Abstract
A thorough DNA sequence analysis reveals that the mouse Cyp1a1 and Cyp1a2 loci are located with coding directions opposite to each other. The two genes are separated by approximately 13.9 kb of genomic DNA containing no open reading frames (mCyp1a1_1a2 junction). Within the mCyp1a1_1a2 junction, eight consensus dioxin responsive elements (DREs) are present and seven of the eight DREs located less than 1.4 kb upstream from the Cyp1a1 transcriptional start site. The genomic structure of mouse Cyp1a1 and Cyp1a2 loci is similar to that of human CYP1A1 and CYP1A2 loci. In the human CYP1A1 and CYP1A2 are also arranged in a head to head orientation and separated by a 23 kb genome junction (hCyp1A1_1A2). Comparative sequence analysis between these two genomic junctions demonstrated that the 1.4 kb upstream region from the transcriptional start site of mouse Cyp1a1 was highly conserved with that of human CYP1A1. In contrast, there are no conserved DREs in the proximal upstream region of Cyp1a2. The “head to head” genomic structure and position of the DREs cluster region near the Cyp1a1 gene on Cyp1a1_1a2 were confirmed in cattle, dog and rat genome. These results suggest that the conservation of genomic structure of Cyp1a1 and Cyp1a2 genes, and the DREs cluster are important in mammalian biology.
Keywords: Cytochrome P450, Cyp1a1, Cyp1a2, DRE, AHR, dioxin
1. INTRODUCTION
The Cytochromes P450 (CYP) are membrane-associated hemoproteins that catalyze mono-oxygenation of endogenous and exogenous substrates such as hormones, fatty acids, drugs and xenobiotics [1]. The Cyp1a1 and Cyp1a2 loci encode major CYP isozymes in liver important in the metabolic activation of polycyclic aromatic hydrocarbons (PAHs) and procarcinogens [2]. These genes are induced by exposure to halogenated hydrocarbons such as 2,3,7,8-tetrachloro-dibenzo-p-dioxin (dioxin) or PAHs such as benzo(a)pyrene and 3-methycholanthrene [3–4]. Induction is mediated by the aryl hydrocarbons receptor (AHR). The ligand-bound AHR translocates to nucleus, where it dimerizes with the AHR nuclear translocator (ARNT). The heterodimeric AHR-ARNT complex then binds to dioxin responsive element (DRE; 5′-TNGCGTG-3′) within the genome, resulting in the transcriptional activation of nearby target gene expression. The reaction leads to transcriptional activation of gene expression [3–4].
The regulation of the Cyp1a1 gene via the AHR signaling pathway has been well characterized. In the mouse, the upstream enhancer region of Cyp1a1 has six consensus DRE sequences within −1.4 kb which mediated transcriptional activation of Cyp1a1 gene by AHR [4–6]. This cluster of DREs in the enhancer region of Cyp1a1 has been confirmed in several species [7–11]. In contrast, Cyp1a2 regulation mechanism is poorly understood, because no consensus DREs are located in the nearby upstream region of mouse Cyp1a2 gene [12]. Although a few AHR response elements, such as X1, X2 and xenobiotics response element II, have been identified in the nearby upstream of human CYP1A2 or rat Cyp1a2 [13–14], the position of these potential AHR response elements are not conserved in other species.
The sequence and genomic organization of CYP1A1 and CYP1A2 loci on human chromosome 15 determined by J. Corchero et. al revealed that the CYP1A1 and CYP1A2 genes are located immediately adjacent to each other in a head-to head orientation [15]. The genes are separated by a 23.3 kb genome junction region, designated CYP1A1_1A2, which possesses a total of 13 DREs and no open reading frames. In contrast to the human CYP1A1 and CYP1A2 loci, there are no reports with regard to genomic structure of the Cyp1a1 and Cyp1a2 in other species. We determined sequence and structure of the mouse Cyp1a1 and Cyp1a2 loci located on mouse chromosome 9. To compare the sequence with that of human, we identified highly conserved elements which should be important for the regulation of Cyp1a1 and Cyp1a2. In addition, we also inquired into the genomic structure of Cyp1a1 and Cyp1a2 loci in cattle, dog and rat for considering the evolutional and biological meaning of the conservation.
2. MATERIALS & METHODS
2.1. Analysis of mouse Cyp1a1 and Cyp1a2 loci
The bacterial artificial chromosome (BAC) clone 17278 (BAC17278) carrying intact mouse Cyp1a1 and Cyp1a2 genes form 129/Sv strain was employed for sequence analysis (Genome Systems. St. Louis, MO). The sequence was determined by employing both shotgun sequencing and PCR-direct sequencing. Construction of BAC shotgun library was prepared with the CloneSmart® system (Lucigen, Middleton, WI). Plasmids from the shotgun library were isolated and sequenced by DYEnamic™ ET dye terminators and megaBACE technology (GE Healthcare Bio-Science, Piscataway, NJ). Based on the resultant sequence, 49 PCR primer pairs (OL5827–5876, OL5899–5946) were designed to amplify approximately 30 kb of intact Cyp1a1, Cyp1a2 and their junction region (supplementary data). PCR was carried out for 35 cycles (95°C for 30 s, 58°C for 45 s, and 72°C for 1m) in a reactionmixture containing 2.5 units of Taq polymerase (Promega, Madison, WI), 50 mM KCl (Sigma-Aldrich, St. Louis, MO), 10 mM Tris-HCl (pH 9.0 at 25°C) (Sigma-Aldrich, St. Louis, MO), 1.5 mM MgCl2 (Sigma-Aldrich, St. Louis, MO), 1% Triton X-100 (Sigma-Aldrich, St. Louis, MO), 0.2 mM dNTPs (Promega, Madison, WI), and 0.2 μM of each primer. The amplified PCR products were subcloned into pGEM®-T easy vector (Promega, Madison, WI) and sequenced by using Bigdye terminator v3.1 (Applied Biosystems, Foster city, CA)
2.2. Southern blot analysis
Either BAC DNA (100 ng) or genomic DNA (10 μg) were digested by AflII, EcoRI, KpnI, PstI, or SacI (Promega, Madison, WI). The digested DNAs were electrophoresed in a 0.8% agarose gel and transferred to Hybond™-N+ membrance (GE Healthcare Life-Science, Piscataway, NJ). The membrane was hybridized with 32P-labeled cDNA probes specific for Cyp1a1 and Cyp1a2. Probe 1 was a segment of mouse Cyp1a2 gene (−622 to +298) and probe 2 was that of Cyp1a1 gene (−246 to +257). Radioactive detection was visualized by Molecular Dynamics Storm® system (GE Healthcare Life-Science, Piscataway, NJ).
2.3. Comparative analysis of genomic sequence
Comparative genomic analysis between mCyp1a1_1a2 and hCyp1A1_1A2 (GenBank accession no. AF253322) was performed by VISTA (http://genome.lbl.gov/vista/index.shtml) [16–17]. Conserved elements are defined as above 70% sequence identity over a 50 bp window. Sequences of Cyp1a1_Cyp1a2 in the cattle, dog and rat genomes were determined by employing genomic contig sequences (GenBank accession no.) NC007319, NC006612 and NW047799, respectively.
3. RESULTS & DISCUSSION
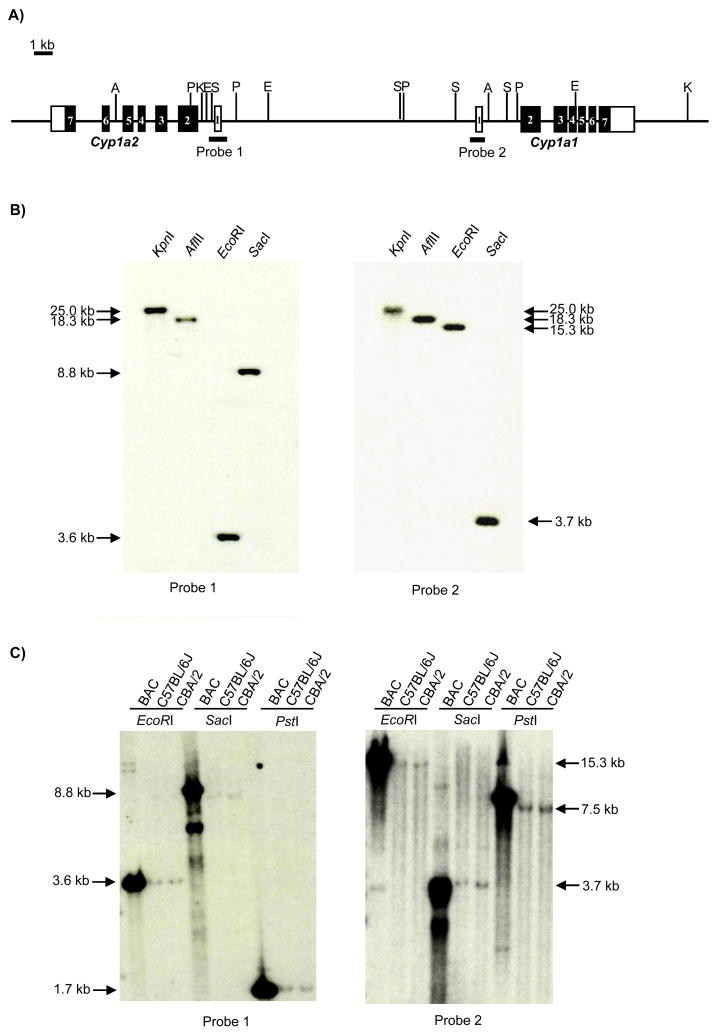
To determine the sequence of the mouse Cyp1a1 and Cyp1a2 loci, we carried out multiple sequence analyses of the BAC17278 clone which contained approximately 192 kb of genomic DNA derived from 129/Sv strain. From analysis of the sequences of approximately 4,000 clones in BAC shotgun library, we determined the sequence for 30 kb of the Cyp1a1 and Cyp1a2 loci. The sequence of the Cyp1a1 and Cyp1a2 loci was reconfirmed by the sequencing of specific PCR fragments using primers designed from the sequence determined by the BAC shotgun sequencing. Fig. 1A presents a schematic diagram and restriction enzyme digestion map of Cyp1a1 and Cyp1a2 loci, showing that the Cyp1a1 gene is located next to the Cyp1a2 gene and the coding directions are opposite to each other. The two genes are separated by 13,927 bp of genomic junction region, designated mCyp1a1_1a2, which contains no known or putative open reading frames. The genomic map was verified by Southern blot analysis employing cDNA probes corresponding to the first exon and proximal promoter regions of Cyp1a2 (probe1) or Cyp1a1 (Probe2) (Fig. 1A–B, C). The size of digested BAC and genomic DNA fragments by restriction enzymes, KpnI, AflII, EcoRI, SacI and PstI, corresponded with predicted sizes estimated from the sequences of the Cyp1a1 and Cyp1a2 loci. The results demonstrate that the sequence of Cyp1a1 and Cyp1a2 loci in native mouse genome would be the same in BAC clone.
Fig. 1. Genomic structure of mouse Cyp1a1 and Cyp1a2 loci.
(A) Schematic diagram and R.E. digestion map of Cyp1a1 and Cyp1a2. Probe 1; −622 to +298 of Cyp1a2 gene, Probe 2; −246 to +257 of Cyp1a1 gene. A; AflII, E; EcoRI, K;KpnI, P; PstI, S; SacI site. Open box; untranslated exon, Solid box; translated exon. (B) Determination of physical distance between Cyp1a1 and Cyp1a2 in BAC17278 by Southern blot analysis. (C) Comparison of restriction enzymes digestion pattern between BAC clone and Genomic DNA (C57BL/6J and CBA/2 mice).
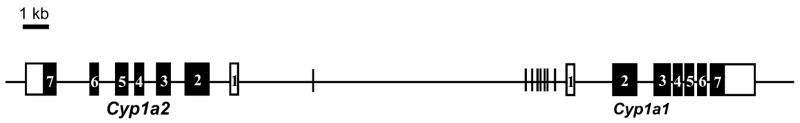
The Cyp1a1 and Cyp1a2 genes are known as AHR inducible genes. Since it is generally considered that induction of the AHR targeted gene is regulated through dioxin responsive elements (5′-TNGCGTC-3′) [3–4], we screened the sequence of mCyp1a1_1a2 to determine the precise number and location of potential DREs (Fig. 2). A total of eight DREs were identified and seven out of eight DREs were located 1.4 kb upstream region from the transcriptional start site of Cyp1a1 (Table 1). In contrast, there is no DRE up to 4.2 kb upstream region from the transcriptional start site of Cyp1a2 gene.
Fig. 2. Location of DREs on mCyp1a1_1a2 region.
Bar line; DRE consensus sequence.
Table 1.
Sequences and positions of DREs on mCyp1a1_1a2 region
| DRE (No.) | Location from Cyp1a1 | Sequence | Location from Cyp1a2 | Reference |
|---|---|---|---|---|
| 1 | −488 | cTcGCGTGaga | −13434 | [6, 19] |
| 2 | −821 | cTcGCGTGgat | −13101 | [5–6, 19] |
| 3 | −892 | cTaGCGTGcgt | −13030 | [5–6, 19] |
| 4 | −981 | gTtGCGTGaga | −12941 | [5–6, 19] |
| 5 | −1058 | cTaGCGTGaca | −12864 | [5–6, 19] |
| 6 | −1203 | tTtGCGTGcga | −12719 | [5–6, 19] |
| 7 | −1379 | tTgGCGTGtct | −12543 | [19] |
| 8 | −9738 | tTgGCGTGgga | −4184 |
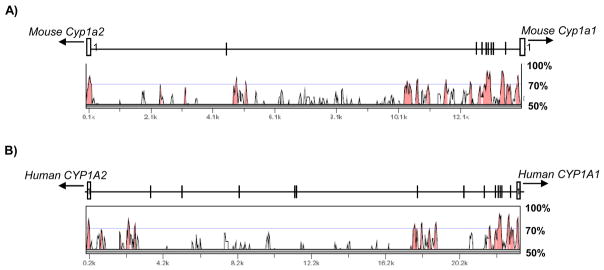
The genomic sequence of human CYP1A1 and CYP1A2 loci was determined by Corchero et al [15]. Interestingly, genomic structure of human CYP1A1 and CYP1A2 loci is also in a head to head orientation and separated by approximately 23.3 kb of genomic junction region (hCYP1A1_1A2). This fact suggests that the genomic structure of Cyp1a1 and Cyp1a2 genes is conserved between human and mouse, although the size of mCyp1a1_Cyp1a2 junction is 9.4 kb smaller than that of the hCYP1A1_1A2 junction. Since induction of Cyp1a1 and Cyp1a2 mediated by AHR are observed in both species [3–4, 18], we predict that the AHR regulatory elements of Cyp1a1 and Cyp1a2 should be conserved between human and mouse. To identify the conserved AHR regulatory elements between mCyp1a1_Cyp1a2 and hCYP1A1_CYP1A2, we compared to the sequences by employing VISTA as comparative pair-wise alignment analysis tool (Fig. 3). Overall, the conserved sequences, defined above 70% sequence identity over 50 bp length, of mCyp1a1_1a2 was only 13% compared to that of hCYP1A1_1A2. However, the sequence conservation of 1.5 kb upstream region immediately proximal the mouse Cyp1a1 gene was 55% compared to that of human CYP1A1 gene. This region has seven DREs in mouse and five DREs in human, respectively. It has been reported that two DREs (mouse DRE1 and DRE4) of them are fully conserved in the position and sequence with the cluster of human DREs (human DRE1 and DRE3), and all of them show AHR binding activity in vitro [19]. The observation suggests that the two conserved DREs could represent significant and universal AHR regulatory elements for Cyp1a1. In contrast, examination of upstream region of Cyp1a2 gene shows little conservation with the exception of the proximal promoter region of Cyp1a2 (+1 to −147 from Cyp1a2) and a part of the upstream regions (−4720 to −5158 from Cyp1a2). Although one consensus DRE is found at −4184 from Cyp1a2 gene, this DRE is not conserved with no counterpart in hCYP1A1_1A2. Interestingly, the conserved upstream region of mouse Cyp1a2 gene from −4720 to −5158 corresponds to that of human CYP1A2 gene from −2,138 to −2,602. Although this conserved region shows AHR-mediated enhancer activity and contains another type of AHR regulatory element, X1 and X2, in humans [13], no counterpart of X1 or X2 is present in the mouse. We assume three possibilities responsible for the AHR regulatory elements of Cyp1a2. First, the non-conserved DRE could act as mouse-specific AHR regulatory element of Cyp1a2. Second, another AHR regulatory element could exist in the conserved upstream region (−4720 to −5158 from Cyp1a2). Finally, Cyp1a1 and Cyp1a2 genes could share the DREs cluster proximal to Cyp1a1 as a common AHR regulatory element.
Fig. 3. Comparative analysis of genomic sequences between mCyp1a1_1a2 and hCYP1A1_1A2 by VISTA.
Curve indicates the conserved percentage of sequences between mCyp1a1_1a2 and hCYP1A1_1A2, (A) mCyp1a1_1a2 vs hCYP1A1_1A2, (B) hCYP1A1_1A2 vs mCyp1a1_1a2. The red area represents conserved sequences (above 70% identity over 50 bp length). The schematic diagrams of (A) mCyp1a1_1a2 and (B) hCYP1A1_1A2 (Upper), and distribution map of conserved sequences (lower) correspond to each other. Open box; exon 1, Bar line; DRE.
Analysis of the genomic structures of the Cyp1a1 and Cyp1a2 loci in cattle, dog and rat (GenBank; NC007319, NC00612, NW047799) showed that the three species also had the “head to head” genomic structure of Cyp1a1 and Cyp1a2 loci. The size of genomic junction region between Cyp1a1 and Cyp1a2 in cattle, dog and rat was 19.1 kb, 18.9 kb and 13.8 kb, and the conservation percentage was 49%, 38% and 11%, respectively, compared to hCYP1A1_1A2. In these species, several DREs were present in 1.5 kb upstream region of Cyp1a1 gene. Although the total number of DREs in the genomic junction region was different, the two of highly conserved DREs confirmed in mouse and human are conserved in the position and sequence among these species (Table 2). Our finding suggest that the genomic structure of Cyp1a1 and Cyp1a2 loci, and the DREs cluster are highly conserved across mammalians, although the size of genomic junction region between the Cyp1a1 and Cyp1a2 genes are different.
Table 2.
Sequences and positions of DREs on Cyp1a1_1a2 in human, cattle, dog and rat
| Organism | DRE (No.) | Location from Cyp1a1 | Sequence |
|---|---|---|---|
| Human | 1a | −497 | cTcGCGTGaga |
| 2 | −892 | cTtGCGTGcgc | |
| 3b | −980 | gTtGCGTGaga | |
| 4 | −1061 | cTcGCGTGact | |
| 5 | −1373 | tTtGCGTGcct | |
| 6 | −2116 | gTgGCGTGatc | |
| 7 | −5604 | gTgGCGTGatc | |
| 8 | −12213 | tTtGCGTGaga | |
| 9 | −12384 | cTgGCGTGagc | |
| 10 | −15061 | aTgGCGTGaac | |
| 11 | −18617 | aTaGCGTGcct | |
| 12 | −20417 | aTgGCGTGatc | |
|
| |||
| Cattle | 1a | −490 | cTcGCGTGaga |
| 2 | −949 | cTaGCGTGcct | |
| 3b | −1040 | gTtGCGTGaga | |
| 4 | −1118 | cTcGCGTGact | |
| 5 | −1273 | tTtGCGTGcag | |
| 6 | −4276 | tTtGCGTGaga | |
|
| |||
| Dog | 1a | −506 | cTcGCGTGaga |
| 2b | −972 | gTtGCGTGaga | |
| 3 | −1036 | tTcGCGTGaca | |
| 4 | −1115 | tTtGCGTGcgg | |
| 5 | −4332 | tTgGCGTGgag | |
| 6 | −4379 | tTtGCGTGaga | |
| 7 | −5152 | tTtGCGTGccc | |
| 8 | −15797 | aTtGCGTGagc | |
| 9 | −17856 | cTgGCGTGcca | |
|
| |||
| Rat | 1a | −538 | cTaGCGTGaga |
| 2 | −898 | tTgGCGTGcac | |
| 3 | −923 | cTgGCGTGcgt | |
| 4b | −1012 | gTtGCGTGaga | |
| 5 | −1089 | cTaGCGTGaca | |
| 6 | −1235 | tTtGCGTGcaa | |
| 7 | −1409 | tTgGCGTGtct | |
Sequences and positions of DREs on human CYP1A1_CYP1A2 were referred to AF253322 (GenBank) and [15].(XRE13 was not included.) Those of DREs on cattle, dog and rat Cyp1a1_Cyp1a2 were determined by analyzing genomic contig sequeneces NC007319, NC006612 and NW047799, respectively.
Highly conserved DREs in the position and sequence among these organisms
We speculate that the structural conservation of Cyp1a1 and Cyp1a2 loci, and different lengths of Cyp1a1_1a2 might be related to differential evolutionary stresses. During the course of evolution, the organism(s) might have experienced selective pressure for a broad range of regulatory elements for Cyp1a1 and Cyp1a2 genes as a response to diverse environments. As a result, the length of Cyp1a1_Cyp1a2 could be extended or reduced in this process. In contrast to the alternation of length of the Cyp1a1_Cyp1a2, the DREs cluster is highly conserved among species. This suggests that the conservation of the DREs cluster in Cyp1a1_Cyp1a2 would be important in biology. It is understood that the AHR plays important roles in dioxin or PAHs-induced toxicity, hepatic vascular development and up-regulation of the adaptive battery of xenobiotics metabolism enzymes [3–4, 18, 20–21]. Our experiments with Ahr and Arnt mutant mouse models demonstrate that the AHR carries out these functions through the binding to DRE [22–23]. Therefore, we prospect that the DREs cluster might be essential to control these AHR functions in all mammalian species. We believe that the analysis of Cyp1a1 and Cyp1a2 loci would help to clarify the regulatory mechanism(s) of Cyp1a1 and Cyp1a2 genes, the role of DREs cluster in AHR biology, evolutional and biological meaning of these conservations among species.
Acknowledgments
This work was supported by National Institutes of Health Grants R01-ES-013566-01, P01-CA-22484-27 and P30-CA-014520-29. We thank Kevin R. Hayes for performing shotgun sequencing and Anna L. Shen for reviewing the manuscript.
The abbreviations
- AHR
aryl hydrocarbon receptor
- ARNT
AHR nuclear translocator
- BAC
bacterial artificial chromosome
- DRE
dioxin responsive element
- CYP
cytochrome P450
- dioxin
2, 3, 7, 8-tetrachlorodibenzo-p-dioxin
- Cyp1a1_1a2
Genomic junction region of Cyp1a1 and Cyp1a2
- kb
kilo basepair(s)
- PAHs
polycyclic aromatic hydrocarbons
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–43. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Yang SK. Stereoselectivity of cytochrome P-450 isozymes and epoxide hydrolase in the metabolism of polycyclic aromatic hydrocarbons. Biochem Pharmacol. 1988;37:61–70. doi: 10.1016/0006-2952(88)90755-1. [DOI] [PubMed] [Google Scholar]
- 3.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–25. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 4.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–40. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 5.Fisher JM, Wu L, Denison MS, Whitlock JP., Jr Organization and function of a dioxin-responsive enhancer. J Biol Chem. 1990;265:9676–81. [PubMed] [Google Scholar]
- 6.Lusska A, Shen E, Whitlock JP., Jr Protein-DNA interactions at a dioxin-responsive enhancer. Analysis of six bona fide DNA-binding sites for the liganded Ah receptor. J Biol Chem. 1993;268:6575–80. [PubMed] [Google Scholar]
- 7.Jaiswal AK, Gonzalez FJ, Nebert DW. Human dioxin-inducible cytochrome P1-450: complementary DNA and amino acid sequence. Science. 1985;228:80–3. doi: 10.1126/science.3838385. [DOI] [PubMed] [Google Scholar]
- 8.Kubota M, Sogawa K, Kaizu Y, Sawaya T, Watanabe J, Kawajiri K, et al. Xenobiotic responsive element in the 5′-upstream region of the human P-450c gene. J Biochem. 1991;110:232–6. doi: 10.1093/oxfordjournals.jbchem.a123562. [DOI] [PubMed] [Google Scholar]
- 9.Fujii-Kuriyama Y, Imataka H, Sogawa K, Yasumoto K, Kikuchi Y. Regulation of CYP1A1 expression. FASEB J. 1992;6:706–10. doi: 10.1096/fasebj.6.2.1537460. [DOI] [PubMed] [Google Scholar]
- 10.Strom DK, Postlind H, Tukey RH. Characterization of the rabbit CYP1A1 and CYP1A2 genes: developmental and dioxin-inducible expression of rabbit liver P4501A1 and P4501A2. Arch Biochem Biophys. 1992;294:707–16. doi: 10.1016/0003-9861(92)90745-i. [DOI] [PubMed] [Google Scholar]
- 11.Zeruth G, Pollenz RS. Isolation and Characterization of a Dioxin-Inducible CYP1A1 Promoter/Enhancer Region from Zebrafish (Danio rerio) Zebrafish. 2005;2:197–210. doi: 10.1089/zeb.2005.2.197. [DOI] [PubMed] [Google Scholar]
- 12.Owens RA, Nebert DW. Expression of the chloramphenicol acetyltransferase (CAT) reporter gene by the murine Cyp1a-2 (cytochrome P3(450)) promoter in hepatoma cell cultures. Biochem Biophys Res Commun. 1990;172:1109–15. doi: 10.1016/0006-291x(90)91561-6. [DOI] [PubMed] [Google Scholar]
- 13.Quattrochi LC, Vu T, Tukey RH. The human CYP1A2 gene and induction by 3-methylcholanthrene. A region of DNA that supports AH-receptor binding and promoter-specific induction. J Biol Chem. 1994;269:6949–54. [PubMed] [Google Scholar]
- 14.Sogawa K, Numayama-Tsuruta K, Takahashi T, Matsushita N, Miura C, Nikawa J, et al. A novel induction mechanism of the rat CYP1A2 gene mediated by Ah receptor-Arnt heterodimer. Biochem Biophys Res Commun. 2004;318:746–55. doi: 10.1016/j.bbrc.2004.04.090. [DOI] [PubMed] [Google Scholar]
- 15.Corchero J, Pimprale S, Kimura S, Gonzalez FJ. Organization of the CYP1A cluster on human chromosome 15: implications for gene regulation. Pharmacogenetics. 2001;11:1–6. doi: 10.1097/00008571-200102000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brudno M, Poliakov A, Minovitsky S, Ratnere I, Dubchak I. Multiple whole genome alignments and novel biomedical applications at the VISTA portal. Nucleic Acids Res. 2007;35:W669–74. doi: 10.1093/nar/gkm279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitlock JP, Jr, Chichester CH, Bedgood RM, Okino ST, Ko HP, Ma Q, et al. Induction of drug-metabolizing enzymes by dioxin. Drug Metab Rev. 1997;29:1107–27. doi: 10.3109/03602539709002245. [DOI] [PubMed] [Google Scholar]
- 19.Kress S, Reichert J, Schwarz M. Functional analysis of the human cytochrome P4501A1 (CYP1A1) gene enhancer. Eur J Biochem. 1998;258:803–12. doi: 10.1046/j.1432-1327.1998.2580803.x. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci USA. 1996;93:6731–6. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahvis GP, Lindell SL, Thomas RS, McCuskey RS, Murphy C, Glover E, et al. Portosystemic shunting and persistent fetal vascular structures in aryl hydrocarbon receptor-deficient mice. Proc Natl Acad Sci USA. 2000;97:10442–7. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunger MK, Moran SM, Glover E, Thomae TL, Lahvis GP, Lin BC, et al. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J Biol Chem. 2003;278:17767–74. doi: 10.1074/jbc.M209594200. [DOI] [PubMed] [Google Scholar]
- 23.Bunger MK, Glover E, Moran SM, Walisser JA, Lahvis GP, Hsu EL, et al. Abnormal Liver Development and Resistance to 2,3,7,8-Tetrachlorodibenzo-p-dioxin Toxicity in Mice Carrying a Mutation in the DNA Binding Domain of the Aryl Hydrocarbon Receptor. Toxicol Sci. 2008 doi: 10.1093/toxsci/kfn149. in printing. [DOI] [PMC free article] [PubMed] [Google Scholar]