comprehensive genome information portal providing an integrated set of genome annotation for chordate, disease vector genomes and a number of selected model organisms.
Protein sequences in TrEMBL are annotated using an automated annotation pipeline of three programs. The first is Rulebase, which uses a number of manually curated annotation rules. For example, looking at eukaryotic protein sequences from the Interpro family IPR000685, it is found that 434 out of the 436 have chloroplast as a UniProt keyword. As a result, one can have good confidence in applying the ‘chloroplast’ keyword to any non-annotated eukaryotic protein sequences which have clustered into this Interpro family. It is a powerful technique. However, the manual generation of rules is time consuming and therefore the second program, Spearmint, generates similar rules automatically (Kretschmann et al. 2001). This program works in conjunction with the third in this suite of programs, Xanthippe, which aims to remove false positives and erroneous imports from other databases by generating rules to predict the absence of annotations, a ‘contradiction’ program rather than a prediction program (Wieser et al. 2004).
3. Annotation of phenotypes
Now that parts of the major genomes are annotated to a high quality, more attention has been turned to the annotation of allele-specific information and the differences between the genomes of individuals of the same species. For example, workers at the WormBase database have recently provided their users with details of all sequenced alleles described in papers published since 2001 (Rogers et al. 2008). Variation occurs by the presence of SNPs, which, in humans, occur every 500–1000 bp and are among the most common types of genetic variation. These are associated with altered response to drug treatment, susceptibility to disease and other phenotypic variation. These types of analyses highlight regions of the genome, which are highly variable between individuals, and could lead to differences in phenotype. This type of information is integrated into the major databases (e.g. UniProt and Ensembl); however, there are also other specific databases, for example dbSNP (Smigielski et al. 2000), a catalogue of variations from the National Center for Biotechnology Information. Other databases focus on the provision of annotation of the effect of these SNPs such as SNPeffect (Reumers et al. 2005).
There are a number of locus-specific databases (LSDBs) conveying such information for particular genes. Some groups have collected variation information for particular genes of interest. This information is particularly important for the elucidation of disease and integration of these sources would allow the creation of a catalogue of variation within the human genome. The federation of LSDBs was set up in order to ascertain the best mode of collecting and curating accurate lists of mutations. This was followed by an analysis of the characteristics of 94 LSDBs on the basis of 80 content criteria (Claustres et al. 2002). In addition, the Human Genome Variation Society aims to promote collection, documentation and free distribution of genomic variation information. With this in mind, work has begun on providing a catalogue of variation within the human genome. The elucidation of two individual human genomes (Levy et al. 2007; Wheeler et al. 2008) and now the beginning of the 1000 genomes project (Siva 2008) aims to cover variation in the human genome.
The study of variation in complex phenotypes, where a phenotype is determined by the expression of several genes, is also being considered. These genes are called quantitative trait loci (QTLs; Reuveni et al. 2007), with each locus contributing only a small amount to the eventual phenotype (Flint & Mott 2001; Mott 2006). Such studies have identified and narrowed the genetic regions that control particular phenotypes involved in metabolic and immunological processes (Valdar et al. 2006; Liu et al. 2007) and particular phenotypes responsible for behavioural responses (Malmanger et al. 2006; Valdar et al. 2006; Liu et al. 2007). Other studies have explored the influence of environment or inbreeding on the phenotypical variations (Solberg et al. 2006). As many of the most common human diseases are caused by a polygenic effect, these sorts of studies will be crucial to find new drug targets (Rollins et al. 2006).
Information conveying genetic mutations and their effect on the phenotype is essential for understanding inherited diseases. In contrast to SNPs, which have tolerated phenotypic effects, some of these mutations are extremely deleterious (e.g. the mutations which go on to cause cancer). There are several approaches to the study of the phenotypic effects of the mutation: using gene information (e.g. presence of TF binding sites or splice junctions; Conde et al. 2004); using artificial intelligence tools to score protein features such as accessibility, secondary structure or number of specific residues (Ferrer-Costa et al. 2004, 2005a,b; Capriotti et al. 2006; Bromberg & Rost 2007); or combining both strategies (Conde et al. 2005; Karchin et al. 2005). In order to increase the amount of data, it has been successfully tested using cross-species prediction. This involves training the tool using data from one species and predicting the effect on another one (Ferrer-Costa et al. 2005b).
In addition, as more genome data are released, there have been initiatives to annotate a catalogue of human cancer genes and mutations (Futreal et al. 2004; Sjöblom et al. 2006; Greenman et al. 2007; Wood et al. 2007). This process typically involves two steps. The first step is known as the discovery screen, in which all coding exons are used to design primers to amplify DNA from tumour and normal samples from the same individual. After assembling the PCR results, the mutations are analysed both computationally and manually, discarding all changes appearing in normal samples and in SNP databases. The remaining genes are resequenced to verify that any changes are not an artefact (Sjöblom et al. 2006; Greenman et al. 2007; Wood et al. 2007). The second stage is the validation screen, which involves using other tumour lines to amplify and sequence genes that have non-synonymous mutations on the discovery screen. Using a similar protocol to the first stage, the discovery screen, the validation screen focuses on a set of genes instead of the whole genome (Sjöblom et al. 2006; Wood et al. 2007). However, these censuses contain driver and passenger mutations, as it is impossible to differentiate which mutations are causative and which ones are the product of speed replication cycles. Some attempts have been made to statistically predict causative genes or mutations. They use the number of mutations per gene and the synonymous/non-synonymous ratio to perform these predictions (Sjöblom et al. 2006; Greenman et al. 2007; Wood et al. 2007). Another successful approach to obtaining a catalogue of human cancer genes has involved a gene census compiled from the literature (Futreal et al. 2004). They collected genes for which at least two independent reports of somatic mutations, chromosomal rearrangements or copy number alterations were available. Finally, this protocol was used to create the Catalogue Of Somatic Mutations In Cancer (COSMIC; Forbes et al. 2008), which in its 38th release contains almost 60 000 mutations. Among the biggest resources covering genomic data are those from the Genomics Institute of the Novartis Research Foundation, which include several databases such as SymAtlas, a database of gene expression (formerly human and mouse genes, but now extended to other species; Su et al. 2004), and SNPview, a database containing SNPs from 48 genotyped mouse strains (Pletcher et al. 2004) and an interface to search human druggable genes (Orth et al. 2004).
4. Integration of annotations
As we have described above, many annotation methods exist for genomic and proteomic data, often spread throughout the world. It is impossible to query all these resources at once and interpretation of results from each site is different. Therefore, the user must learn to query and interpret each method separately. Furthermore, these methods often change locations as laboratories change their geographical sites. Accordingly, a new challenge has arisen: to integrate all these different sources of data into a single comprehensive source. In order for this to be possible, it is important that all databases communicate using the same language. For example, methods need to use the same term names to describe their annotations. In addition to this, there needs to be an appropriate infrastructure to facilitate the provision and display of annotations from these disparate laboratories.
4.1 Infrastructure
One such method that provides the infrastructure is the distributed annotation system (DAS; Dowell et al. 2001), a method which comprises a reference server that contains the information for other servers to ‘refer’ to. DAS is a client–server system in which clients are configured to read and interpret data from multiple servers. An example in this case would be a UniProt sequence. Each partner site then supplies their annotations, which refer to that sequence in a particular XML format. Interpretation and display of the information from all servers (reference and annotation) is done by a DAS client, e.g. Dasty2 (Jimenez et al. 2008), Spice (Prlic et al. 2005), the Ensembl genome browser (Spudich et al. 2007) and the Pfam DAS alignment viewer (Finn et al. 2008). The information that the client interprets is provided by the servers organized into ‘reference servers’ that hold specific information such as the sequence to relate one entry to another on a physical map and multiple sites then act as ‘annotation servers’ providing as few or as many annotations on a segment of the reference. This method allows great flexibility in information provided and not only can information be integrated from a number of different laboratories into one central site but also any research group can view annotations provided in conjunction with their own data. Contrary to more traditional set-ups, in DAS, it is the client that does all of the interpretation of information (the smart system), and the servers merely provide the information in a computer-readable format. This contrasts with the more widely used format where the data source provides and interprets the data. The third compartment is the registry (Prlic et al. 2007), which provides a catalogue of all servers available. After its invention in 2001, the system was widely adopted by the genomic annotation community and forms a major part of the Ensembl infrastructure. It was then adopted by the proteomics world in 2005 and there now exist approximately 400 DAS servers currently registered with the DAS registry. Today, this method is heavily used in the popular genome browsers such as Ensembl, which provides approximately 200 different DAS sources, WormBase (Stein et al. 2001) and GBrowse (Stein et al. 2002). This circumvents the need for centralized control over the data and centralized database archives do not need to set aside time and resources to resolve contradictions between different third-party annotations as all information is reported, leaving the user to interpret the results.
Integration can also be facilitated by providing annotation programs in such a way that others are able to run them remotely on their own datasets; this can be done by providing software as a Web service (Curcin et al. 2005; Labarga et al. 2007). As a Web service, the user is able to use the program however they like and even incorporate it into a workflow (Kappler 2008; a concatenation of several annotation tools, each one using the results from the last as the input to the next, examples include Triana and Taverna; Hull et al. 2006). These tools comprise a WSDL file (Web service definition language, an XML schema), which describes the Web service and how to access it, and a CGI script returning results via Simple Object Access Protocol (SOAP) or Representational State Transfer (REST) information transference protocols. The Web service is based on a ‘computer-to-computer’ communication, and therefore it does not need any Web interface and can be used on the command line, requiring only a client script (e.g. a PERL script) by the user side providing a greatly portable and accessible tool.
4.2 Integration of terms
An important step of integration is the need for the development of a common language by which all methods can communicate. As for both genomic and proteomic annotations, standardization of the nomenclature is fundamental to the success of integration. Without a common language, it is impossible for users to interpret the data manually or computationally. This can be illustrated by looking at the term ‘function’, which can have a variety of meanings. It can be used to describe the biochemical role of the residues carrying out the function, i.e. hydrolysis, or it can mean the overall function of the domain (ATP binding) or, in fact, the full function of the protein, e.g. glutathione reductase. The overall function of the protein can also be related to its biological process or cellular location. The need for standardization has been recognized by centrally managed databases as an important feature. For example, in UniProt, curators are highly trained in the format of the entry. Each one is controlled by a number of predefined descriptions or headings, which head free-text fields to allow easy retrieval of specific topics; there are 941 keywords (by early September 2008) for curators to pick from which summarize the content of an entry. This allows uniformity in the curation of different entries and also the ability to manage the data computationally.
Vega has been instrumental in the classification and standardization of annotation terms used by the community. This is particularly important when comparing haplotypes or syntenic regions. The standardization aids comparative analysis of orthologues across the different finished regions. In order to provide the platform for integration and comparison, Vega communicates with the nomenclature committees from the Human Genome Organisation (HGNC; Bruford et al. 2008), ZFIN (Sprague et al. 2008) and MGD (Eppig et al. 2007). This collaboration has standardized the nomenclature associated with transcribed regions allowing the user to interpret the evidence (cDNA, EST or protein sequences) associated with the data. These transcribed regions are associated with one of five categories that range from the most to least confidence: ‘known genes’, which are identical to human cDNA or protein sequences; ‘novel genes’, which have an open reading frame (ORF) and are identical or homologous to known cDNAs (vertebrates) and/or proteins (all species); ‘novel transcripts’, which are similar to novel genes but it has not been possible to assign an ORF; ‘putative genes’, which are homologous to spliced ESTs (vertebrates) but do not have a significant ORF/CDS; and ‘pseudogenes’, which are sequences homologous to proteins (over 50% of the subject length) with a disrupted CDS and for which an active gene can generally be found at another locus.
4.2.1 Ontologies
A further step is to provide a common language with a standardized relationship between the terms in the language in the form of an ontology. This need for a unified language to describe many different areas within biology has now been widely recognized with the development of a number of ontologies; there are now over 60 ontologies listed on the EBI Ontology Lookup Service (http://www.ebi.ac.uk/ontology-lookup/). An ontology comprises a unique alphanumerical identifier, a common name, synonyms (if applicable) and a definition. These terms and definitions are clustered together by drawing relationships between them; such relationships can be described as ‘is_a’ or ‘part_of’ providing users with not only a controlled vocabulary with common terms and meanings but also relationships that can be computationally inferred.
The gene ontology (GO; Ashburner et al. 2000) provides a comprehensive controlled vocabulary to describe gene product attributes. It is divided into three major sections: the molecular function of the gene product; the role it has in multi-step biological processes; and the location within the cellular components. There are now 25 264 terms in this ontology, and it has been used for a variety of purposes in approximately 530 publications in 2007 alone. In 2001, UniProt became a member of the GO consortium and initiated the GOA project (Camon et al. 2004). They provided a dedicated database curation team for the assignment of GO terms to well-characterized proteins in UniProtKB/SwissProt. For example, the 941 keywords in UniProt have been manually mapped to GO terms.
A project within the GO Consortium is the Sequence Ontology (Eilbeck et al. 2005). This is an ontology describing the parts of a genomic annotation, which has been developed to facilitate exchange, analysis and management of genomic data. This standard has been used to support the features stored in the sequence databases of model organisms (Mungall & Emmert 2007) and to standardize the annotation exchange formats (GFF3 specification). Many model organism communities such as WormBase (Rogers et al. 2008), FlyBase (Grumbling & Strelets 2006), SGD (Christie et al. 2004) and DictyBase (Chisholm et al. 2006) use it to annotate their sequences, and a recent addition to this ontology are the terms that describe features on proteomic sequences and structures (Reeves et al. 2008), which is being used to standardize annotations provided by the BioSapiens NoE (The BioSapiens Network of Excellence 2005). Ontologies have also been created to provide a similar role for other aspects of biology, an ontology for molecular interactions (Kerrien et al. 2007a,b), pathways (Twigger et al. 2007), post-translational modifications (Montecchi-Palazzi 2008), to name a few.
4.3 Collaborations for the integration of annotations
When the concept of e-Science was first introduced, the need for collaboration within a number of scientific disciplines was recognized (bioinformatics, chemistry, engineering, healthcare, particle physics and astronomy) in order to provide greater integration (Hey & Trefethen 2003). For bioinformatics, the myGrid project was launched by a consortium comprising the Universities of Manchester, Southampton, Nottingham, Newcastle, Sheffield, the European Bioinformatics Institute and industrial partners GSK, AstraZeneca, IBM and SUN. It was launched in order to develop an integrated infrastructure to provide tools for automatic annotation and provenance tracking. Since then, a huge number of collaborative projects in all areas of bioinformatics have been taken on.
One particular project, the BioSapiens Network of Excellence, is currently nearing its end and, as a result, its benefits and achievements can be examined. The project was heavily influenced by two other projects: e-protein (http://www.e-protein.org/), which aimed to provide an automated and distributed pipeline for structural and functional annotation of all major proteomes using GRID technologies; the use of a large number of computers, often in varied geographical locations, in concert to perform very large tasks and pioneered the use of DAS for protein sequences. The BioSapiens Network of Excellence was a direct follow on and its main goal was to create a ‘Virtual Institute of Annotation’. This has been achieved through the use of the DAS infrastructure with the integration of 69 different distributed annotation sources from 19 partner sites. In addition to this, they have tackled scientific and data integration from a different angle.
Members of the consortium have come together to provide a joint analysis of a number of different datasets. One particular example is their involvement in the ENCODE project (The ENCODE Project Consortium 2004; Birney et al. 2007). This ongoing project aims to identify all functional elements in the human genome sequence and was designed in three stages: pilot phase in which methods and combinations of methodologies can be evaluated on a released dataset of 1 per cent of the genome (some random and some manually picked regions); technology development phase in which new laboratory and computational methods will be designed; and the production phase. The pilot phase was launched in September 2003 initially with the funding of eight groups with expertise in existing technologies for the detection of a variety of functional elements including gene promoter repressors and exons. However, as an open consortium, results on the pilot phase were collated from 35 groups including the BioSapiens Network of Excellence. This analysis has provided more than 200 experimental and computational datasets in unprecedented detail of annotation on this 30 Mb dataset of the human genome. The overall findings of this pilot phase have been published and include some major findings. A major conclusion of this study challenges the view that the genome could be annotated as a ‘dictionary of conserved genomic elements each with an annotation about their biochemical function’ (The ENCODE Project Consortium 2004; Birney et al. 2007). Instead, it was found by numerous groups that intercalated transcripts spanning the majority of the genome existed (Tress et al. 2007). Previous analysis has shown a similar broad amount of transcription across the human (Bertone et al. 2004; Cheng et al. 2005) and mouse (Carninci et al. 2005) genomes. There were mixed opinions about the biological importance and the question remains unanswered. However, the presence of these transcribed elements was indeed established.
5. Future directions
Although the annotation field has advanced very fast in the last decade, there are still many remaining challenges, such as the role of the expressed non-coding RNA (Huttenhofer et al. 2005; Mendes Soares & Valcarcel 2006; Yazgan & Krebs 2007; Yazaki et al. 2007; Amaral et al. 2008), de novo biological functional prediction of proteins (Watson et al. 2005) or the systemic integration of annotated components (Ge et al. 2003; Reed et al. 2006). All these areas clearly cross the traditional borders of the genome and proteome annotation and go further through the systems biology field. Probably, in the following years, it will be necessary to merge the purely annotation work and the more basic research in order to succeed.
Furthermore, the integrative part of the annotation must continue on progress, inside the bioinformatics area and influencing the experimentalists. Consequently, some things could be done to facilitate the integration of experimental results: (i) in addition to ontologies, some other technical standards should be agreed to facilitate the data linkage between resources, (ii) simple and unequivocal forms should be created for the introduction of data from biological experiments, and (iii) data deposition should be a condition for publication.
6. Conclusions
Given the plethora of annotation tools that have been developed over the past few years (figure 1 and table 1), the next goal has been to try to find ways in which the information that these annotation tools provides can be integrated. This has begun to be achieved with the advent of tools to help infrastructure (DAS, Web services and widgets) and increased funding into integrative projects (such as e-Protein and BioSapiens). To further this, we need to extend our collaborations between the genomic and proteomic disciplines.
Acknowledgments
We gratefully thank Daniel Andrews, Roman Laskowski and Daniela Wieser for their helpful hints and comments. This work was completed as part of the BioSapiens Network of Excellence, funded by the European Commission within its FP6 Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health’, contract number LHSG-CT-2003-503265.
References
- Adams M.D., et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science (NY) 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F., et al. Babelomics: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral P.P., Dinger M.E., Mercer T.R., Mattick J.S. The eukaryotic genome as an RNA machine. Science (NY) 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Andreeva A., Howorth D., Brenner S.E., Hubbard T.J.P., Chothia C., Murzin A.G. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., et al. Gene ontology: tool for the unification of biology. Gene Ontol. Consort. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwood T.K. The Prints database: a resource for identification of protein families. Brief. Bioinform. 2002;3:252–263. doi: 10.1093/bib/3.3.252. [DOI] [PubMed] [Google Scholar]
- Barthelmes J., Ebeling C., Chang A., Schomburg I., Schomburg D. Brenda, Amenda and Frenda: the enzyme information system in 2007. Nucleic Acids Res. 2007;35:D511–D514. doi: 10.1093/nar/gkl972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashton M., Nobeli I., Thornton J.M. Procognate: a cognate ligand domain mapping for enzymes. Nucleic Acids Res. 2008;36:D618–D622. doi: 10.1093/nar/gkm611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bendtsen J.D., Kiemer L., Fausbøll A., Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005a;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J.D., Nielsen H., Widdick D., Palmer T., Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinform. 2005b;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Ostell J., Wheeler D.L. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., et al. The protein Data Bank. Acta Crystallogr. D: Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/S0907444902003451. [DOI] [PubMed] [Google Scholar]
- Berman H., Henrick K., Nakamura H. Announcing the worldwide protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Bertone P., et al. Global identification of human transcribed sequences with genome tiling arrays. Science (NY) 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Birney E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Blom N., Hansen J., Blaas D., Brunak S. Cleavage site analysis in picornaviral polyproteins: discovering cellular targets by neural networks. Protein Sci. 1996;5:2203–2216. doi: 10.1002/pro.5560051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Boeckmann B., et al. The Swiss-Prot protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S.E. A tour of structural genomics. Nat. Rev. 2001;2:801–809. doi: 10.1038/35093574. [DOI] [PubMed] [Google Scholar]
- Brent M.R. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat. Rev. 2008;9:62–73. doi: 10.1038/nrg2220. [DOI] [PubMed] [Google Scholar]
- Brett D., Pospisil H., Valcárcel J., Reich J., Bork P. Alternative splicing and genome complexity. Nat. Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- Bromberg Y., Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru C., Courcelle E., Carrère S., Beausse Y., Dalmar S., Kahn D. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruford E.A., Lush M.J., Wright M.W., Sneddon T.P., Povey S., Birney E. The HGNC database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K. An overview of structural genomics. Nat. Struct. Biol. 2000;7:932–934. doi: 10.1038/80697. [DOI] [PubMed] [Google Scholar]
- Camon E., et al. The gene ontology annotation (GOA) database: sharing knowledge in UniProt with gene ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Calabrese R., Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics (Oxf.) 2006;22:2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- Carninci P., et al. The transcriptional landscape of the mammalian genome. Science (NY) 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Casadio R., Martelli P.L., Pierleoni A. The prediction of protein subcellular localization from sequence: a shortcut to functional genome annotation. Brief. Funct. Genom. Proteom. 2008;7:63–73. doi: 10.1093/bfgp/eln003. [DOI] [PubMed] [Google Scholar]
- Castrignano T., et al. ASPicDB: a database resource for alternative splicing analysis. Bioinformatics (Oxf.) 2008;24:1300–1304. doi: 10.1093/bioinformatics/btn113. [DOI] [PubMed] [Google Scholar]
- Caudevilla C., Serra D., Miliar A., Codony C., Asins G., Bach M., Hegardt F.G. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc. Natl Acad. Sci. USA. 1998;95:12 185–12 190. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science (NY) 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Chisholm R.L., Gaudet P., Just E.M., Pilcher K.E., Fey P., Merchant S.N., Kibbe W.A. DictyBase, the model organism database for Dictyostelium discoideum. Nucleic Acids Res. 2006;34:D423–D427. doi: 10.1093/nar/gkj090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Lesk A.M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie K.R., et al. Saccharomyces genome database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark F., Thanaraj T.A. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Genet. 2002;11:451–464. doi: 10.1093/hmg/11.4.451. [DOI] [PubMed] [Google Scholar]
- Claustres M., Horaitis O., Vanevski M., Cotton R.G.H. Time for a unified system of mutation description and reporting: a review of locus-specific mutation databases. Genome Res. 2002;12:680–688. doi: 10.1101/gr.217702. [DOI] [PubMed] [Google Scholar]
- Cohen K.B., Hunter L. Getting started in text mining. PLoS Comput. Biol. 2008;4:e20. doi: 10.1371/journal.pcbi.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde L., Vaquerizas J.M., Santoyo J., Al-Shahrour F., Ruiz-Llorente S., Robledo M., Dopazo J. PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res. 2004;32:W242–W248. doi: 10.1093/nar/gkh438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde L., Vaquerizas J.M., Ferrer-Costa C., de la Cruz X., Orozco M., Dopazo J. PupasView: a visual tool for selecting suitable SNPs, with putative pathological effect in genes, for genotyping purposes. Nucleic Acids Res. 2005;33:W501–W505. doi: 10.1093/nar/gki476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde L., Vaquerizas J.M., Dopazo H., Arbiza L., Reumers J., Rousseau F., Schymkowitz J., Dopazo J. PupaSuite: finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res. 2006;34:W621–W625. doi: 10.1093/nar/gkl071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcin V., Ghanem M., Guo Y. Web services in the life sciences. Drug Discov. Today. 2005;10:865–871. doi: 10.1016/S1359-6446(05)03481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curwen V., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M.J., Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69(Suppl. 8):118–128. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- Degtyarenko K., et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R., Reuter G., Loewendorf A. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc. Natl Acad. Sci. USA. 2001;98:9724–9729. doi: 10.1073/pnas.151268698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell R.D., Jokerst R.M., Day A., Eddy S.R., Stein L. The distributed annotation system. BMC Bioinform. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckert P., Brunak S., Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 2004;17:107–112. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- Dunham I., et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Hidden Markov models. Curr. Opin. Struct. Biol. 1996;6:361–365. doi: 10.1016/S0959-440X(96)80056-X. [DOI] [PubMed] [Google Scholar]
- Eilbeck K., Lewis S.E., Mungall C.J., Yandell M., Stein L., Durbin R., Ashburner M. The sequence ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Eppig J.T., Blake J.A., Bult C.J., Kadin J.A., Richardson J.E. The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35:D630–D637. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Costa C., Orozco M., de la Cruz X. Sequence-based prediction of pathological mutations. Proteins. 2004;57:811–819. doi: 10.1002/prot.20252. [DOI] [PubMed] [Google Scholar]
- Ferrer-Costa C., Gelpí J.L., Zamakola L., Parraga I., de la Cruz X., Orozco M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics (Oxf.) 2005a;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- Ferrer-Costa C., Orozco M., de la Cruz X. Use of bioinformatics tools for the annotation of disease-associated mutations in animal models. Proteins. 2005b;61:878–887. doi: 10.1002/prot.20664. [DOI] [PubMed] [Google Scholar]
- Finn R.D., et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W.M. Homology a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/S0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Fleming K., Kelley L.A., Islam S.A., MacCallum R.M., Muller A., Pazos F., Sternberg M.J. The proteome: structure, function and evolution. Phil. Trans. R. Soc. B. 2006;361:441–451. doi: 10.1098/rstb.2005.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P., et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Mott R. Finding the molecular basis of quantitative traits: successes and pitfalls. Nat. Rev. 2001;2:437–445. doi: 10.1038/35076585. [DOI] [PubMed] [Google Scholar]
- Forbes, S. A. et al 2008 The catalogue of somatic mutations in cancer (COSMIC). Current Protocols in Human Genetics, ch. 10, unit 10.11. Somerset, NJ: John Wiley & Sons. [DOI] [PMC free article] [PubMed]
- Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattiker A., et al. Automated annotation of microbial proteomes in Swiss-Prot. Comput. Biol. Chem. 2003;27:49–58. doi: 10.1016/S1476-9271(02)00094-4. [DOI] [PubMed] [Google Scholar]
- Ge H., Walhout A.J., Vidal M. Integrating ‘omic’ information: a bridge between genomics and systems biology. Trends Genet. 2003;19:551–560. doi: 10.1016/j.tig.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Goñi J.R., Pérez A., Torrents D., Orozco M. Determining promoter location based on DNA structure first-principles calculations. Genome Biol. 2007;8:R263. doi: 10.1186/gb-2007-8-12-r263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J., Karplus K., Hughey R., Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Gray J.J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C.A., Baker D. Protein–protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 2003;331:281–299. doi: 10.1016/S0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- Greene L.H., et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C., et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman A.P. Site specific mutagenesis. Prog. Clin. Biol. Res. A. 1990;340:61–70. [PubMed] [Google Scholar]
- Grumbling G., Strelets V. FlyBase: anatomical data, images and queries. Nucleic Acids Res. 2006;34:D484–D488. doi: 10.1093/nar/gkj068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Brunak S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002;7:310–322. [PubMed] [Google Scholar]
- Gupta R., Jung E., Gooley A.A., Williams K.L., Brunak S., Hansen J. Scanning the available Dictyostelium discoideum proteome for O-linked GlcNAc glycosylation sites using neural networks. Glycobiology. 1999;9:1009–1022. doi: 10.1093/glycob/9.10.1009. [DOI] [PubMed] [Google Scholar]
- Gupta S., Zink D., Korn B., Vingron M., Haas S.A. Strengths and weaknesses of EST-based prediction of tissue-specific alternative splicing. BMC Genom. 2004;5:72. doi: 10.1186/1471-2164-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D.H., Selengut J.D., White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi H., Gerstein M. Annotation transfer for genomics: measuring functional divergence in multi-domain proteins. Genome Res. 2001;11:1632–1640. doi: 10.1101/gr.183801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey T., Trefethen A. e-Science and its implications. Phil. Trans. R. Soc. A. 2003;361:1809–1825. doi: 10.1098/rsta.2003.1224. [DOI] [PubMed] [Google Scholar]
- Hiller M., Huse K., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- Hiller M., Huse K., Platzer M., Backofen R. Creation and disruption of protein features by alternative splicing—a novel mechanism to modulate function. Genome Biol. 2005;6:R58. doi: 10.1186/gb-2005-6-7-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday G.L., Almonacid D.E., Bartlett G.J., O'Boyle N.M., Torrance J.W., Murray-Rust P., Mitchell J.B.O., Thornton J.M. MACiE (mechanism, annotation and classification in enzymes): novel tools for searching catalytic mechanisms. Nucleic Acids Res. 2007;35:D515–D520. doi: 10.1093/nar/gkl774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M.T., Cole S.P.C. Down-regulation of Na+/H+ exchanger regulatory factor 1 increases expression and function of multidrug resistance protein 4. Cancer Res. 2008;68:4802–4809. doi: 10.1158/0008-5472.CAN-07-6778. [DOI] [PubMed] [Google Scholar]
- Horiuchi T., Aigaki T. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol. Cell (under the auspices of the European Cell Biology Organization) 2006;98:135–140. doi: 10.1042/BC20050002. [DOI] [PubMed] [Google Scholar]
- Hull D., Wolstencroft K., Stevens R., Goble C., Pocock M.R., Li P., Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N., Bairoch A., Bulliard V., Cerutti L., De Castro E., Langendijk-Genevaux P.S., Pagni M., Sigrist C.J.A. The Prosite database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A., Schattner P., Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Ingrell C.R., Miller M.L., Jensen O.N., Blom N. NetPhosYeast: prediction of protein phosphorylation sites in yeast. Bioinformatics (Oxf.) 2007;23:895–897. doi: 10.1093/bioinformatics/btm020. [DOI] [PubMed] [Google Scholar]
- Jimenez R.C., Quinn A.F., Garcia A., Labarga A., O'Neill K., Martinez F., Salazar G.A., Hermjakob H. Dasty2, an Ajax protein DAS client. Bioinformatics (Oxf.) 2008;24:2119–2121. doi: 10.1093/bioinformatics/btn387. [DOI] [PubMed] [Google Scholar]
- Johansen M.B., Kiemer L., Brunak S. Analysis and prediction of mammalian protein glycation. Glycobiology. 2006;16:844–853. doi: 10.1093/glycob/cwl009. [DOI] [PubMed] [Google Scholar]
- Jones D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics (Oxf.) 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- Jones P., Côté R. The PRIDE proteomics identifications database: data submission, query, and dataset comparison. Methods Mol. Biol. 2008;484:287–303. doi: 10.1007/978-1-59745-398-1_19. [DOI] [PubMed] [Google Scholar]
- Jones S.J.M. Prediction of genomic functional elements. Annu. Rev. Genom. Hum. Genet. 2006;7:315–338. doi: 10.1146/annurev.genom.7.080505.115745. [DOI] [PubMed] [Google Scholar]
- Julenius K. NetCGlyc 1.0: prediction of mammalian C-mannosylation sites. Glycobiology. 2007;17:868–876. doi: 10.1093/glycob/cwm050. [DOI] [PubMed] [Google Scholar]
- Julenius K., Mølgaard A., Gupta R., Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Juncker A.S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z., Rouchka E.C., Gish W.R., States D.J. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 2001;11:889–900. doi: 10.1101/gr.155001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N., Sasson O., Inbar U., Friedlich M., Fromer M., Fleischer H., Portugaly E., Linial N., Linial M. ProtoNet 4.0: a hierarchical classification of one million protein sequences. Nucleic Acids Res. 2005;33:D216–D218. doi: 10.1093/nar/gki007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T., Friedman N., Margalit H. Ab initio prediction of transcription factor targets using structural knowledge. PLoS Comput. Biol. 2005;1:e1. doi: 10.1371/journal.pcbi.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappler M.A. Software for rapid prototyping in the pharmaceutical and biotechnology industries. Curr. Opin. Drug Discov. Dev. 2008;11:389–392. [PubMed] [Google Scholar]
- Karchin R., Diekhans M., Kelly L., Thomas D.J., Pieper U., Eswar N., Haussler D., Sali A. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics (Oxf.) 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- Karolchik D., et al. The UCSC Genome browser database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S., et al. IntAct-open source resource for molecular interaction data. Nucleic Acids Res. 2007a;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrien S., et al. Broadening the horizon-level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007b;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemer L., Lund O., Brunak S., Blom N. Coronavirus 3CLpro proteinase cleavage sites: possible relevance to SARS virus pathology. BMC Bioinform. 2004;5:72. doi: 10.1186/1471-2105-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiemer L., Bendtsen J.D., Blom N. NetAcet: prediction of N-terminal acetylation sites. Bioinformatics (Oxf.) 2005;21:1269–1270. doi: 10.1093/bioinformatics/bti130. [DOI] [PubMed] [Google Scholar]
- Kim N., Alekseyenko A.V., Roy M., Lee C. The ASAP II database: analysis and comparative genomics of alternative splicing in 15 animal species. Nucleic Acids Res. 2007;35:D93–D98. doi: 10.1093/nar/gkl884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmann E., Fleischmann W., Apweiler R. Automatic rule generation for protein annotation with the C4.5 data mining algorithm applied on Swiss-Prot. Bioinformatics (Oxf.) 2001;17:920–926. doi: 10.1093/bioinformatics/17.10.920. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kuhlman B., Dantas G., Ireton G.C., Varani G., Stoddard B.L., Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science (NY) 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Kulikova T., et al. EMBL nucleotide sequence database in 2006. Nucleic Acids Res. 2007;35:D16–D20. doi: 10.1093/nar/gkl913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labarga A., Valentin F., Anderson M., Lopez R. Web services at the European bioinformatics institute. Nucleic Acids Res. 2007;35:W6–11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Lander E.S., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Chistyakov V.V., Thornton J.M. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005a;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Watson J.D., Thornton J.M. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005b;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S., et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.J., Doherty G.P., Clarke J. Transcription factor dynamics. Microbiology. 2008;154:1837–1844. doi: 10.1099/mic.0.2008/018549-0. [DOI] [PubMed] [Google Scholar]
- Liolios K., Mavromatis K., Tavernarakis N., Kyrpides N.C. The genomes on line database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–D479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Vikis H., Lu Y., Wang D., You M. Large-scale in silico mapping of complex quantitative traits in inbred mice. PLoS ONE. 2007;2:e651. doi: 10.1371/journal.pone.0000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G., Valencia A., Tress M. FireDB—a database of functionally important residues from proteins of known structure. Nucleic Acids Res. 2007a;35:D219–D223. doi: 10.1093/nar/gkl897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G., Valencia A., Tress M.L. Firestar-prediction of functionally important residues using structural templates and alignment reliability. Nucleic Acids Res. 2007b;35:W573–W577. doi: 10.1093/nar/gkm297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmanger B., Lawler M., Coulombe S., Murray R., Cooper S., Polyakov Y., Belknap J., Hitzemann R. Further studies on using multiple-cross mapping (MCM) to map quantitative trait loci. Mamm. Genome. 2006;17:1193–1204. doi: 10.1007/s00335-006-0070-2. [DOI] [PubMed] [Google Scholar]
- Mardis E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Street S.A., Bryson K., Sørensen S.-A., Jones D.T. The genomic threading database: a comprehensive resource for structural annotations of the genomes from key organisms. Nucleic Acids Res. 2004;32:D196–D199. doi: 10.1093/nar/gkh043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Giglione C. Tools for analyzing and predicting N-terminal protein modifications. Proteomics. 2008;8:626–649. doi: 10.1002/pmic.200700592. [DOI] [PubMed] [Google Scholar]
- Mendes Soares L.M., Valcarcel J. The expanding transcriptome: the genome as the ‘Book of sand’. EMBO J. 2006;25:923–931. doi: 10.1038/sj.emboj.7601023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B., Resch A., Grasso C., Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaner D., et al. Next station in microarray data analysis: GEPAS. Nucleic Acids Res. 2006;34:W486–W491. doi: 10.1093/nar/gkl197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecchi-Palazzi L., Beavis R., Binz P.-A., Chalkley R.J., Cottrell J., Creasy D., Seymour S.L., Garavelli J.S. PSI-MOD: a community standard for representation of protein modification data. Nat. Biotechnol. 2008;26:864–866. doi: 10.1038/nbt0808-864. [DOI] [PubMed] [Google Scholar]
- Mott R. Finding the molecular basis of complex genetic variation in humans and mice. Phil. Trans. R. Soc. B. 2006;361:393–401. doi: 10.1098/rstb.2005.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder N.J., et al. New developments in the Interpro database. Nucleic Acids Res. 2007;35:D224–D228. doi: 10.1093/nar/gkl841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall C.J., Emmert D.B. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics (Oxf.) 2007;23:i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- Neu-Yilik G., Gehring N.H., Hentze M.W., Kulozik A.E. Nonsense-mediated mRNA decay: from vacuum cleaner to Swiss army knife. Genome Biol. 2004;5:218. doi: 10.1186/gb-2004-5-4-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofran Y., Punta M., Schneider R., Rost B. Beyond annotation transfer by homology: novel protein-function prediction methods to assist drug discovery. Drug Discov. Today. 2005;10:1475–1482. doi: 10.1016/S1359-6446(05)03621-4. [DOI] [PubMed] [Google Scholar]
- Okazaki Y., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Okubo K., Sugawara H., Gojobori T., Tateno Y. DDBJ in preparation for overview of research activities behind data submissions. Nucleic Acids Res. 2006;34:D6–D9. doi: 10.1093/nar/gkj111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/S0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Orth A.P., Batalov S., Perrone M., Chanda S.K. The promise of genomics to identify novel therapeutic targets. Expert Opin. Ther. Targets. 2004;8:587–596. doi: 10.1517/14728222.8.6.587. [DOI] [PubMed] [Google Scholar]
- Parkinson H., et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsko G.A. Homologuephobia. Genome Biol. 2001;2:1–2. doi: 10.1186/gb-2001-2-2-comment1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U., et al. ModBase: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher M.T., et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop M., Salzberg S.L. Bioinformatics challenges of new sequencing technology. Trends Genet. 2008;24:142–149. doi: 10.1016/j.tig.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter C.T., Bartlett G.J., Thornton J.M. The catalytic site atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic A., Down T.A., Hubbard T.J. Adding some SPICE to DAS. Bioinformatics (Oxf.) 2005;21(Suppl. 2):ii40–ii41. doi: 10.1093/bioinformatics/bti1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic A., Down T.A., Kulesha E., Finn R.D., Kähäri A., Hubbard T.J.P. Integrating sequence and structural biology with DAS. BMC Bioinform. 2007;8:333. doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Maglott D.R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T., et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebholz-Schuhman D., et al. SYMBIOmatics: synergies in medical informatics and bioinformatics—exploring current scientific literature for emerging topics. BMC Bioinform. 2007;8(Suppl. 1):S18. doi: 10.1186/1471-2105-8-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.L., Famili I., Thiele I., Palsson B.O. Towards multidimensional genome annotation. Nat. Rev. 2006;7:130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- Reeves G.A., et al. The protein feature ontology: a tool for the unification of protein feature annotations. Bioinformatics (Oxf.) 2008;24:2767–2772. doi: 10.1093/bioinformatics/btn528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumers J., Schymkowitz J., Ferkinghoff-Borg J., Stricher F., Serrano L., Rousseau F. SNPeffect: a database mapping molecular phenotypic effects of human non-synonymous coding SNPs. Nucleic Acids Res. 2005;33:D527–D532. doi: 10.1093/nar/gki086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuveni E., Ramensky V.E., Gross C. Mouse SNP Miner: an annotated database of mouse functional single nucleotide polymorphisms. BMC Genom. 2007;8:24. doi: 10.1186/1471-2164-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A., et al. WormBase 2007. Nucleic Acids Res. 2008;36:D612–D617. doi: 10.1093/nar/gkm975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins J., Chen Y., Paigen B., Wang X. In search of new targets for plasma high-density lipoprotein cholesterol levels: promise of human–mouse comparative genomics. Trends Cardiovasc. Med. 2006;16:220–234. doi: 10.1016/j.tcm.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Rueda M., Ferrer-Costa C., Meyer T., Pérez A., Camps J., Hospital A., Gelpí J.L., Orozco M. A consensus view of protein dynamics. Proc. Natl Acad. Sci. USA. 2007;104:796–801. doi: 10.1073/pnas.0605534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M.C. Swiss-Model: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva N. 1000 Genomes project. Nat. Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- Sjöblom T., et al. The consensus coding sequences of human breast and colorectal cancers. Science (NY) 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Smigielski E.M., Sirotkin K., Ward M., Sherry S.T. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg L.C., et al. A protocol for high-throughput phenotyping, suitable for quantitative trait analysis in mice. Mamm. Genome. 2006;17:129–146. doi: 10.1007/s00335-005-0112-1. [DOI] [PubMed] [Google Scholar]
- Southan C. Has the yo-yo stopped? An assessment of human protein-coding gene number. Proteomics. 2004;4:1712–1726. doi: 10.1002/pmic.200300700. [DOI] [PubMed] [Google Scholar]
- Sprague J., et al. The zebrafish information network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich G., Fernandez-Suarez X.M., Birney E. Genome browsing with Ensembl: a practical overview. Brief. Funct. Genom. Proteom. 2007;6:202–219. doi: 10.1093/bfgp/elm025. [DOI] [PubMed] [Google Scholar]
- Stamm S., Riethoven J.J., Le T.V., Gopalakrishnan C., Kumanduri V., Tang Y., Barbosa-Morais N.L., Thanaraj T.A. ASD: a bioinformatics resource on alternative splicing. Nucleic Acids Res. 2006;34:D46–D55. doi: 10.1093/nar/gkj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L., Sternberg P., Durbin R., Thierry-Mieg J., Spieth J. WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 2001;29:82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L.D., et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetefeld J., Ruegg M.A. Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem. Sci. 2005;30:515–521. doi: 10.1016/j.tibs.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Stoesser G., Moseley M.A., Sleep J., McGowran M., Garcia-Pastor M., Sterk P. The EMBL nucleotide sequence database. Nucleic Acids Res. 1998;26:8–15. doi: 10.1093/nar/26.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck D., et al. The arabidopsis information resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagari M., et al. E-MSD: improving data deposition and structure quality. Nucleic Acids Res. 2006;34:D287–D290. doi: 10.1093/nar/gkj163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The BioSapiens Network of Excellence. Research networks: BioSapiens: a European network for integrated genome annotation. Eur. J. Hum. Genet. 2005;13:994–997. doi: 10.1038/sj.ejhg.5201470. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. The ENCODE (encyclopedia of DNA elements) project. Science (NY) 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- The UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. Panther: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrance J.W., Bartlett G.J., Porter C.T., Thornton J.M. Using a library of structural templates to recognise catalytic sites and explore their evolution in homologous families. J. Mol. Biol. 2005;347:565–581. doi: 10.1016/j.jmb.2005.01.044. [DOI] [PubMed] [Google Scholar]
- Tress M.L., et al. The implications of alternative splicing in the ENCODE protein complement. Proc. Natl Acad. Sci. USA. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tringe S.G., et al. Comparative metagenomics of microbial communities. Science (NY) 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Twigger S.N., Shimoyama M., Bromberg S., Kwitek A.E., Jacob H.J. The Rat genome database, update 2007—easing the path from disease to data and back again. Nucleic Acids Res. 2007;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami S., et al. The localization of proteins encoded by CRYM, KIAA1199, UBA52, COL9A3, and COL9A1, genes highly expressed in the cochlea. Neuroscience. 2008;154:22–28. doi: 10.1016/j.neuroscience.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Valdar W., et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Venter J.C., et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science (NY) 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Watson J.D., Laskowski R.A., Thornton J.M. Predicting protein function from sequence and structural data. Curr. Opin. Struct. Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Wen F., Li F., Xia H., Lu X., Zhang X., Li Y. The impact of very short alternative splicing on protein structures and functions in the human genome. Trends Genet. 2004;20:232–236. doi: 10.1016/j.tig.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Wheeler D.L., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.A., et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Wieser D., Kretschmann E., Apweiler R. Filtering erroneous protein annotation. Bioinformatics (Oxf.) 2004;20(Suppl. 1):i342–i347. doi: 10.1093/bioinformatics/bth938. [DOI] [PubMed] [Google Scholar]
- Wilming L.G., Gilbert J.G.R., Howe K., Trevanion S., Hubbard T., Harrow J.L. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.A., Kreychman J., Gerstein M. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J. Mol. Biol. 2000;297:233–249. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]
- Wong G.K., Passey D.A., Yu J. Most of the human genome is transcribed. Genome Res. 2001;11:1975–1977. doi: 10.1101/gr.202401. [DOI] [PubMed] [Google Scholar]
- Wood L.D., et al. The genomic landscapes of human breast and colorectal cancers. Science (NY) 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Wu C.H., Huang H., Nikolskaya A., Hu Z., Barker W.C. The iProClass integrated database for protein functional analysis. Comput. Biol. Chem. 2004;28:87–96. doi: 10.1016/j.compbiolchem.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Wu Q., Gaddis S.S., MacLeod M.C., Walborg E.F., Thames H.D., DiGiovanni J., Vasquez K.M. High-affinity triplex-forming oligonucleotide target sequences in mammalian genomes. Mol. Carcinog. 2007;46:15–23. doi: 10.1002/mc.20261. [DOI] [PubMed] [Google Scholar]
- Yazaki J., Gregory B.D., Ecker J.R. Mapping the genome landscape using tiling array technology. Curr. Opin. Plant Biol. 2007;10:534–542. doi: 10.1016/j.pbi.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazgan O., Krebs J.E. Noncoding but nonexpendable: transcriptional regulation by large noncoding RNA in eukaryotes. Biochem. Cell Biol. 2007;85:484–496. doi: 10.1139/O07-061. [DOI] [PubMed] [Google Scholar]
- Yeats C., Lees J., Reid A., Kellam P., Martin N., Liu X., Orengo C. Gene3D: comprehensive structural and functional annotation of genomes. Nucleic Acids Res. 2008;36:D414–D418. doi: 10.1093/nar/gkm1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., He Y. Extracting interactions between proteins from the literature. J. Biomed. Inform. 2008;41:393–407. doi: 10.1016/j.jbi.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Zweigenbaum P., Demner-Fushman D., Yu H., Cohen K.B. Frontiers of biomedical text mining: current progress. Brief. Bioinform. 2007;8:358–375. doi: 10.1093/bib/bbm045. [DOI] [PMC free article] [PubMed] [Google Scholar]