Abstract
Plasmodium falciparum thymidylate synthase-dihydrofolate reductase (TS-DHFR) is an essential enzyme in nucleotide biosynthesis, and a validated molecular drug target in malaria. Because P. falciparum TS and DHFR are highly homologous to their human counterparts, existing active-site antifolate drugs can have dose-limiting toxicities. In humans, TS and DHFR are two separate proteins. In P. falciparum, however, TS-DHFR is bifunctional, with both TS and DHFR active sites on a single polypeptide chain of the enzyme. Consequently, P. falciparum TS-DHFR contains unique distant or ‘non-active’ regions which might modulate catalysis: 1) an N-terminal tail; and 2) a ‘linker’ region tethering DHFR to TS, and encoding a ‘crossover helix’ that forms critical electrostatic interactions with the DHFR active site. The role of these non-active sites in the bifunctional P. falciparum TS-DHFR is unknown. We report the first in-depth, pre-steady state, kinetic characterization of the full-length, WT P. falciparum TS-DHFR enzyme, and probe the role of distant, non-active regions through mutational analysis. We show that the overall rate-limiting step in the WT P. falciparum TS-DHFR enzyme is TS catalysis. We further show that if TS is in an ‘activated’ (liganded) conformation, the DHFR rate is 2-fold activated, from 60 s−1 to 130 s−1 in the WT enzyme. The TS rate is also reciprocally activated by ~1.5-fold if DHFR is in an activated, ligand-bound conformation. Mutations to the linker region affect neither catalytic rate nor domain-domain communication. Deletion of the N-terminal tail – although in a location remote to the active site - decreases DHFR single and the bifunctional TS-DHFR rate by a factor of 2. The two-fold activation of the DHFR rate by TS ligands remains intact, although even the activated N-terminal mutant has just half the DHFR activity of the WT enzyme. However, the reciprocal communication – between TS active site and DHFR ligands - is impaired in N-terminal mutants. Surprisingly, deletion of the analogous N-terminal tail in Leishmania major TS-DHFR causes a 3-fold enhancement of the DHFR rate from ~14 s−1 to ~40 s−1. In summary, our results demonstrate a complex interplay of domain-domain communication and non-active site modulation of catalysis in P. falciparum TS-DHFR. Furthermore, each parasitic TS-DHFR is activated by unique mechanisms, modulated by their non-active site regions. Finally, our studies suggest the N-terminal tail of P. falciparum TS-DHFR is a highly selective, novel target for potential antifolate development in malaria.
Keywords: Dihydrofolate reductase-thymidylate synthase (DHFR-TS), Plasmodium falciparum, antifolates, presteady state kinetics, non-active site
Malaria is a parasitic disease that kills over 3,000 a day and infects some 300 million people per year. The causative agents of malaria are Plasmodium spp. parasites, bred in warm stagnant waters, and spread in humans by the bite of the Anopheles mosquito. While many species of Plasmodium are endemic to different regions, it is Plasmodium falciparum which accounts for over 90% of malaria-related deaths. Despite advances in healthcare, mortality from malaria has increased by nearly 25% over the last decade in sub-Saharan Africa, due primarily to an increase in drug-resistant parasites. High-dose therapy is often limited by the toxicity of the anti-malarials [1]. New, targeted therapies are urgently needed.
Thymidylate synthase-dihydrofolate reductase (TS-DHFR) is a critical metabolic enzyme and validated drug target in P. falciparum [2]. As illustrated in Figure 1a, TS catalyzes the conversion of methylene tetrahydrofolate (CH2H4-folate) to dihydrofolate (H2-folate) while methylating 2’-deoxyuridine monophosphate (dUMP) to 2’-deoxythymidine monophosphate (dTMP) [2]. DHFR subsequently converts H2-folate to H4-folate while oxidizing NADPH to NADP+. Both H2-folate and H4-folate are important metabolites in the nucleotide biosynthesis pathway; consequently, active-site inhibitors of P. falciparum DHFR (e.g. pyrimethamine, cycloguanil) have been used successfully in antifolate therapy in malaria [3].
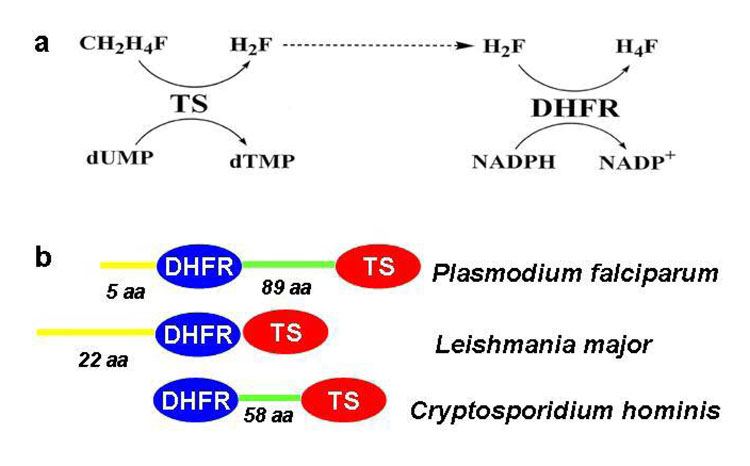
Figure 1. The Reaction Scheme and the Non-active Site Regions of Bifunctional Parasitic TS-DHFR enzymes.
a) TS catalyzes the conversion of CH2H4folate and dUMP to dTMP and H2folate. H2-folate is converted to H4-folate at the DHFR active site while an NADPH is oxidized to NADP+.
b) Diagrammatic representation of organism-specific differences in non-active site regions of TS-DHFR. The N-terminal tail (yellow) of the DHFR active site (blue) can be of variable length, or completely absent. This is also true of the linker or junctional region (green), which links the DHFR and TS (red) active sites.
TS-DHFR is a bifunctional enzyme. Most higher eukaryotes, including humans and prokaryotes, express TS and DHFR as two discrete enzymes. However, several human parasites like P. falciparum encode TS and DHFR on a single polypeptide chain, with DHFR N-terminal to TS [2–4]. Previous studies have highlighted novel characteristics of these bifunctional enzymes, with significant variation in different parasites [5–7]. One such interesting property is domain-domain communication; i.e. if the TS active site is liganded in the Leishmania major TS-DHFR, a ten-fold activation of the DHFR rate results [6]. Conformational changes in TS and DHFR are thought to modulate this domain-domain communication [4]. Notably, this activation is absent in the Cryptosporidium hominis enzyme [5], and has not previously been explored in P. falciparum TS-DHFR.
Because TS and DHFR are evolutionarily well-conserved, there is high homology with the corresponding enzymes from other eukaryotes (including humans). However, flanking the TS and DHFR active sites in parasitic TS-DHFR’s are distinct, distant, structural or “non-active site” regions, which have no homology between organisms [8–10]. Our work thus focuses on two such non-active site regions in P. falciparum TS-DHFR: the crossover helix of the linker region, and the N-terminal tail (Figure 1b).
The linker, a long region connecting TS and DHFR, is 89 and 52 aa in P. falciparum and C. hominis, respectively. The L. major TS-DHFR has no linker (Figure 1b). Although there is no sequence homology between the linkers of P. falciparum and C. hominis, both encode a 15-aa crossover helix, one face of which forms electrostatic interactions with the backside of the DHFR active site [8–10]. The crossover helices in C. hominis and P. falciparum TS-DHFR vary in their amino acid compositions and interactions. In C. hominis, the face of the crossover is composed of hydrophilic and hydrophobic residues (like S195, D198, L202, I206, R210), which interact with several important residues in the DHFR backbone (like Y132, N42, F35, F172 and E31). However, remarkably, the crossover helix in P. falciparum TS-DHFR (residues 283 to 295) has numerous acidic residues (residues 283 to 289 are either Asp or Glu, followed by F290, V291, Y292, F293, N294, and F295), which form electrostatic interactions with the many positively charged residues (primarily Lys) in the backside of the DHFR active site (shown in Figure 2b). Mutational disruption of the electrostatic interactions of the crossover helix in C. hominis TS-DHFR diminishes DHFR catalytic rates by 10-fold if the entire crossover helix is substituted with Alanines, or by 3-fold with Alanine-substitution of only the face of the helix which forms interactions with the DHFR active site [11]. However, the kinetic consequences of disrupting the electrostatic interactions crossover helix in P. falciparum TS-DHFR have not yet been studied. The interactions between the crossover helix face and the DHFR active site that can potentially be disrupted by mutagenesis are shown in Figure 2b.
Figure 2. The Non-active Sites Regions of P. falciparum TS-DHFR enzymes, highlighting the interactions disrupted in the Ala-FACE and D4 mutants.
a) The Pf TS-DHFR (PDB entry: 1J3I) structure is colored with DHFR domains in blue, TS in red, with the linker region highlighted in green, and the N-terminal tail in yellow.
b) The interactions between the crossover helix (green) and the backside DHFR active site (burgundy) disrupted by the Ala-FACE mutant. The residues shown are Asp 284, Glu 285, Asp 288, Asp 289 and Tyr 292 in the crossover helix, and Lys 69, Lys 72, Tyr 158, Lys 160, Lys 180, Lys 181 and Tyr 183 in the DHFR backbone. The substrates WR99210 and NADPH are shown in navy blue to mark the active site. For clarity, only a single DHFR monomer and its adjacent TS domain are shown, and also the residues Asn 231 to Asp 222 and Lys 227 are not shown.
c) Interactions of the residues of the N-terminal tail (shown in yellow) with Insert II (shown in orange) and with the αE-βE loop (shown in purple). The residues highlighted are Glu 3 and Val 5 in the N-terminal tail, Tyr 90 in Insert II and Ile 50 in the αE-βE loop. The substrates WR99210 [23] and NADPH are shown in navy blue to mark the active site. Only a single DHFR domain is shown for clarity.
The N-terminal tail comprises the first few amino acids preceding the DHFR domains in certain TS-DHFR’s. P. falciparum TS-DHFR has a 6 aa long N-terminal tail, oriented away from the DHFR, in a position remote from active site [9] (Figures 2a and c). The N-terminal tail is absent in C. hominis TS-DHFR, and in L. major, it is 22 aa long and is of unknown function (Figure 1b). The L. major N-terminal tail has no homology to its counterpart in P. falciparum, and wraps around the surface of the bifunctional enzyme making extensive direct contacts with the TS domain [10]. Studies done on the monofunctional P. falciparum DHFR and TS enzymes suggest that deletions of the N-terminal region decrease activity of DHFR and co-expressed TS [12, 13]. However, the role of the N-terminal tail in modulating catalysis in the full-length, bifunctional TS-DHFR enzyme has not yet been studied.
To investigate the kinetics of the WT P. falciparum TS-DHFR and the role of its non-active site regions, we used pre-steady state kinetic analysis. Transient kinetic methodology allows one to follow chemical catalysis individually at both TS and DHFR active sites, to investigate chemical rates of all three reactions (TS, DHFR and TS-DHFR) individually, to monitor intermediates as they shift from one active site to another, and to investigate the importance of communication between these active sites.
Our first objective was a detailed transient kinetic analysis of the wildtype (WT) P. falciparum TS-DHFR enzyme. Specifically, using rapid chemical quench and stopped flow fluorescence, we asked whether communication occurs between the TS and DHFR domains of the enzyme. Our second objective was to determine whether the electrostatic interactions of the crossover helix play an important role in catalysis and communication in P. falciparum TS-DHFR. The “face” of the crossover helix which interacts with the DHFR active site is comprised of five residues; we substituted only these five residues with Alanines to form the “Ala-FACE” helix mutant, or replaced the entire crossover helix with Alanines to form the “all-Ala” helix mutant of P. falciparum TS-DHFR. Finally, our third objective was to understand the effects of an N-terminal tail mutation on the pre-steady state kinetics the bifunctional, full-length P. falciparum TS-DHFR enzyme. The results of deleting residues 2 to 5 of N-terminal tail in P. falciparum (“D4”) were compared to those of an analogous deletion of residues 2 to 22 of the N-terminal tail in L. major (“N22”). We used the same nomenclature for the P. falciparum N-terminal mutants as described previously in the literature [12].
This is the first systematic, transient kinetic characterization of the WT P. falciparum TS-DHFR enzyme, and of the non-active site regions of any bifunctional parasitic TS-DHFR. We demonstrate that elegant communication exists between the TS and DHFR active sites of P. falciparum TS-DHFR. Also, we show that mutations of the linker crossover helix do not disrupt the catalytic rates in this enzyme. A four amino-acid deletion of the N-terminal tail, however, affects the chemical rates of the TS, DHFR and TS-DHFR reactions in P. falciparum TS-DHFR. Surprisingly, analogous mutations in the L. major N-terminal tail increase the DHFR chemical rate of the enzyme, suggesting differential regulation between bifunctional TS-DHFR’s from different organisms. Our studies of the P. falciparum TS-DHFR enzyme highlight the unique regulation of the kinetic mechanisms of bifunctional parasitic TS-DHFR enzymes, especially since P. falciparum encodes both an N-terminal tail and a linker with crossover helix. Since TS-DHFR is also a validated drug target, understanding the role of these distinct structural regions may also aid in rational drug design for the development of more targeted and less toxic therapies for this deadly disease.
METHODS
Reagents, Chemicals and Bacterial strains
We used reagents of the highest available commercial grade. Millipore ultrapure water was used in the preparation of all solutions. β-nicotinamine adenine dinucleotide 2’-phosphate (NADPH), dUMP (2’-deoxyuridine 5’-monophosphate disodium salt), 5-Fluoro-2-‘-deoxyuridine 5’-monophosphate sodium salt (FdUMP), sodium hydrosulfite, chloramphenicol and methotrexate agarose were purchased from Sigma Aldrich (St Louis, MO). 1,4-Dithitothreitol, isopropyl β-D-thiogalactopyranoside (IPTG) and ampicillin (D-α-aminobenzylpenicillin) were purchased from American Bioanalytical. Luria Broth (LB) and Terrific Broth (TB) were obtained from Invitrogen (Carlsbad, Ca) and VWR International (WestChester, PA), respectively.
The tritiated H2folate was prepared also by reducing commercially available [3’,5’,7,9-3H]folic acid (Moravek Biochemicals, Brea, CA) with sodium hydrosulfite, and purified using a triethylammonium bicarbonate (TEAB) gradient on a DE-52 resin [14, 15]. CH2H4folate (methylene) was prepared first by enzymatic conversion of H2folate to (6R,S)-5,6,7,8-tetrahydrofolate, and then by a condensation reaction in the presence of formaldehyde. Radiolabeled CH2H4folate was prepared from tritiated-H2 folate, with the synthesis otherwise identical to that of the unlabelled CH2H4folate. Both unlabelled and tritiated CH2H4folate were purified using a TEAB gradient on a DE-52 resin (Whatman) [14, 15]. The purity of the radiolabelled substrates was confirmed using HPLC analysis (see below). Calcium-competent Escherichia coli strains BL21 (DE3) pLysS were purchased from Invitrogen (Carlsbad, CA).
Enzyme Mutagenesis, Expression and Purification
The plasmid expressing the TM4/8.2 strain of wildtype P. falciparum TS-DHFR is a generous gift from Drs. Penchit Chitnumsub and Yongyuth Yuthavong (BIOTEC, National Science and Technology Development Agency, Thailand) [16]. Using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), we mutated P. falciparum TS-DHFR residues 283 to 295 to Ala to create an “all-Alanine” crossover helix, and residues 284, 285, 288, 289, and 292 to Ala to make the “Ala-FACE” crossover helix. We also deleted residues 2 through 5 to make the N-terminal tail mutant “D4”, and residues 2 through 6 to make the mutant “D5”. Primers were synthesized and gel purified by the Yale Keck Facility according to protocols available online. The mutated plasmids were amplified using calcium-competent DH5α Escherichia coli cells, and then purified using the Qiaprep Spin Maniple Kit (Qiagen, Valencia, CA). Plasmids were sequenced by the Yale Keck Facility (New Haven, CT) to confirm the mutagenesis, and stored at −20°C. Concentration of DNA was determined spectrophotometrically using absorbance at 260nm. The plasmid expressing the TS-DHFR enzyme from L. major was the generous gift of C.-C.Kan and D. Matthews (Agouron Pharmaceuticals, La Jolla, CA). Its N-terminal residues 2 to 22 were deleted using QuikChange (Stratagene, La Jolla, CA) to create the N-terminal mutant “N22”. The plasmid was cloned into a pET11b for expression in E. coli BL21 cells. All plasmids were sequenced by the Yale Keck Facility to confirm mutagenesis.
The P. falciparum WT TS-DHFR was expressed and purified as previously described [16]. The protocol for expression of Ala-FACE and all-Ala helix mutants was identical to that for wild type P. falciparum TS-DHFR. The protocols for expression and purification of the D4 and D5 N-terminal mutants were also identical to that for the WT enzyme, except that 350 mM KCl (instead of 1 M KCl) was used to wash the unbound enzyme off of the methotrexate affinity resin. The active-site concentrations of wild type and mutant enzymes were equivalent, as determined by pre-steady state burst amplitudes of their DHFR reactions (see below). The method for expression and purification of L. major TS-DHFR WT and N23 deletion mutant were as described previously [6].
Assessment of Enzyme Concentration and Activity
Enzyme concentration was determined spectrophotometrically by following absorbance at 280nm. The extinction coefficient for P. falciparum TS-DHFR is 83740 M−1cm−1, and for L. major TS-DHFR is 69955 M−1 cm−1. DHFR steady-state activity was assessed by reacting enzyme with H2folate and NADPH, and following absorbance at 340 nm, which decreases as NADPH is depleted to form NADP+. An extinction coefficient of −12.8 mM−1 cm−1 was used for the DHFR reaction. TS steady-state activity was assayed by incubating the enzyme with CH2H4folate and dUMP, and following the increase in absorbance at 340nm with the production of dTMP and H4folate. An extinction coefficient of −6.4 mM−1 cm−1 was used for the TS reaction [17].
Rapid Chemical Quench Experiments
We performed rapid chemical quench experiments on a Kintek RFQ-3 Rapid Chemical Quench Apparatus (Kintek Instruments, Austin, TX) to enable us to study enzymatic experiments on a millisecond time scale. All reactions were performed at 25°C. The reactions involved mixing 15 µL of enzyme solution with 15 µL of substrate solution. The substrates were tritiated (as described above), which enabled subsequent high performance liquid chromatography (HPLC) analysis to follow the reaction substrates and products (see below for details). The enzyme solution consisted of a mixture of enzyme, non-limiting substrate, and 2x reaction buffer (50 mM MgCl2, 1 mM EDTA, 50 mM Tris-HCl pH=7.8). The substrate solution consisted of approximately 20,000 dpm of the appropriate tritiated substrate. The DHFR single turnover experiment consisted of mixing an “enzyme solution” of enzyme, NADPH and 2x reaction buffer with a “substrate solution” of tritiated H2folate. The TS single turnover experiment consisted of mixing an “enzyme solution” of enzyme, dUMP, and 2x reaction buffer with a “substrate solution” of tritiated CH2H4folate. Finally, the TS-DHFR bifunctional single turnover experiment consisted of mixing an “enzyme solution” of enzyme, dUMP, NADPH and 2x reaction buffer with a “substrate solution” of tritiated CH2H4folate. All reactions were quenched by addition of 67 µL of 0.78M KOH solution. In each reaction, the final concentration of reaction buffer was 1x. All concentrations for enzyme and substrate listed in the text are final concentrations after mixing.
High Performance Liquid Chromatography Analysis
The tritiated reaction products from the rapid chemical quench reactions were analyzed by high performance liquid chromatography (HPLC) with radioactivity and ultraviolet flow detectors. A BDS-Hypersil C18 reverse phase column (250 × 4.6 mm2, Keystone Scientific, Bellefonte, PA) was used for separation. We used a solvent consisting of 10% Methanol in 180 mM triethylammonium bicarbonate (pH=7.8), in isocratic separation mode at a flow rate of 1 mL/min, for optimal separation of our substrates and products. Under these conditions, tritiated H4folate eluted at 7 minutes, tritiated H2folate at 14 minutes, and tritiated CH2H4folate at 15 minutes.
Stopped Flow Experiments
We used a Kintek SF-2001 Stopped Flow apparatus (Kintek Instruments, Austin, TX) to probe communication between the TS and DHFR domains. Reactions were performed under burst conditions, in which substrate concentration is in slight excess over enzyme concentration. For the DHFR reaction, the “enzyme solution” – consisting of enzyme, NADPH, +/− CH2H4folate, +/− FdUMP, and 2x reaction buffer - was mixed with the “substrate solution” of H2folate. For the TS reaction, the “enzyme solution” - consisting of enzyme, dUMP, +/− NADPH – was mixed with the “substrate solution” of CH2H4folate. For the DHFR reaction, we excited the reaction at 287 nm and followed the FRET (fluorescent resonance energy transfer) at 450 nm. For the TS reaction, we excited the reaction at 287 nm and followed the fluorescent emission at 340 nm.
RESULTS
Expression of P. falciparum WT and Mutant TS-DHFR enzymes
We were able to produce 0.28 mg of WT enzyme, 0.29 mg of Ala-FACE enzyme and 0.14 mg of D4 pure protein per liter of E. coli culture. The DHFR steady-state rates for the WT, Ala-FACE and D4 enzymes were 16.7 ± 0.164 s−1, 17.6 ± 0.258 s−1 and 0.496 ± 0.00421 s−1, respectively. The bifunctional TS-DHFR steady-state rates for the WT, Ala-FACE and D4 enzymes were 1.77 ± 0.0795 s−1, 1.875 ± 0.0693 s−1 and 2.53 ± 0.537 × 10−2 s−1, respectively. We also mutated the P. falciparum TS-DHFR plasmid to encode an All-Ala crossover helix. The full-length, all-Ala enzyme consistently co-purified with a 39 kDa band, which was confirmed by mass spectrometry to be a fragment of P. falciparum TS-DHFR (and not an E. coli TS or DHFR enzyme). Despite using high concentrations of protease inhibitors and expediting the protein purification, degradation of the All-Ala mutant into this 39 kDa fragment could not be prevented. Under these conditions, only 0.01 mg of pure all-Ala helix mutant could be expressed per liter of E. coli culture. When we tried to express a mutant with the crossover helix completely deleted, the protein was consistently degraded intracellularly. The interactions disrupted by the Ala-FACE and N-terminal deletion mutants are highlighted in Figure 2b and c.
We also tried to express the “D5” mutant of the N-terminal tail in which residues 2 through 6 of P. falciparum WT TS-DHFR were deleted. Protein expression was too low to allow a detailed kinetic characterization of the protein; however, the expressed enzyme reproducibly showed DHFR activity by spectrophotometric analysis.
Single Turnover Experiments for DHFR Reaction for P. falciparum TS-DHFR
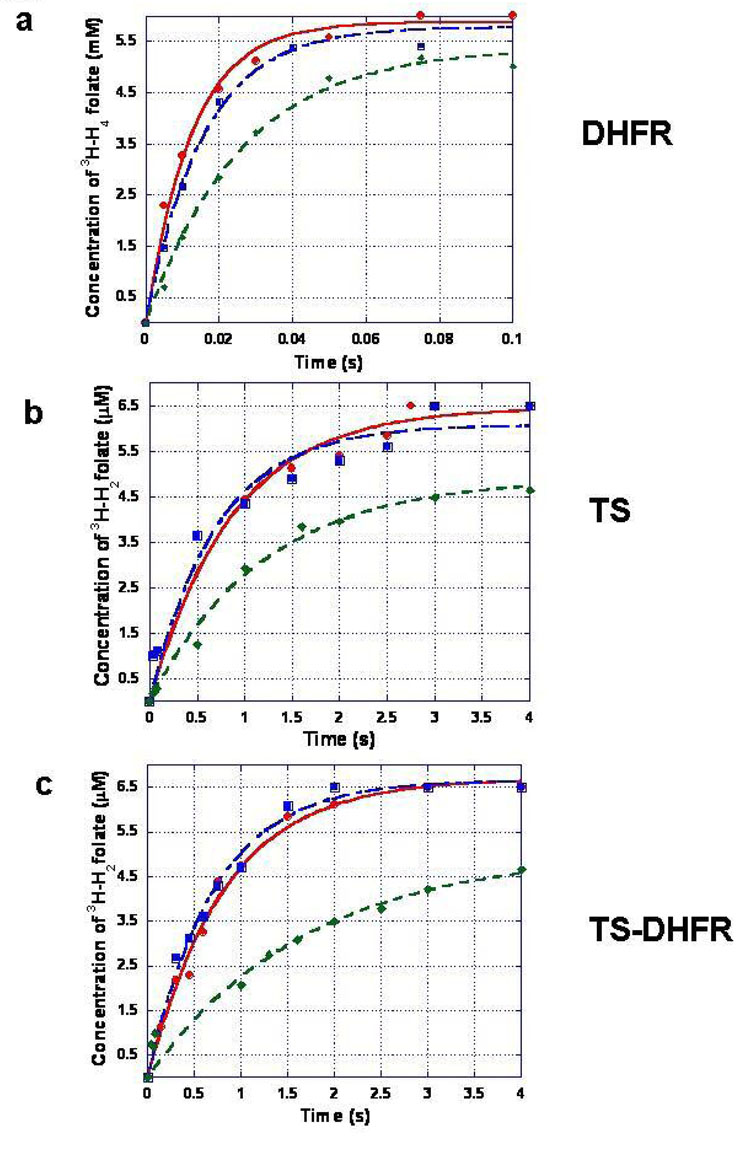
To examine the DHFR activity of the WT, Ala-FACE and D4 mutants, single enzyme turnover experiments were conducted using rapid chemical quench approach. The formation of tritiated H4-folate was monitored over time by HPLC analysis of the reaction products. We pre-incubated 80 µM of the appropriate enzyme and a saturating concentration of NADPH (500 µM), and then mixed with 6.5 µM tritiated H2-folate. All reactions were carried out to completion but only the earlier time points are shown here for clarity (Figure 3a). The data were fit to a single exponential equation, and the rate constants for the WT and Ala-FACE mutant were similar at 80 ± 5.2 s−1 and 64 ± 4.4 s−1, respectively. The rate constant for the D4 mutant was ~ 2-fold slower than WT at 38 ± 2.4 s−1.
Figure 3. Single turnover experiments for WT, Ala-FACE and D4 mutants of P. falciparum TS-DHFR show the N-terminal tail mutant slows the all three reactions, TS, DHFR and TS-DHFR.
The data for WT enzyme are (red, circle), for Ala-FACE enzyme (blue, square) and for the D4 N-terminal mutant (green, diamond) are plotted by reaction. The reactions were conducted using rapid chemical quench apparatus under single turnover conditions at 25°C.
a) The DHFR experiment was conducted with 80 µM enzyme and 500 µM NADPH mixed with 6.5 µM tritiated H2-folate.
b) The TS experiment was conducted with 80 µM enzyme and 500 µM dUMP mixed with 6.5 µM tritiated CH2H4-folate.
c) The bifunctional TS-DHFR experiment was conducted with 80 µM enzyme, 500 µM NADPH, and 500 µM dUMP mixed with 6.5 µM tritiated H2-folate.
In all cases, the data were fit to a single exponential equation, and the rate constants from these are summarized in Table 1.
Single Turnover Experiments for the TS Reaction for P. falciparum TS-DHFR
To examine the TS activity for the WT, Ala-FACE and D4 mutants, single enzyme turnover experiments were conducted again using a rapid chemical quench approach. The formation of tritiated H2-folate was followed over time by HPLC analysis of the reaction products. In these experiments, 80 µM of the appropriate enzyme was preincubated with a saturating concentration of dUMP (500 µM), and then mixed with 6.5 µM tritiated CH2H2-folate. All reactions were carried out to completion but only a subset of the data is shown here (Figure 3b). The data were fit to a single exponential equation, and the rate constants for the WT and Ala-FACE mutant were similar at 1.2 ± 0.22 s−1 and 1.4 ± 0.28 s−1, respectively. The rate constant for the D4 N-terminal tail mutant was 0.84 ± 0.083 s−1, demonstrating that the TS activity of the enzyme for the D4 is very similar to that of the WT enzyme. These results are summarized in Table 1.
Table 1.
Summary of Single turnover results for DHFR, TS and TS-DHFR reactions from Rapid Chemical Quench Experimentsa
| Enzyme | DHFR rate constant (s−1) | TS rate constant (s−1) | Bifunctional TS-DHFR rate constant (s−1) |
|---|---|---|---|
| WT | 80 ± 5.2 | 1.2 ± 0.22 | 1.2 ± 0.085 |
| Ala-FACE | 64 ± 4.4 | 1.4 ± 0.28 | 1.4 ± 0.091 |
| D4 | 38 ± 2.4 | 0.84 ± 0.083 | 0.60 ± 0.15 |
The data were fit to a single-exponential equation, and the conditions used were as summarized in the legend for Figure 3.
Single Turnover Experiments for the Bifunctional Reaction for P. falciparum TS-DHFR
To examine the TS-DHFR bifunctional activity for the WT, Ala-FACE and D4 mutants, single enzyme turnover experiments were conducted with a similar rapid chemical quench approach. In this case, the formation of tritiated H4-folate was monitored over time by HPLC analysis of the reaction products. We pre-incubated 80 µM of the appropriate enzyme with a saturating concentration of dUMP (500 µM) and NADPH (500 µM), and then mixed with 6.5 µM tritiated CH2H2-folate. The reactions were carried out to completion, but only a subset of the data is shown here for clarity (Figure 3c). The data were fit to a single exponential equation, and the rate constants for the WT and Ala-FACE mutant were similar at 1.2 ± 0.085 s−1 and 1.4 ± 0.091 s−1, respectively. The rate constant for the D4 N-terminal tail mutant for this same reaction is only half that, at 0.60 ± 0.15 s−1. These results are summarized in Figure 3c and Table 1.
Pre-steady state burst experiments of the DHFR reaction for P. falciparum TS-DHFR
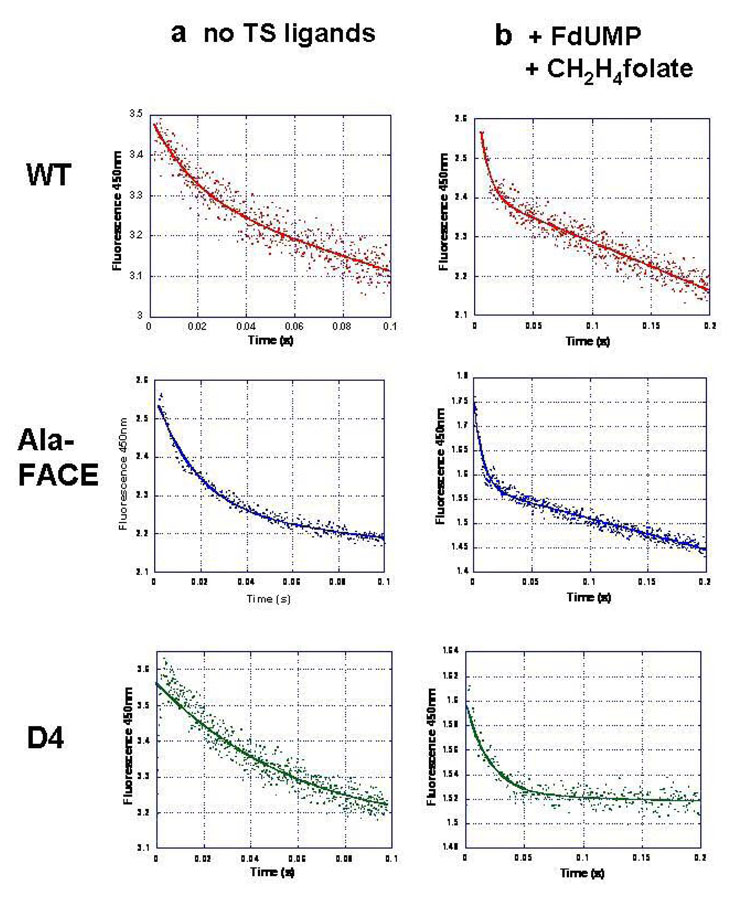
NADPH bound to the DHFR active site produces a fluorescent resonant energy transfer (FRET) at 450nm. As H2-folate is mixed, the NADPH bound at the active site is oxidized to NADP+, producing a decrease in fluorescence. Thus, stopped flow fluorescence studies can be used to follow consumption of NADPH at the DHFR active site on a millisecond time scale. The purpose of these particular studies was to confirm the rapid chemical quench DHFR chemical rates determined by following the production of H4-folate. In these reactions, 7.5 µM of the corresponding enzyme were preincubated with 500 µM NADPH, and then mixed with 50 µM H2-folate in the stopped flow. Excitation was at 287nm and emission at 450nm. The data were fit to a single exponential burst equation (Figure 4 a), and the rate constants are reported in Table 2. The chemical rates determined from the stopped flow burst experiments are very similar to those obtained by rapid chemical quench single turnover. Importantly, these results also confirm that the D4 mutant has a DHFR rate constant that is about half that of the WT and Ala-FACE enzymes.
Figure 4. DHFR rate in P. falciparum TS-DHFR is activated in the presence of TS ligands, dUMP and CH2H4-folate.
DHFR burst experiments for WT, Ala-FACE and D4 N-terminal mutants conducted in the presence or absence of TS ligands. Using stopped flow fluorescence, we performed these experiments under burst conditions, with fluorescence excitation at 287 nm and the emission at 450 nm.
Column a) shows data from DHFR burst experiments conducted in the absence of TS ligands. The conditions were used were 7.5 µM enzyme and 500 µM NADPH, mixed with 50 µM H2-folate.
Column b) shows data from DHFR burst experiments conducted in the presence of TS ligands, FdUMP and CH2H4-folate. The conditions were used were 7.5 µM enzyme, 500 µM NADPH, 100 µM FdUMP, and 25 µM CH2H4folate, mixed with 50 µM H2-folate.
WT data points are shown in red, Ala-FACE data points, in blue and D4 data points, in green. In all cases, the data were fit to a single exponential equation, and the rate constants from these are summarized in Table 2.
Table 2.
Summary of DHFR rate constants from Stopped Flow FRET experiments under burst conditions for WT, Ala-FACE and D4 mutantsa
| + no TS ligands | + FdUMP | |
|---|---|---|
| + CH2H4 folate | ||
| WT DHFR burst rate (s−1) | 61.0 ± 7.52 | 135 ± 9.18 |
| Ala-FACE DHFR burst rate (s−1) | 68.0 ± 8.14 | 161 ± 7.09 |
| D4 DHFR burst rate (s−1) | 28.9 ± 4.56 | 63.2 ± 2.93 |
The data were fit to a single-exponential burst equation, and the conditions used were as summarized in the legend for Figure 4.
DHFR Activation reaction in the presence of TS ligands FdUMP and CH2H4-folate
We also monitored the DHFR activity in the presence of the two TS ligands, CH2H4-folate and FdUMP (a dead-end inhibitor of TS), for WT and mutant P. falciparum TS-DHFR. We preincubated 7.5 µM of the corresponding enzyme with 500 µM NADPH, 100 µM FdUMP and 25 µM CH2H4folate, and then mixed with 50 µM H2-folate in the stopped flow. Excitation was at 287nm and emission at 450nm. The data were fit to a single exponential burst equation (Figure 4b) and summarized in Table 2. In the presence of TS ligands, the chemical rate is almost doubled in all three WT, Ala-FACE and D4 enzymes, and the D4 rate remains about half that of the WT enzyme.
Pre-steady state burst experiments of the TS reaction in the absence and presence of the DHFR ligand NADPH
We also asked the reciprocal question, of whether the TS rate is activated by the DHFR ligand NADPH. Using the stopped flow, we followed the change in intrinsic fluorescence at the TS active site upon substrate binding and conversion to product. Excitation was at 287nm and emission at 340nm. We preincubated 7.5 µM of the corresponding enzyme with 100 µM dUMP and mixed this with 25 µM CH2H4folate. For the experiment in the presence of NADPH, 7.5 µM of the corresponding enzyme were preincubated with 100 µM dUMP and 500 µM NADPH, and then mixed with 25 µM CH2H4folate in the stopped flow. The data were fit to a single exponential burst equation. The TS rates, in the absence of DHFR ligand NADPH, for WT, Ala-FACE and D4 mutants were 9.94 ± 0.203 s−1, 8.58 ± 0.249 s−1 and 10.1 ± 0.507 s−1, respectively. However, the TS rates in the presence of NADPH were 15.3 ± 0.428 s−1, 13.2 ± 0.324 s−1 and 10.3 ± 0.541 s−1, for the WT, Ala-FACE and D4 mutants, respectively. Thus, the DHFR ligand NADPH seems to seem activate the TS burst rate in WT and Ala-FACE enzymes, but not in the D4 mutant.
L. major N22 DHFR single turnover and Pre-steady state DHFR Burst
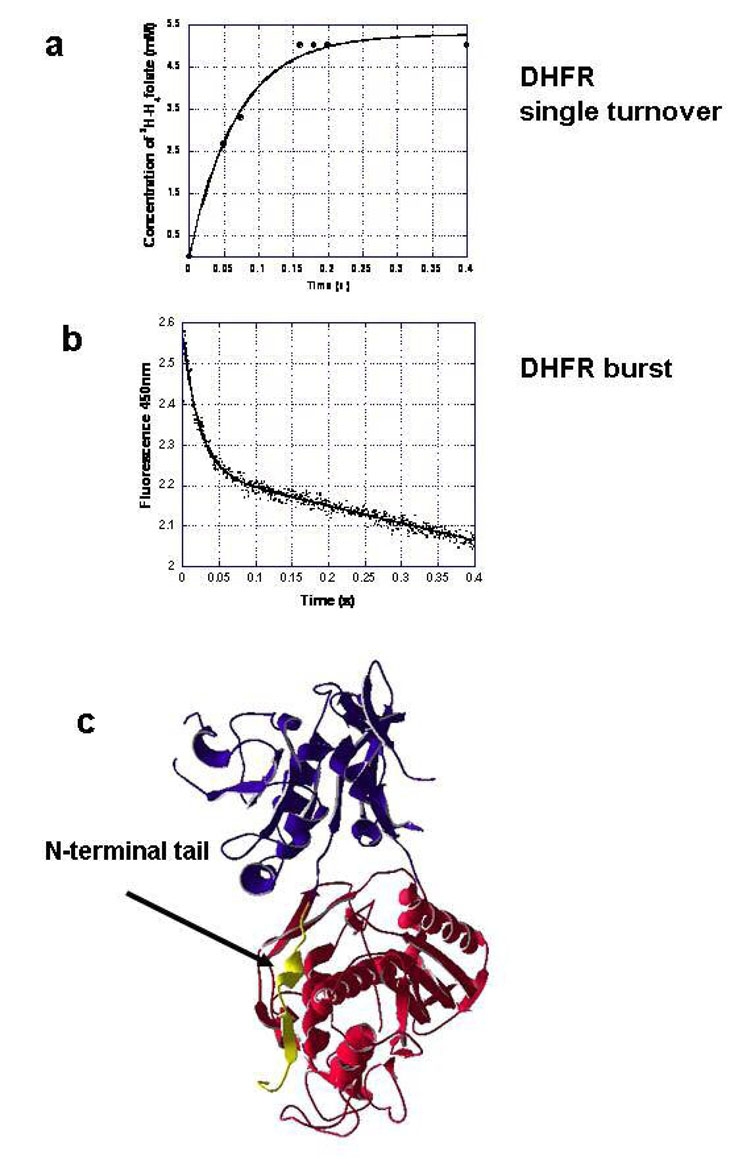
We also expressed the L. major WT bifunctional TS-DHFR enzyme, and a mutation in which only its N-terminal tail is deleted (N22). Though expression of N22 was about 0.07 mg pure protein per liter of E. coli culture, DHFR single turnover and pre-steady burst experiments could be performed. As described above, 45 µM L. major WT or N22 TS-DHFR were preincubated with 500 µM NADPH, and then mixed with 5.5 µM tritiated H2-folate in the rapid chemical quench. The production of tritiated H4-folate was monitored over time by HPLC analysis. The rate constant for DHFR is 40.2 ± 4.60 s−1 in the N22 mutant (Figure 5a), compared to the WT L. major DHFR catalytic rate of 14.5 ± 1.37 s−1 (data not shown). The DHFR rate constant for the WT L. major enzyme is similar to previously published data [6]. However, the fast chemical rate was unexpected in the N22 mutant. To confirm this fast rate, a presteady state, DHFR burst experiment of the N22 enzyme was also performed using stopped flow analysis, as outlined above for the P. falciparum enzyme. We preincubated 7.5 µM enzyme and 500 µM NADPH, and mixed with 50 µM H2-folate. Excitation was at 287 nm and emission at 450 nm, and the data were fit to a single exponential. The DHFR burst rate constant for the N22 mutant was 45.21 ± 0.853 s−1 (Figure 5 b), which is in good agreement with our single turnover rate.
Figure 5. DHFR reactions of the L. major N-terminal deletion mutant, N22, show that the N-terminal tail is autoinhibitory in the L. major TS-DHFR enzyme.
a) Single turnover, rapid chemical quench experiments for N22 conducted using 45 µM enzyme and 500 µM NADPH mixed with 5.5 µM tritiated H2-folate.
The data were fit to a single exponential equation to provide a rate constant of 40.2 ± 4.6 s−1. In comparison, the DHFR single turnover rate for WT L. major TS-DHFR had a rate constant of 14.5 ± 1.37 s−1 (data not shown).
b) N22 mutant DHFR burst experiment, performed on the stopped flow, was conducted using 7.5 µM enzyme and 500 µM NADPH, mixed with 50 µM H2-folate. The fluorescence excitation was at 287 nm and the emission was at 450 nm. The data were fit to a single exponential burst equation to provide a rate constant of 45.21 ± 0.853 s−1.
c) The L. major TS-DHFR structure is colored with DHFR domains in blue, TS in red, and the N-terminal tail in yellow, which wraps around and makes extensive contacts with the TS domain. The substrates, methotrexate, NADPH, FdUMP and 10-propargyl-5,8-dideazafolate (PDDF) are shown in gray.
DISCUSSION
Kinetics of the Wildtype P. falciparum TS-DHFR enzyme
This is the first report of the single turnover rate constants for the TS, DHFR and bifunctional TS-DHFR reactions for P falciparum TS-DHFR. We have shown that, in WT P. falciparum TS-DHFR, the DHFR chemical rate (70s−1) is significantly faster than TS. TS is the rate-limiting step in the both the TS reaction and the bifunctional TS-DHFR reaction with a chemical rate ~1 s−1 (Table 1), an observation consistent with the known kinetic mechanisms for other parasitic TS-DHFR’s [5, 6].
This is also the first report of domain-domain communication in P. falciparum TS-DHFR. We show that, if the TS active site is liganded in the P. falciparum TS-DHFR, the DHFR chemical rate is nearly doubled to ~130s−1 (Table 2). This communication between TS and DHFR active sites is important for three reasons. First, the activation of P. falciparum DHFR is analogous to the regulation of DHFR activity in the L. major TS-DHFR enzyme, but very different from that of C. hominis and T. gondii TS-DHFR’s, which are activated regardless of whether ligands are bound at the TS active site. Second, the rate of the activated P. falciparum and L. major DHFR’s are about 130 s−1 and 120 s−1 [6], respectively, while the rate constants of DHFR from C. hominis and T. gondii are about 130 s−1 and 180 s−1, respectively. This suggests that these four parasitic TS-DHFR enzymes have similar in vitro DHFR chemical rates, but different methods of activation. And third, since presumably the P. falciparum TS-DHFR is in a liganded state in the cell, this “activated” DHFR rate is probably a more accurate representation of the chemical rate. Our data also show a reciprocal communication occurs between TS and DHFR in the WT enzyme, as TS rate is activated in the presence of the DHFR ligand, NADPH.
This communication between active sites is likely mediated by conformational changes to the enzyme active site initiated by ligand-binding, a well-studied phenomenon in both the E. coli TS and DHFR enzymes [18–22]. In fact, when we performed the DHFR reactions in the presence of only FdUMP (without CH2H4-folate), the DHFR chemical rates were not activated (data not shown). The published crystal structure of P. falciparum TS-DHFR has a doubly-liganded DHFR active site (bound with NADPH and WR99210, a triazine DHFR inhibitor [23, 24]) but only a singly-liganded TS active site (bound with dUMP but no folate ligand) [9]. It would be interesting to see if our kinetic data on activation and communication are reflected in significant structural differences in a TS-DHFR where the TS is doubly liganded. If the DHFR active site is conformationally different in the presence of a fully-liganded TS, then these activation data could have considerable implications for structure-based drug design against P. falciparum TS-DHFR.
The crossover helix mutant of P. falciparum TS-DHFR enzyme
Our results show that the crossover helix does not play a role in modulating catalysis in P. falciparum TS-DHFR. We designed several mutations of the crossover helix (residues 284 to 295) in the linker region of the P. falciparum TS-DHFR enzyme, and probed these mutants for changes in the chemical rates of the TS, DHFR and bifunctional TS-DHFR reactions. The sequence encoding the crossover helix has no homology to the linker region of other parasitic TS-DHFR’s, but the helix itself is conserved in the C. hominis and T. gondii TS-DHFR enzymes. In P. falciparum, the crossover helix is very negatively charged, and interacts with positively charged residues of the backside of the active site of DHFR (as shown in Figure 2b) [9]. The importance of this helix, already suggested by its numerous interactions with the DHFR domain, was compounded by unpublished data from W. Sirawaraporn demonstrating that the shortest region of a linker-TS construct that could interact with DHFR started at residue 282, immediately N-terminal to the crossover helix. In C. hominis, mutations of the crossover helix have been shown to reduce DHFR activity [11].
Thus, we designed three mutations of the crossover helix: 1) deletion of the entire helix (Δ-helix); 2) substitution of all residues of the helix with Alanines (All-ALA); and 3) substitution of only those residues on the face of the helix which interact with the backside of the DHFR active site (Ala-FACE). The Δ-helix and All-ALA mutations underwent significant intracellular proteolysis, and could not be expressed in sufficient quantity for detailed kinetic analysis.
The Ala-FACE enzyme had TS, DHFR, and TS-DHFR chemical rates that were very similar to WT, under both single turnover and pre-steady state burst conditions (Table I–Table II). Also, the Ala-FACE enzyme also had a similar activation of the DHFR chemical rate by TS ligands (Table 2), and the TS rate by the DHFR ligand NADPH. Taken together, these results suggest that the crossover helix perhaps plays a role in stabilizing the interaction between TS and DHFR in the P. falciparum bifunctional enzyme, but does not affect catalytic rate directly. Significant disruption of the electrostatic interactions of the helix, or deletion of the helix, might expose previously internalized regions of the enzyme to cytosolic proteases. Since disruption of crossover helix interactions in C. hominis significantly decreases DHFR catalytic rate, similar results for the P. falciparum enzyme had initially been expected. However, in P. falciparum, the much higher prevalence of acidic residues in, and the larger number of electrostatic interactions made by, the helix with the DHFR backbone perhaps renders this crossover helix more critical in maintaining structural integrity in this large enzyme.
The D4 N-terminal mutant of P. falciparum TS-DHFR enzyme
The N-terminal tail of the P, falciparum protein comprises only residues 1 to 6, and yet two previous studies have suggested it plays an important role in DHFR activity and the interaction between TS and DHFR domains [12, 13]. Shallom et al. demonstrated that the integrity of the N-terminal region of P. falciparum DHFR is essential for TS to be active [13]. Wattanarangsan et al. made sequential deletions of the N-terminal region of the DHFR domain, and found that the number of amino acids deleted from the N-terminal tail corresponded with decreasing DHFR activity. They also suggest that TS will remain active in the presence of a conformationally-intact DHFR, even though the DHFR enzyme has significantly impaired activity [12]. One must note that neither of these previous studies was carried out on the full-length, bifunctional P. falciparum TS-DHFR enzyme, or involved pre-steady state kinetic analysis. They involved expressing monofunctional P. falciparum DHFR and linker-TS constructs, and monitoring their interaction with complementation assays in E. coli.
Our results confirm that deleting the N-terminal tail in P. falciparum TS-DHFR decreases DHFR catalytic rate (Figure 3 and Figure IV). Furthermore, our data newly suggest that the rate of the bifunctional TS-DHFR reaction is 2-fold slower in the D4 mutant than in wildtype, although TS chemical rate in D4 mutant is about two-thirds that of the wildtype enzymes (Table 2, Figure 3c). We also show that 2-fold activation of DHFR rate by TS ligands remains intact in the D4 mutant (Table 2), but is impaired in the activation of TS by the DHFR ligand NADPH.
It is imperative to note that the bifunctional D4 mutant is significantly more active than might have been suggested by the previous studies on the monofunctional DHFR [12]. Wattanarangsan et al. show the D4 DHFR mutant to be 80-fold less active than WT DHFR when activity is determined spectrophotometrically from crude extract, and over 400-fold less active when DHFR activity is determined by bacterial complementation assay [12]. In our studies, the D4 mutant DHFR rate is only 2-fold decreased compared to WT under single turnover conditions, and about 30-fold decreased under steady state conditions. Also, we show that the full-length D4 mutant has impaired bifunctional TS-DHFR activity under both single turnover and steady state conditions (Figure 3b). We attribute the differences between the monofunctional and bifunctional D4 enzyme studies to the elegant communication between TS and DHFR domains in the intact enzyme. Our results also suggest that the DHFR ligand NADPH does not activate the TS activity, which might explain why the bifunctional TS-DHFR chemical rate is somewhat slower than the TS single turnover rate alone. The differences in our pre-steady state in vitro data, compared to previous studies on N-terminal mutants, may also be accounted for by the different microenvironment of the intracellular environment.
Why does the N-terminal tail, although remote from the active site, affect catalysis? Since D4DHFR can still be activated in the presence of TS ligands (Figure 4), and since substrate channeling from TS to DHFR domains remains unperturbed in the D4 mutant (data not shown), we suggest that the D4 mutant is affecting co-ordinated, domain-domain motions which occur during catalysis and ligand-binding in DHFR. Such movements have been clearly demonstrated in E. coli DHFR [18, 22] and TS enzymes [19–21]. Close examination of the structure of P. falciparum TS-DHFR shows that the N-terminal tail is distant from the active site, but can interact with helices in Insert II (a 35aa region unique to Plasmodial DHFR’s) and the αβ loop which encodes part of the active site [9] (Figure 2c). Thus, the interaction between these loops and the N-terminal tail may be essential in the co-ordinated motions of DHFR during catalysis. Since the structure of the fully-liganded, P. falciparum TS-DHFR enzyme has not yet been solved, there may be additional, or distinct, interactions between the N-terminal tail and regions linked to the DHFR active site, which are not fully appreciable with the present structure.
It should be noted, however, that our data cannot exclude solvation differences between wildtype and mutant enzymes to account for the difference in their activities. We would expect such differences to be negligible, given that only four amino acids were deleted to make the D4 mutant. However, such potential solvation differences may especially become important in ligand design if the N-terminal tail of P. falciparum TS-DFHR is targeted in rational drug design [25–27].
A comparison of mutant forms of the enzyme with WT requires normalization of the active functional protein. Our previous studies on other enzymes have established that equivalent active site concentrations using pre-steady-state burst experiments offers a reliable means for comparing WT and mutant bifunctional TS-DHFR proteins. The steady state determination of TS specific activity has also been suggested as a reliable indicator of functional protein [28], but cannot be used to compare mutants with impaired TS function.
The N22 N-terminal mutant of the L. major TS-DHFR enzyme
We wanted to determine whether deleting the 22aa N-terminal tail in L. major TS-DHFR would also decrease the DHFR catalytic rate. Surprisingly, we found that the N22 mutation had a faster single turnover DHFR rate, an observation confirmed with pre-steady state burst studies on the stopped flow (Figure 5a–b). Since the N-terminal tail in L. major makes extensive contacts with the TS domain (Figure 5c) [10], perhaps it is playing an autoinhibitory role, and is dislocated during the co-ordinated motions which accompany catalysis in TS and DHFR enzymes. The pre-steady state DHFR burst experiment in the presence of TS ligands for the N22 mutant shows that the enzyme has lost its DHFR burst when TS is activated (data not shown), although it is already known that the WT L. major enzyme demonstrates clear domain-domain communication between DHFR and TS [6]. Therefore, we conclude that deleting the L. major TS-DHFR N-terminal tail leads to a faster DHFR rate of chemistry, and a disruption in the activation of DHFR by TS. Furthermore, the N-terminal tail plays opposite roles in L. major and P. falciparum TS-DHFR’s. Perhaps because of the extensive contacts it makes with the TS domain, the N-terminal may play an autoinhibitory role, and must be dislocated for the enzyme to be active (Figure 5c).
Summary and Implications for Parasitic Bifunctional TS-DHFR’s and Inhibitor Design
This is the first systematic, in-depth, pre-steady state kinetic analysis of the non-active site regions of any bifunctional TS-DHFR enzyme, especially one from P. falciparum. Our results show that, in the WT enzyme, TS catalyzes the rate-limiting step, and communication occurs between both the TS and the DHFR domains. Mutations to the face of the crossover helix in the linker region affect neither the rate of catalysis at either active site, nor inter-domain communication. Deletion of the N-terminal tail of P. falciparum TS-DHFR decreases the DHFR single turnover rate by one-half, and the bifunctional TS-DHFR rate by about one-third. While DHFR rate can still be activated 2-fold in the presence of TS ligands, reciprocal activation of TS by DHFR ligands is impaired in the D4 mutant. However, an analogous deletion in the N-terminal tail of the L. major TS-DHFR N-terminal tail leads to a three-fold activated DHFR rate and impaired DHFR-to-TS communication.
Even though there is overall structural similarity in parasite TS-DHFR’s, general conclusions about enzyme mechanism cannot be drawn: TS-DHFR enzymes from different parasites are regulated very differently; and there is minimal kinetic basis for previous phylogenetic classification of these TSDHFR enzymes by homologous structural features [29]. The N-terminal tail can have both an activating and an auto-inhibitory role. The crossover helix in the linker may increase catalytic activity or have minimal effect on rate of chemistry at DHFR [7]. Each one of these enzymes is unique and elegant in its design. The impact of these non-active mutations needs to be further elucidated in cell culture models of P. falciparum and L. major parasites. More studies are needed to dissect the interaction of the DHFR N-terminal tail with its neighboring helices in the DHFR domain.
Implications for Novel Therapeutics
In many well-characterized enzymes like the HIV reverse transcriptase and the HIV-1 protease, the importance of non-active site regions in drug design and drug resistance is already well-appreciated [30, 31]. Even in human and E. coli DHFR’s, the non-active site regions have long been known to play an important role in therapeutics and regulation of expression [32, 33]. Antifolates are used in the clinic for treatment of malaria and leishmaniasis, and, in both cases, toxicity is a limiting factor in high-dose treatment [34]. Thus, a better understanding of the non-active site regions, with no homologous counterparts in the corresponding human enzymes, could lead to the development of more targeted therapy. In P. falciparum TS-DHFR, inhibitors directed towards the N-terminus could be used in combination with active-site inhibitors for a synergistic inhibition of the malaria TS-DHFR. Our study, examining the functional contributions of non-active site regions in P. falciparum and L. major TS-DHFR, thus has implications in the design of novel, selective, less toxic therapies for these parasitic diseases.
ACKNOWLEDGEMENTS
We would like to thank Dr. Melissa Vargo, Walter (Eddie) Martucci and Dr. Lanxuan (Cindy) Doan for helpful discussions in the preparation of this manuscript.
FUNDING INFORMATION: This work was supported in part by NIH Grant AI 44630 (to KSA), by an NIH Medical Student Training Program grant to the Yale M.D.-Ph.D. Program (to TD), and by a Doctoral Research Award from the Canadian Institutes of Health Research (to TD).
The following abbreviations were used
- TS-DHFR
thymidylate synthase-dihydrofolate reductase (this is a functional designation as dihydrofolate is produced at TS and utilized at DHFR; this enzyme is also commonly referred to as DHFR-TS in the literature because the DHFR domain is N-terminal to TS)
- dUMP
deoxyuridine monophosphate
- dTMP
deoxythymidine monophosphate
- CH2H4folate
methylene tetrahydrofolate
- H2folate
dihydrofolate
- H4folate
tetrahydrofolate
- NADPH
nicotinamide adenine dinucleotide phosphate
- HPLC
high-performance liquid chromatography
REFERENCES
- 1.Greenwood BM, et al. Malaria. Lancet. 2005;365(9469):1487–1498. doi: 10.1016/S0140-6736(05)66420-3. [DOI] [PubMed] [Google Scholar]
- 2.Ivanetich KM, Santi DV. Thymidylate synthase-dihydrofolate reductase in protozoa. Exp Parasitol. 1990;70(3):367–371. doi: 10.1016/0014-4894(90)90119-w. [DOI] [PubMed] [Google Scholar]
- 3.Yuthavong Y. Basis for antifolate action and resistance in malaria. Microbes Infect. 2002;4(2):175–182. doi: 10.1016/s1286-4579(01)01525-8. [DOI] [PubMed] [Google Scholar]
- 4.Ivanetich KM, Santi DV. Bifunctional thymidylate synthase-dihydrofolate reductase in protozoa. Faseb J. 1990;4(6):1591–1597. doi: 10.1096/fasebj.4.6.2180768. [DOI] [PubMed] [Google Scholar]
- 5.Atreya CE, Anderson KS. Kinetic characterization of bifunctional thymidylate synthasedihydrofolate reductase (TS-DHFR) from Cryptosporidium hominis: a paradigm shift for ts activity and channeling behavior. J Biol Chem. 2004;279(18):18314–18322. doi: 10.1074/jbc.M400009200. [DOI] [PubMed] [Google Scholar]
- 6.Liang PH, Anderson KS. Substrate channeling and domain-domain interactions in bifunctional thymidylate synthase-dihydrofolate reductase. Biochemistry. 1998;37(35):12195–12205. doi: 10.1021/bi9803168. [DOI] [PubMed] [Google Scholar]
- 7.Doan LT, et al. Nonconserved Residues Ala287 and Ser290 of the Cryptosporidium hominis Thymidylate Synthase Domain Facilitate Its Rapid Rate of Catalysis(,) Biochemistry. 2007;46(28):8379–83791. doi: 10.1021/bi700531r. [DOI] [PubMed] [Google Scholar]
- 8.O'Neil RH, et al. The crystal structure of dihydrofolate reductase-thymidylate synthase from Cryptosporidium hominis reveals a novel architecture for the bifunctional enzyme. J Eukaryot Microbiol. 2003;50 Suppl:555–556. doi: 10.1111/j.1550-7408.2003.tb00627.x. [DOI] [PubMed] [Google Scholar]
- 9.Yuvaniyama J, et al. Insights into antifolate resistance from malarial DHFR-TS structures. Nat Struct Biol. 2003;10(5):357–365. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]
- 10.Knighton DR, et al. Structure of and kinetic channelling in bifunctional dihydrofolate reductase-thymidylate synthase. Nat Struct Biol. 1994;1(3):186–194. doi: 10.1038/nsb0394-186. [DOI] [PubMed] [Google Scholar]
- 11.Vargo MA, Anderson KS. Exploring interactions of the cross-over helix of C. hominis -Thymidylate Synthase-Dihydrofolate Reductase. Manuscript in preparation. 2004 [Google Scholar]
- 12.Wattanarangsan J, et al. Effect of N-terminal truncation of Plasmodium falciparum dihydrofolate reductase on dihydrofolate reductase and thymidylate synthase activity. Mol Biochem Parasitol. 2003;126(1):97–102. doi: 10.1016/s0166-6851(02)00240-2. [DOI] [PubMed] [Google Scholar]
- 13.Shallom S, et al. Essential protein-protein interactions between Plasmodium falciparum thymidylate synthase and dihydrofolate reductase domains. J Biol Chem. 1999;274(53):37781–37786. doi: 10.1074/jbc.274.53.37781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathews CK, Huennekens FM. Enzymic preparation of the 1,L-diastereoisomer of tetrahydrofolic acid. J Biol Chem. 1960;235:3304–3308. [PubMed] [Google Scholar]
- 15.Curthoys NP, Scott JM, Rabinowitz JC. Folate coenzymes of Clostridium acidi-urici. The isolation of (l)-5,10-methenyltetrahydropteroyltriglutamate, its conversion to (l)-tetrahydropteroyltriglutamate and (l)-10-( 14 C)formyltetrahydropteroyltriglutamate, and the synthesis of (l)-10-formyl-(6,7- 3 H 2 )tetrahydropteroyltriglutamate and (l)-(6,7- 3 H 2)tetrahydropteroyltriglutamate. J Biol Chem. 1972;247(7):1959–1964. [PubMed] [Google Scholar]
- 16.Chitnumsub P, et al. Characterization, crystallization and preliminary X-ray analysis of bifunctional dihydrofolate reductase-thymidylate synthase from Plasmodium falciparum. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 4):780–783. doi: 10.1107/S0907444904001544. [DOI] [PubMed] [Google Scholar]
- 17.Meek TD, Garvey EP, Santi DV. Purification and characterization of the bifunctional thymidylate synthetase-dihydrofolate reductase from methotrexate-resistant Leishmania tropica. Biochemistry. 1985;24(3):678–686. doi: 10.1021/bi00324a021. [DOI] [PubMed] [Google Scholar]
- 18.Rod TH, Radkiewicz JL, Brooks CL., 3rd Correlated motion and the effect of distal mutations in dihydrofolate reductase. Proc Natl Acad Sci U S A. 2003;100(12):6980–6985. doi: 10.1073/pnas.1230801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- 20.Stroud RM, Finer-Moore JS. Stereochemistry of a multistep/bipartite methyl transfer reaction: thymidylate synthase. Faseb J. 1993;7(8):671–677. doi: 10.1096/fasebj.7.8.8500692. [DOI] [PubMed] [Google Scholar]
- 21.Matthews DA, et al. Stereochemical mechanism of action for thymidylate synthase based on the X-ray structure of the covalent inhibitory ternary complex with 5-fluoro-2'-deoxyuridylate and 5,10-methylenetetrahydrofolate. J Mol Biol. 1990;214(4):937–948. doi: 10.1016/0022-2836(90)90347-O. [DOI] [PubMed] [Google Scholar]
- 22.McElheny D, et al. Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc Natl Acad Sci U S A. 2005;102(14):5032–5037. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edstein MD, et al. In vitro activities of the biguanide PS-15 and its metabolite, WR99210, against cycloguanil-resistant Plasmodium falciparum isolates from Thailand. Antimicrob Agents Chemother. 1997;41(10):2300–2301. doi: 10.1128/aac.41.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canfield CJ, et al. PS-15: a potent, orally active antimalarial from a new class of folic acid antagonists. Am J Trop Med Hyg. 1993;49(1):121–126. doi: 10.4269/ajtmh.1993.49.121. [DOI] [PubMed] [Google Scholar]
- 25.Lafont V, et al. Compensating enthalpic and entropic changes hinder binding affinity optimization. Chem Biol Drug Des. 2007;69(6):413–422. doi: 10.1111/j.1747-0285.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Larrea D, et al. Role of solvation barriers in protein kinetic stability. J Mol Biol. 2006;360(3):715–724. doi: 10.1016/j.jmb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Whitesides GM, Krishnamurthy VM. Designing ligands to bind proteins. Q Rev Biophys. 2005;38(4):385–395. doi: 10.1017/S0033583506004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mudeppa DG, et al. Cell-free production of functional Plasmodium falciparum dihydrofolate reductase-thymidylate synthase. Mol Biochem Parasitol. 2007;151(2):216–219. doi: 10.1016/j.molbiopara.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 29.O'Neil RH, et al. Phylogenetic classification of protozoa based on the structure of the linker domain in the bifunctional enzyme, dihydrofolate reductase-thymidylate synthase. J Biol Chem. 2003;278(52):52980–52987. doi: 10.1074/jbc.M310328200. [DOI] [PubMed] [Google Scholar]
- 30.Olsen DB, et al. Non-active site changes elicit broad-based cross-resistance of the HIV-1 protease to inhibitors. J Biol Chem. 1999;274(34):23699–23701. doi: 10.1074/jbc.274.34.23699. [DOI] [PubMed] [Google Scholar]
- 31.Muzammil S, Ross P, Freire E. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry. 2003;42(3):631–638. doi: 10.1021/bi027019u. [DOI] [PubMed] [Google Scholar]
- 32.Dicker AP, et al. Methotrexate resistance in an in vivo mouse tumor due to a non-active-site dihydrofolate reductase mutation. Proc Natl Acad Sci U S A. 1993;90(24):11797–11801. doi: 10.1073/pnas.90.24.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skacel N, et al. Identification of amino acids required for the functional up-regulation of human dihydrofolate reductase protein in response to antifolate Treatment. J Biol Chem. 2005;280(24):22721–22731. doi: 10.1074/jbc.M500277200. [DOI] [PubMed] [Google Scholar]
- 34.Sands M, Kron MA, Brown RB. Pentamidine: a review. Rev Infect Dis. 1985;7(5):625–634. doi: 10.1093/clinids/7.5.625. [DOI] [PubMed] [Google Scholar]