Abstract
Background
Atrial fibrillation (AF) is heterogeneous at the clinical and molecular levels. Association studies have reported that common single nucleotide polymorphisms (SNPs) in KCNE1 and SCN5A may predispose to AF. In this study, we tested the hypothesis that specific AF-associated genotypes confer variation on the appearance of AF assessed by analysis of fibrillatory rate of the atria.
Methods and Results
Twenty-six non-related patients (21 male, mean age 55 ± 12 years) with persistent lone AF (median AF duration 5 weeks) not taking class I or III antiarrhythmic drugs were studied. Fibrillatory rate was obtained by spatiotemporal QRST cancellation and time-frequency analysis of the index surface ECG. Genotypes at the AF-associated loci in KCNE1 (S38G) and SCN5A (H558R) were determined by direct DNA sequencing. The atrial fibrillatory rate was 418±50 fibrillations per minute (fpm, range 336–521) in the study cohort. Carriers of the 38GG KCNE1 genotype (n=13) had significantly lower fibrillatory rates (392±36 vs 443±49 fpm, p=.006) compared to those with GS or SS genotype (n=13). Six patients (23 %) with fibrillatory rates > 450 fpm, all had either the GS or SS genotype (chi2 p=.008). In contrast, both the heterozygeous and homozygeous SCN5A H558R polymorphism had no effect on fibrillatory rate.
There were no significant associations between fibrillatory rate and clinical (age, gender, AF duration, drug treatment) or echocardiographic (left atrial diameter, LVEF) variables. In multivariate analysis, KCNE1 (S38G) was the only independent predictor of fibrillatory rate (R=.528, B=45.091, p=.006).
Conclusions
Atrial fibrillatory rate obtained from the surface ECG is at least in part determined by KCNE1 (S38G) genotype, suggesting this variant exerts functional effects on atrial electrophysiology. This intermediate ECG phenotype may be useful for elaborating genetic influences on AF mechanisms and identifying subsets of patients for variability in AF susceptibility or response to therapies.
Keywords: Atrial fibrillation, gene variants, ECG
Introduction
Atrial fibrillation (AF) is a heterogeneous arrhythmia at both the clinical and molecular level. Association studies have reported that common single nucleotide polymorphisms (SNPs) in genes encoding cardiac ion channels may predispose to AF development.1–3 For instance, the 38G allele of KCNE1 encoding the beta-subunit of IKs potassium channels 1,2 and the R558 allele of the cardiac sodium channel gene SCN5A 3 have been found to constitute risk factors for lone AF.
Experimental studies have revealed modulation of atrial refractoriness 4 or conduction velocity 5 by the KCNE1 (S38G) and SCN5A (H558R) polymorphisms, respectively. However, their association with an intermediate ECG-phenotype in humans is unknown. One ECG-phenotype is atrial fibrillatory rate that can reliably be assessed from the surface ECG using spatiotemporal QRST cancellation and time frequency analysis. Fibrillatory rate of ECG lead V1 corresponds closely with fibrillatory rates of the high right atrium, coronary sinus and pulmonary veins and is a reproducible marker of atrial refractoriness.6
It is a common observation that fibrillatory waves have various appearances from fine to coarse and from disorganized to organized. This inter-individual variance is also reflected by fibrillatory rates ranging from 240 to 540 fibrillations per minute (fpm). However, clinical and echocardiographic patient characteristics explain fibrillatory rate variance only in part.7,8
In this study, we tested the hypothesis that specific AF-associated genotypes confer variation on the ECG appearance of AF assessed by analysis of fibrillatory rate.
Methods
Study population
The study was performed in patients prospectively enrolled in our AF registry, which consists of a clinical, a genetic and a digital ECG registry. Inclusion criteria include age ≥18 years, and a documented history of AF or atrial flutter. At enrollment into the registry, patients give informed consent, a detailed medical and drug history is obtained in all patients in addition to a standard 12-lead ECG and a transthoracic echocardiogram.
For this study, patients were selected if they had persistent lone AF > 7 days with or without mild hypertension, not taking class I or III antiarrhythmic drugs at the time of ECG acquisition.
Left atrial and left ventricular measurements from the M-mode echocardiograms were performed by an experienced physician blinded to the genotype status of the patient. The echocardiograms were evaluated according to the recommendations of the American Society of Echocardiography.
Molecular analysis
Genomic DNA was extracted according to standard protocols, and the KCNE1 (S38G) and SCN5A (H558R) polymorphisms were detected by direct sequencing blinded to the ECG findings.
ECG analysis
Standard 10-s, 12-lead surface ECG recordings were acquired in all patients with the subject relaxed in a supine position. Digital ECG (500 Hz sampling rate) were retrieved from the hospital ECG database for further signal processing.
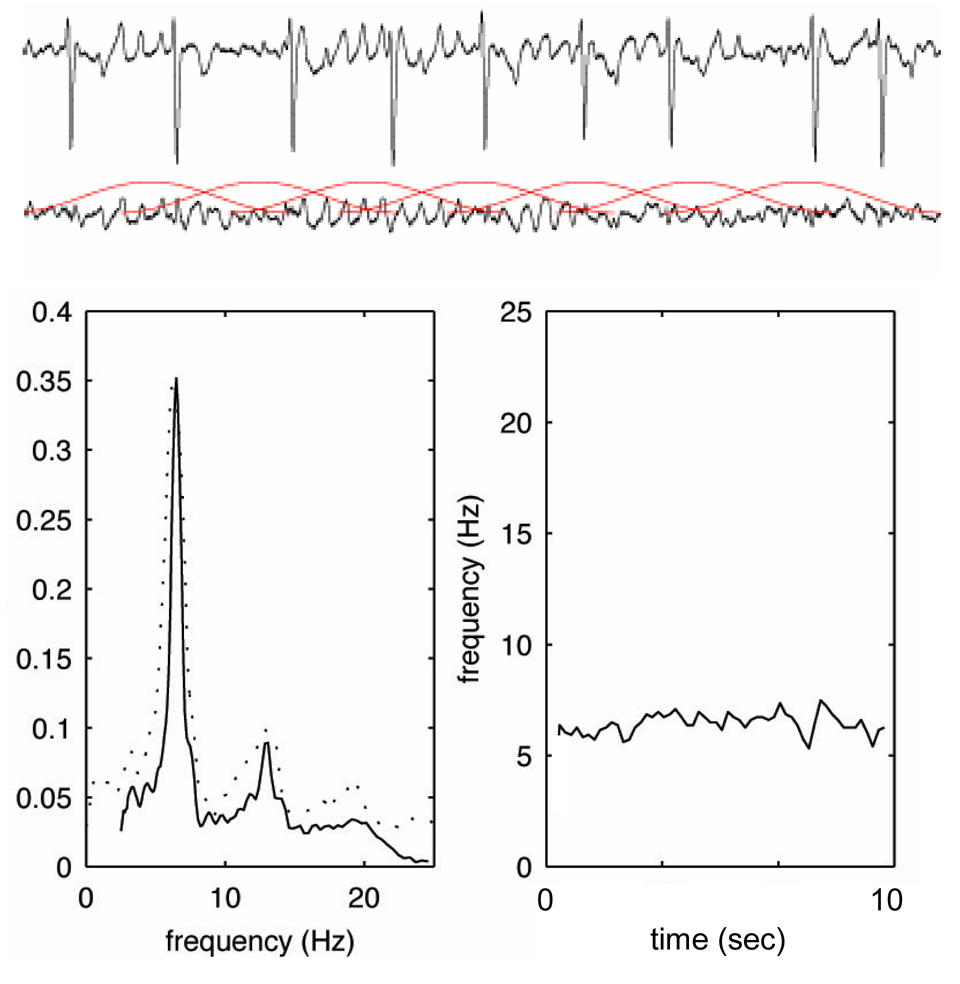
After high-pass filtering to remove baseline wander, atrial fibrillatory activity was extracted in lead V1 using spatiotemporal QRST cancellation. Since the dominant frequency component of interest is within the 4–9 Hz range, the resulting fibrillatory baseline signal was downsampled to 50 Hz and subjected to spectral analysis. The time–frequency distribution of the atrial signal (obtained by short-term Fourier transform) was decomposed such that each spectrum can be modelled as a frequency-shifted and amplitude-scaled version of the spectral profile. This procedure is based on a spectral profile, dynamically updated from previous spectra, which was matched to each new spectrum using weighted least squares estimation. The frequency shift needed to achieve optimal matching then yields a measure of instantaneous fibrillatory rate of a 2.5-s ECG segment (overlapping with one segment each second) and was trended as a function of time.6
Frequencies were converted to fibrillatory rates with its unit fibrillations per minute (fpm) as advocated previously (rate = frequency * 60).9 Mean fibrillatory rate (in fpm) defined as average of instantaneous fibrillatory rates over the 10-second ECG segment was determined (Figure 1).
Figure 1.
Time-frequency analysis of AF. ECG recording of lead V1 from a patient with AF (upper panel), and the same interval after spatiotemporal QRST cancellation (middle panel). Please note the overlapping 2.5-second ECG windows, which are then subjected to short-term Fourier transform resulting in a power frequency spectrum (bottom left), and a frequency trend (bottom right).
Statistical analysis
Data are presented as mean ± one standard deviation for normally distributed continuous variables.
The possible relation between continuous clinical variables and fibrillatory rate was analyzed using bivariate correlations (Pearson). Student’s t-test for unpaired data or ANOVA was used to compare fibrillatory rates among categorical clinical variables and genotypes. With a sample size of 26 patients and a predicted standard deviation of fibrillatory rate of 60 fpm in variant carriers, fibrillatory rate differences of ±60 fpm between patients with a wild type and variant carriers would have been detectable with a power of 80 %. This difference is below the spontaneous fibrillatory rate variability and would have been considered a clinically relevant genotype – phenotype correlation. Multivariate analysis that included variables with a p-value < .1 found in univariate analysis was used to identify independent predictors of fibrillatory rate.
A p value of < .05 was considered statistically significant.
Results
Patient characteristics
Patient characteristics are summarized in Table 1. The atrial fibrillatory rate was 418±50 fibrillations per minute (fpm, range 336–521).
Table 1.
Patient characteristics (n=26).
| Age, years | 55±12 (22 – 82) |
| Male/Female | 21/5 |
| AF duration, weeks (median) | 5 (1 – 52) |
| Mild hypertension | 15 |
| LAD, mm | 43±8 (31 – 49) |
| LVEF, % | 54±8 (45 – 65) |
| Digitalis | 6 |
| Beta and/or calcium channel blocker | 8 |
| Atrial fibrillatory rate, fpm | 418±50 (336 – 521) |
| Ventricular rate, bpm | 88±21 (61 – 123) |
LAD = left atrial diameter, LVEF = left ventricular ejection fraction
The frequencies of the KCNE1 genotypes were SS in 4 %, GS in 46 %, and GG in 50 % and of the SCN5A genotypes HH in 57 %, HR in 31 %, and RR in 12 %. Patient characteristics according to genotype are summarized in Table 2.
Table 2.
Patient characteristics according to genotype.
| KCNE1 | SCN5A | |||||
|---|---|---|---|---|---|---|
| SS | GS | GG | HH | HR | RR | |
| N=1 | N=12 | N=13 | N=15 | N=8 | N=3 | |
| Age, years | 50 | 58±14 | 54±11 | 56±13 | 55±14 | 60±5 |
| Male/Female | 1/0 | 8/4 | 12/1 | 12/3 | 7/1 | 2/1 |
| AF duration, weeks (median) | 5 | 4 | 8 | 4 | 5 | 7 |
| Mild hypertension | 0 | 6 | 9 | 5 | 7 | 3 |
| LAD, mm | 43 | 43±9 | 44±7 | 42±7 | 46±10 | 42±3 |
| LVEF, % | 45 | 54±7 | 53±9 | 54±9 | 54±7 | 50±2 |
| Digitalis | 0 | 5 | 1 | 4 | 2 | 0 |
| Beta and/or calcium channel blocker | 0 | 4 | 4 | 5 | 2 | 1 |
| Atrial fibrillatory rate, fpm | 470 | 431±52* | 392±36* | 403±50 | 434±50 | 408±28 |
| Ventricular rate, bpm | 115 | 82±12 | 91±27 | 91±18 | 79±13 | 96±40 |
LAD = left atrial diameter, LVEF = left ventricular ejection fraction
p<.05
Genotype – ECG phenotype correlations
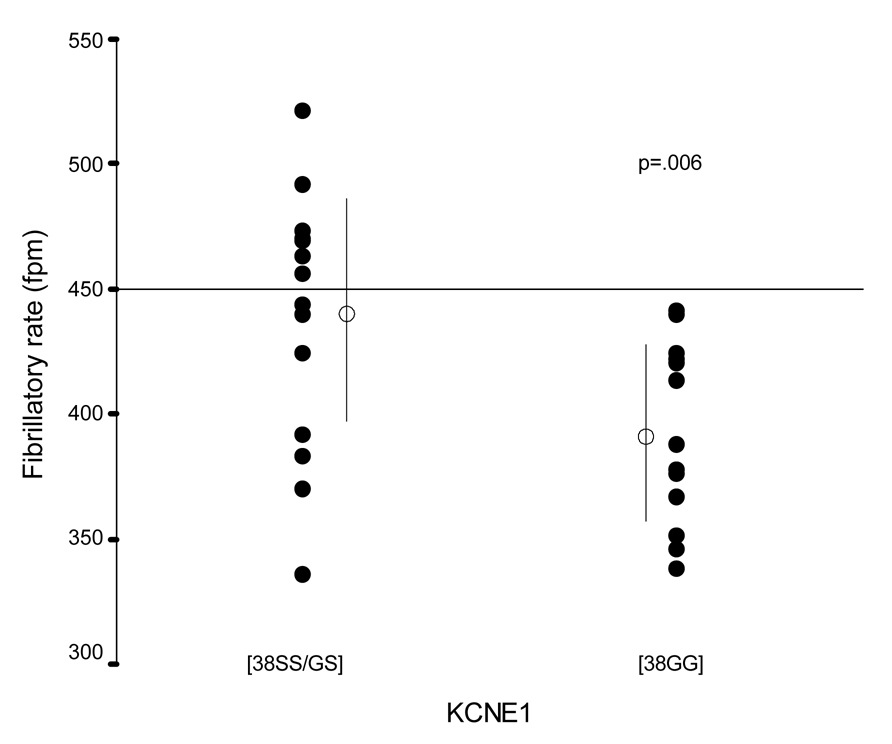
Carriers of the 38GG KCNE1 genotype had significantly lower fibrillatory rates (392±36 vs 443±49 fpm, p=.006) compared to those with GS or SS genotype (Table 2). Six patients (23 %) with fibrillatory rates > 450 fpm, all had either the GS or SS genotype (chi2 p=.008; Figure 2). Their clinical characteristics were similar to patients with lower fibrillatory rates. In particular, their AF duration ranged from 2 to 42 weeks (median 5 weeks), similar to the remaining patients (1 to 52 weeks, median 6 weeks). In contrast, the SCN5A H558R polymorphism was not associated with fibrillatory rate (Table 2).
Figure 2.
KCNE1 S38G and fibrillatory rate.
There were no significant associations between fibrillatory rate and clinical (age, gender, AF duration, drug treatment) or echocardiographic (left atrial diameter, LVEF) variables. In particular, the use of digoxin (426±67 vs. 409±42 fpm, p=.454) or beta blockers (394±52 vs. 418±47 fpm, p=.319) did not affect fibrillatory rate.
In multivariate analysis, KCNE1 genotype (S38G) was the only independent predictor of fibrillatory rate (R=.528, B=45.091, p=.006).
Discussion
Main finding
To the best of our knowledge, this study is the first in patients with persistent lone AF to assess possible genotype –phenotype correlations using atrial fibrillatory rate of the surface ECG. In contrast to clinical and echocardiographic variables, KCNE1 (S38G) but not SCN5A (H558R) genotype was associated with fibrillatory rate. It was demonstrated that carriers of the 38GG KCNE1 genotype had significantly lower rates than those with the GS or SS genotype. Of interest, in none of the patients with the GG genotype did fibrillatory rate exceed 450 fpm.
Modulation of atrial electrophysiology by gene variants
Few previous studies 4,5,10 have assessed in vitro modulatory effects of gene variants on atrial electrophysiological properties such atrial refractoriness or conduction velocity. Of special importance for our study are findings on KCNE1 (S38G) and SCN5A (H558R) polymorphisms. Ehrlich and co-workers found that the KCNE1 38G isoform is associated with reduced IKs likely due to decreased KvLQT1 membrane expression. This in turn leads to longer atrial action potential duration and refractoriness. Since longer refractory periods are associated with lower fibrillatory rates,11 our observation that lower rates not exceeding 450 fpm are present in patients with the GG genotype is in agreement with these findings. It needs to be pointed out that the increased risk for AF initiation with this genotype due to early afterdepolarizations is not in conflict with our observations that refer to AF persistence.
Although the SCN5A H558R polymorphism has been suggested to be associated with non-functioning or poorly functioning sodium channels and a subsequently reduced conduction velocity,5 we have found no association between this genotype and slower fibrillatory rates. On the one hand, it needs to be emphasized, however, that atrial refractoriness is the main determinant of fibrillatory rate, while conduction velocity seems to be similar in a narrow range among patients with persistent AF.12 On the other hand, the sodium channel function is also affected by the amino acid splice variant 5 that is unknown in the clinical setting.
Very limited in vivo data is available showing that certain gene variants may indeed exert functional effects on atrial electrophysiology. For instance, the promoter polymorphism −44G>A of the connexin 40 (Cx40) gene is associated with reduced transcriptional activity thereby influencing atrial Cx40 expression.13 Conceptually similar to our study, Firouzi and colleagues 10 have shown that this Cx40 polymorphism was associated with atrial refractoriness dispersion. In their study, the prevalence of the minor Cx40 allele (−44A) and −44AA genotype was significantly higher in subjects with increased dispersion. Interestingly, as with our study, this difference was revealed already in a small population (n=30).
Previous studies have shown that a lower fibrillatory rate is associated with a better response to antiarrhythmic drugs 14 and less AF recurrence following cardioversion.15,16 Similarly, certain genotypes such as the ACE I/D polymorphism may modulate the response to antiarrhythmic therapy via increased ACE and consequently angiotensin II levels.17 The latter is known to promote atrial fibrosis and electrophysiological remodeling.18 In consequence, the presence of the ACE ID or DD polymorphisms may indicate a more severe remodeling process in AF that is refractory to class I and III antiarrhythmics.
In summary, only limited data is available on few polymorphisms that point to a variety of mechanisms and pathways involved in the electrical and structural remodeling during AF. The elaboration of their individual contribution that may, moreover, vary over time is in its infancy. Nevertheless, the findings of our study are hypothesis generating and suggest that fibrillatory rate is an intermediate genotype dependent ECG phenotype which - together with its underlying genotype - may be useful for predicting response to therapy.
Limitations
This study is limited by a small but homogenous patient population. However, in agreement with a previous study 10 our current approach assessing genotype – ECG phenotype correlations among patients with AF requires a much smaller sample size than classic association studies.
Using the candidate gene approach, this study was limited to the investigation of two SNPs of two cardiac ion channel genes. However, as discussed above, other genes may also influence atrial electrophysiology 10 and consequently the ECG-phenotype.
This study was limited to the analysis of ECG lead V1. It can be argued that this is a closer reflection of right atrial but not left atrial activity. However, fibrillatory rates are similar among different sites in patients with persistent AF 19 as in our population. Consequently, it is not surprising that ECG lead V1 closely reflects right atrial, but is also related to pulmonary venous/left atrial rates,6 especially when AF is persistent and the left to right atrial frequency gradient diminishes.19
Aside from P wave parameters and ventricular rate, fibrillatory rate during AF can be considered as one specific ECG phenotype. Although there are several theories about AF sustenance including multiple wavelets, focal activation, spiral wave; in the individual patient, the underlying mechanism is currently difficult (if not impossible) to assess. Consequently, the association between fibrillatory rate and the dominant underlying AF mechanisms has not been explored. But even with this limited knowledge, fibrillatory rate of ECG lead V1 can be considered as a general marker of atrial remodeling as it is also associated with spontaneous and drug-induced AF termination and recidivism after cardioversion.9
ECG recordings in this study were limited to 10 sec and variability over time was not assessed. However, previous studies have shown that fibrillatory rate obtained from the surface ECG is reproducible over 24 hours in clinically stable patients with persistent AF.20
Finally, although we did not invasively assess ERP in our population, previous studies have shown the association between the KCNE1 (S38G) genotype and atrial refractoriness 4 as well as between atrial refractoriness and fibrillaytory rate.11
Conclusions
Atrial fibrillatory rate obtained from the surface ECG is at least in part determined by KCNE1 (S38G) genotype, suggesting this variant exerts functional effects on atrial electrophysiology. This intermediate ECG phenotype may be useful for elaborating genetic influences on AF mechanisms and identifying subsets of patients for variability in AF susceptibility or response to therapies.
Acknowledgments
The authors wish to thank Christie Ingram, Shannon Carter, Vanderbilt University, and Andreu Climent, Polytechnic University of Valencia, Spain for their help with this study.
Grant support:
Drs. Husser and Stridh were supported by the Volkswagen Foundation, Germany. Dr. Husser was supported by the Riksbanken Jubileumsfond, Sweden through the Nils-Eric Svenssons Award and research grants (HU 1679/1-1, DFG and NBL Formel.1-109, University Leipzig). This study was supported by VOI HL65962, The Pharmacogenomics of Arrhythmia Therapy.
Footnotes
Parts of this study were presented at the Annual Scientific Sessions of the American Heart Association 2007 and at the Annual Scientific Sessions of the Heart Rhythm Society 2008.
Disclosures
None
References
- 1.Lai LP, Su MJ, Yeh HM, Lin JL, Chiang FT, Hwang JJ, Hsu KL, Tseng CD, Lien WP, Tseng YZ, Huang SK. Association of the human minK gene 38G allele with atrial fibrillation: evidence of possible genetic control on the pathogenesis of atrial fibrillation. Am Heart J. 2002;144:485–490. doi: 10.1067/mhj.2002.123573. [DOI] [PubMed] [Google Scholar]
- 2.Fatini C, Sticchi E, Genuardi M, Sofi F, Gensini F, Gori AM, Lenti M, Michelucci A, Abbate R, Gensini GF. Analysis of minK and eNOS genes as candidate loci for predisposition to non-valvular atrial fibrillation. Eur Heart J. 2006;27:1712–1718. doi: 10.1093/eurheartj/ehl087. [DOI] [PubMed] [Google Scholar]
- 3.Chen LY, Ballew JD, Herron KJ, Rodeheffer RJ, Olson TM. A common polymorphism in SCN5A is associated with lone atrial fibrillation. Clin Pharmacol Ther. 2007;81:35–41. doi: 10.1038/sj.clpt.6100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrlich JR, Zicha S, Coutu P, Hebert TE, Nattel S. Atrial fibrillation-associated minK38G/S polymorphism modulates delayed rectifier current and membrane localization. Cardiovasc Res. 2005;67:520–528. doi: 10.1016/j.cardiores.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Makielski JC, Ye B, Valdivia CR, Pagel MD, Pu J, Tester DJ, Ackerman MJ. A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circ Res. 2003;93:821–828. doi: 10.1161/01.RES.0000096652.14509.96. [DOI] [PubMed] [Google Scholar]
- 6.Husser D, Stridh M, Cannom DS, Bhandari AK, Girsky MJ, Kang S, Sörnmo L, Olsson SB, Bollmann A. Validation and clinical application of time-frequency analysis of atrial fibrillation electrocardiograms. J Cardiovasc Electrophysiol. 2007;18:41–46. doi: 10.1111/j.1540-8167.2006.00683.x. [DOI] [PubMed] [Google Scholar]
- 7.Xi Q, Sahakian AV, Frohlich TG, Ng J, Swiryn S. Relationship between pattern of occurrence of atrial fibrillation and surface electrocardiographic fibrillatory wave characteristics. Heart Rhythm. 2004;1:656–663. doi: 10.1016/j.hrthm.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Husser D, Cannom DS, Bhandari AK, Stridh M, Sornmo L, Olsson SB, Bollmann A. Electrocardiographic characteristics of fibrillatory waves in new-onset atrial fibrillation. Europace. 2007;9:638–642. doi: 10.1093/europace/eum074. [DOI] [PubMed] [Google Scholar]
- 9.Bollmann A, Husser D, Mainardi L, Lombardi F, Langley P, Murray A, Rieta JJ, Millet J, Olsson SB, Stridh M, Sörnmo L. Analysis of surface electrocardiograms in atrial fibrillation: techniques, research, and clinical applications. Europace. 2006;8:911–926. doi: 10.1093/europace/eul113. [DOI] [PubMed] [Google Scholar]
- 10.Firouzi M, Ramanna H, Kok B, Jongsma HJ, Koeleman BP, Doevendans PA, Groenewegen WA, Hauer RN. Association of human connexin40 gene polymorphisms with atrial vulnerability as a risk factor for idiopathic atrial fibrillation. Circ Res. 2004;95:e29–e33. doi: 10.1161/01.RES.0000141134.64811.0a. [DOI] [PubMed] [Google Scholar]
- 11.Capucci A, Biffi M, Boriani G, Ravelli F, Nollo G, Sabbatani P, Orsi C, Magnani B. Dynamic electrophysiological behavior of human atria during paroxysmal atrial fibrillation. Circulation. 1995;92:1193–1202. doi: 10.1161/01.cir.92.5.1193. [DOI] [PubMed] [Google Scholar]
- 12.Holm M, Johansson R, Brandt J, Luhrs C, Olsson SB. Epicardial right atrial free wall mapping in chronic atrial fibrillation. Documentation of repetitive activation with a focal spread--a hitherto unrecognised phenomenon in man. Eur Heart J. 1997;18:290–310. doi: 10.1093/oxfordjournals.eurheartj.a015233. [DOI] [PubMed] [Google Scholar]
- 13.Juang JM, Chern YR, Tsai CT, Chiang FT, Lin JL, Hwang JJ, Hsu KL, Tseng CD, Tseng YZ, Lai LP. The association of human connexin 40 genetic polymorphisms with atrial fibrillation. Int J Cardiol. 2007;116:107–112. doi: 10.1016/j.ijcard.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 14.Bollmann A, Binias KH, Toepffer I, Molling J, Geller C, Klein HU. Importance of left atrial diameter and atrial fibrillatory frequency for conversion of persistent atrial fibrillation with oral flecainide. Am J Cardiol. 2002;90:1011–1014. doi: 10.1016/s0002-9149(02)02690-5. [DOI] [PubMed] [Google Scholar]
- 15.Bollmann A, Husser D, Steinert R, Stridh M, Sörnmo L, Olsson SB, Polywka D, Molling J, Geller C, Klein HU. Echo- and electrocardiographic predictors for atrial fibrillation recurrence following cardioversion. J Cardiovasc Electrophysiol. 2003;14:S162–S165. doi: 10.1046/j.1540.8167.90306.x. [DOI] [PubMed] [Google Scholar]
- 16.Holmqvist F, Stridh M, Waktare JE, Sornmo L, Olsson SB, Meurling CJ. Atrial fibrillatory rate and sinus rhythm maintenance in patients undergoing cardioversion of persistent atrial fibrillation. Eur Heart J. 2006;27:2201–2207. doi: 10.1093/eurheartj/ehl098. [DOI] [PubMed] [Google Scholar]
- 17.Darbar D, Motsinger AA, Ritchie MD, Gainer JV, Roden DM. Polymorphism modulates symptomatic response to antiarrhythmic drug therapy in patients with lone atrial fibrillation. Heart Rhythm. 2007;4:743–749. doi: 10.1016/j.hrthm.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li D, Shinagawa K, Pang L, Leung TK, Cardin S, Wang Z, Nattel S. Effects of angiotensin-converting enzyme inhibition on the development of the atrial fibrillation substrate in dogs with ventricular tachypacing-induced congestive heart failure. Circulation. 2001;104:2608–2614. doi: 10.1161/hc4601.099402. [DOI] [PubMed] [Google Scholar]
- 19.Lazar S, Dixit S, Marchlinski FE, Callans DJ, Gerstenfeld EP. Presence of left-to-right atrial frequency gradient in paroxysmal but not persistent atrial fibrillation in humans. Circulation. 2004;110:3181–3186. doi: 10.1161/01.CIR.0000147279.91094.5E. [DOI] [PubMed] [Google Scholar]
- 20.Xi Q, Sahakian AV, Ng J, Swiryn S. Atrial fibrillatory wave characteristics on surface electrogram: ECG to ECG repeatability over twenty-four hours in clinically stable patients. J Cardiovasc Electrophysiol. 2004;15:911–917. doi: 10.1046/j.1540-8167.2004.03577.x. [DOI] [PubMed] [Google Scholar]