Abstract
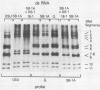
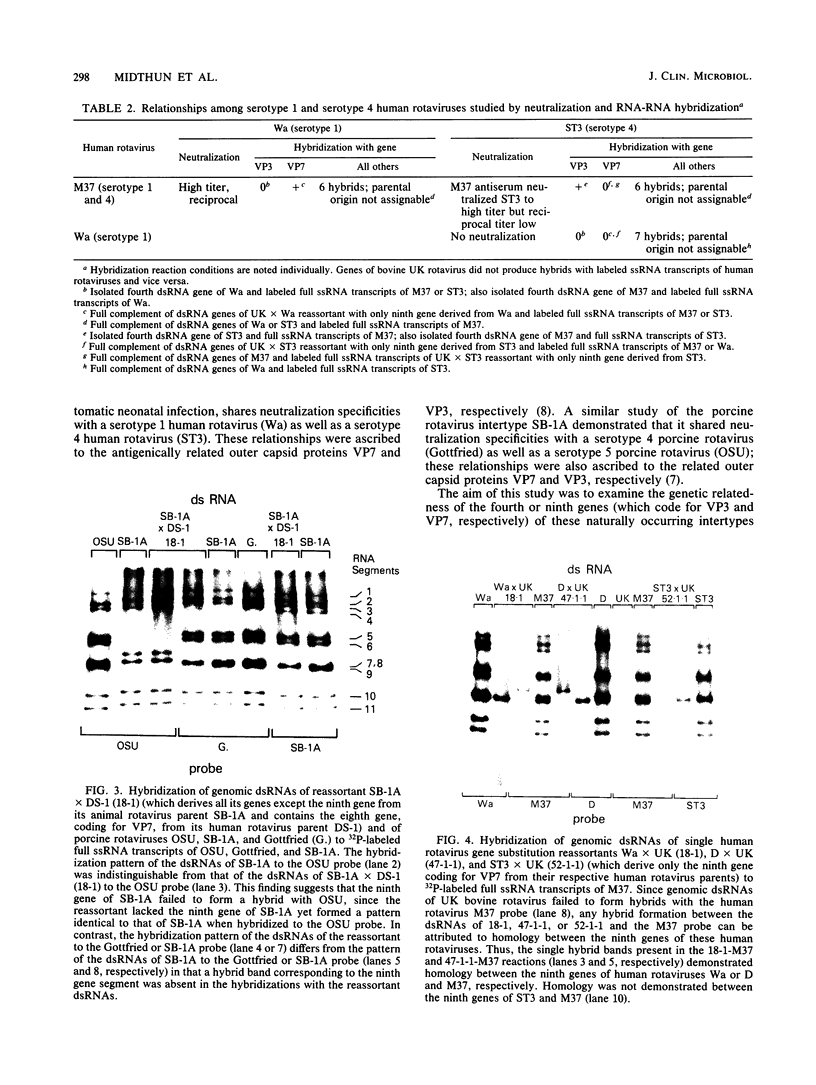
Antigenic characterization of human and animal rotaviruses by the plaque reduction neutralization assay has shown the existence of naturally occurring intertypes. Antiserum to M37, a rotavirus strain isolated from an asymptomatic neonate, neutralizes both Wa and ST3 strains, which are classified as serotype 1 and serotype 4 human rotaviruses, respectively. Likewise, antiserum to SB-1A, a porcine rotavirus, neutralizes rotavirus strains belonging to serotype 4 or 5. Plaque reduction neutralization assay of reassortant rotaviruses produced in vitro from these intertypes indicates that these viruses share one antigenically related outer capsid protein, VP3, with one serotype and another antigenically related outer capsid protein, VP7, with the other serotype. Thus, M37 is related to ST3 on the basis of its fourth-gene product, VP3, and to Wa on the basis of its ninth-gene product, VP7, whereas SB-1A is related to Gottfried (serotype 4 porcine rotavirus) via VP7 and to OSU (serotype 5 porcine rotavirus) via VP3. RNA-RNA hybridization studies revealed a high degree of homology between the VP3 or VP7 gene segments responsible for shared serotype specificity. Thus, the fourth gene segments of M37 and ST3 were highly homologous, while M37 and Wa had homology between their ninth gene segments. SB-1A and Gottfried were homologous not only with respect to the ninth gene but had complete homology in all other genes except the fourth gene. The fourth gene of SB-1A was highly homologous with the fourth gene of OSU. These observations suggested that SB-1A was a naturally occurring reassortant between Gottfried-like and OSU-like porcine rotavirus strains. Our observations also suggested that intertypes may result from genetic reassortment in nature.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Estes M. K., Palmer E. L., Obijeski J. F. Rotaviruses: a review. Curr Top Microbiol Immunol. 1983;105:123–184. doi: 10.1007/978-3-642-69159-1_3. [DOI] [PubMed] [Google Scholar]
- Flores J., Greenberg H. B., Myslinski J., Kalica A. R., Wyatt R. G., Kapikian A. Z., Chanock R. M. Use of transcription probes for genotyping rotavirus reassortants. Virology. 1982 Sep;121(2):288–295. doi: 10.1016/0042-6822(82)90168-4. [DOI] [PubMed] [Google Scholar]
- Flores J., Midthun K., Hoshino Y., Green K., Gorziglia M., Kapikian A. Z., Chanock R. M. Conservation of the fourth gene among rotaviruses recovered from asymptomatic newborn infants and its possible role in attenuation. J Virol. 1986 Dec;60(3):972–979. doi: 10.1128/jvi.60.3.972-979.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass R. I., Keith J., Nakagomi O., Nakagomi T., Askaa J., Kapikian A. Z., Chanock R. M., Flores J. Nucleotide sequence of the structural glycoprotein VP7 gene of Nebraska calf diarrhea virus rotavirus: comparison with homologous genes from four strains of human and animal rotaviruses. Virology. 1985 Mar;141(2):292–298. doi: 10.1016/0042-6822(85)90260-0. [DOI] [PubMed] [Google Scholar]
- Gorziglia M., Aguirre Y., Hoshino Y., Esparza J., Blumentals I., Askaa J., Thompson M., Glass R. I., Kapikian A. Z., Chanock R. M. VP7 serotype-specific glycoprotein of OSU porcine rotavirus: coding assignment and gene sequence. J Gen Virol. 1986 Nov;67(Pt 11):2445–2454. doi: 10.1099/0022-1317-67-11-2445. [DOI] [PubMed] [Google Scholar]
- Greenberg H. B., Flores J., Kalica A. R., Wyatt R. G., Jones R. Gene coding assignments for growth restriction, neutralization and subgroup specificities of the W and DS-1 strains of human rotavirus. J Gen Virol. 1983 Feb;64(Pt 2):313–320. doi: 10.1099/0022-1317-64-2-313. [DOI] [PubMed] [Google Scholar]
- Hoshino Y., Sereno M. M., Midthun K., Flores J., Chanock R. M., Kapikian A. Z. Analysis by plaque reduction neutralization assay of intertypic rotaviruses suggests that gene reassortment occurs in vivo. J Clin Microbiol. 1987 Feb;25(2):290–294. doi: 10.1128/jcm.25.2.290-294.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y., Sereno M. M., Midthun K., Flores J., Kapikian A. Z., Chanock R. M. Independent segregation of two antigenic specificities (VP3 and VP7) involved in neutralization of rotavirus infectivity. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8701–8704. doi: 10.1073/pnas.82.24.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y., Wyatt R. G., Flores J., Midthun K., Kapikian A. Z. Serotypic characterization of rotaviruses derived from asymptomatic human neonatal infections. J Clin Microbiol. 1985 Mar;21(3):425–430. doi: 10.1128/jcm.21.3.425-430.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y., Wyatt R. G., Greenberg H. B., Flores J., Kapikian A. Z. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J Infect Dis. 1984 May;149(5):694–702. doi: 10.1093/infdis/149.5.694. [DOI] [PubMed] [Google Scholar]
- Kalica A. R., Flores J., Greenberg H. B. Identification of the rotaviral gene that codes for hemagglutination and protease-enhanced plaque formation. Virology. 1983 Feb;125(1):194–205. doi: 10.1016/0042-6822(83)90073-9. [DOI] [PubMed] [Google Scholar]
- Kalica A. R., Greenberg H. B., Wyatt R. G., Flores J., Sereno M. M., Kapikian A. Z., Chanock R. M. Genes of human (strain Wa) and bovine (strain UK) rotaviruses that code for neutralization and subgroup antigens. Virology. 1981 Jul 30;112(2):385–390. doi: 10.1016/0042-6822(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Midthun K., Greenberg H. B., Hoshino Y., Kapikian A. Z., Wyatt R. G., Chanock R. M. Reassortant rotaviruses as potential live rotavirus vaccine candidates. J Virol. 1985 Mar;53(3):949–954. doi: 10.1128/jvi.53.3.949-954.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midthun K., Hoshino Y., Kapikian A. Z., Chanock R. M. Single gene substitution rotavirus reassortants containing the major neutralization protein (VP7) of human rotavirus serotype 4. J Clin Microbiol. 1986 Nov;24(5):822–826. doi: 10.1128/jcm.24.5.822-826.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R. G., James H. D., Jr, Pittman A. L., Hoshino Y., Greenberg H. B., Kalica A. R., Flores J., Kapikian A. Z. Direct isolation in cell culture of human rotaviruses and their characterization into four serotypes. J Clin Microbiol. 1983 Aug;18(2):310–317. doi: 10.1128/jcm.18.2.310-317.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]