Abstract
Guanylate kinase (GMK) is an essential nucleoside monophosphate kinase that catalyzes the phosphorylation of guanine-monophosphate (GMP) and dGMP to yield GDP and dGDP, respectively, important precursors for nucleotide synthesis. GMK is also responsible for the activation of 6-thioguanine (6-TG), a drug widely used as chemotherapeutic agent to treat leukemia. Several mechanisms of resistance to 6-TG have been reported but a subset of drug resistant cells cannot be explained by these mechanisms. We propose that mutations in GMK could result in drug resistance. Because cells require the presence of a functional GMK for viability, mutations that arise that lead to 6-TG resistance must retain activity toward GMP. We report three amino acid substitutions at serine 37 (S37) in mouse GMK that display activity toward GMP by conferring genetic complementation to a conditional GMK-deficient Escherichia coli and in enzyme assays. When 6-TG is included in complementation studies, cells expressing wild-type GMK are sensitive whereas all S37 mutants examined are able to effectively discriminate against 6-TG and display a drug resistance phenotype. Activity of the three S37 mutant enzymes toward clinically relevant concentrations of 6-TGMP is undetectable. Mutations in GMK, therefore, represent a previously undescribed mechanism for 6-TG resistance.
Keywords: 6-thioguanine, drug resistance, guanylate kinase, mutagenesis, selection
Introduction
Guanylate kinase (GMK, ATP:GMP phosphotransferase, EC 2.7.4.8) is an essential enzyme involved in purine biosynthesis and is responsible for the phosphorylation of guanine-monophosphate (GMP) and dGMP. In addition, GMK functions in the recovery of cGMP and is therefore thought to regulate the supply of guanine nucleotides to signal transduction pathway components (Woods and Bryant, 1991; Gaidarov et al., 1993). From a medical perspective, GMK performs a key step in the activation of several important antiviral and anti-neoplastic agents. Following initial activation by the viral-encoded thymidine kinase in herpes-infected cells, the prodrugs acyclovir and gancyclovir require further activation by GMK (Miller and Miller, 1980; Boehme, 1984). Another major therapeutic role of GMK is in the activation of the guanine analog, 6-thioguanine (6-TG), which has been primarily used as a treatment for childhood and adult leukemia since the 1950s (Elion, 1989). 6-TG also has the ability to kill proliferating T cells, contributing to its immunosuppressive properties. This has led to its use as an immunosuppressant in transplant surgery and as an anti-inflammatory agent in autoimmune diseases such as inflammatory bowel disease (Karran, 2006). Despite the widespread use of 6-TG to treat a variety of diseases, the mechanism of 6-TG toxicity and drug resistance is not fully understood (Karran, 2006).
Structurally similar to guanine, 6-TG differs only at the carbon 6 position where the oxygen in guanine is replaced by a thiol group. Upon entering the cell, hypoxanthine–guanine phosphoribosyltransferase (HGPRT) initiates the activation of 6-TG by transferring a phosphoribosyl group to 6-TG forming 6-thioguanine-monophosphate (6-TGMP) (Karran, 2006). 6-TGMP is then further phosphorylated to 6-thioguanine diphosphate (6-TGDP) and 6-thioguanine triphosphate (6-TGTP) by GMK and nucleoside diphosphokinase, respectively (Sekulic et al., 2002). Once 6-TG enters the cell, the process of 6-TG degradation can be initiated by thiopurine S-methyltransferase (TPMT) to form inactive methylated products (Karran, 2006). The presence of TPMT serves to protect patients from negative effects of 6-TG, such as myelosuppression and low whole blood cell counts (Karran, 2006). Indeed, patients who inherit two low-activity variant alleles of the TPMT gene are more prone to 6-TG-induced hematopoietic toxicity from the accumulation of the inactive form and usually suffer intense myelosuppression (Karran, 2006).
Incorporation of 6-TGMP into DNA during synthesis has been reported to cause a plethora of DNA-damaging events including single- and double-strand breaks, errors during sister-chromatid exchange events, protein cross-links and large-scale chromosomal damage (Bohon and de los Santos, 2003; Yan et al., 2003; Karran and Attard, 2008). 6-TGMP-containing DNA has also been shown to be a poor template for replication by DNA polymerase and a poor substrate for DNA ligases, thereby interfering with both leading and lagging strand DNA replication (Ling et al., 1992). Furthermore, once 6-TGMP is incorporated into DNA, methylation by S-adenosylmethionine (SAM) results in the formation of S6-methylguanine (Me-6-TG). During DNA replication, Me-6-TG acts as a mutagenic lesion due to its preference to base pair with thymine, rather than cytosine. The abnormal base pairing subsequently activates the mismatch repair (MMR) system, which results in lethal processing and leads to cell death. The general outcome is that while incorporation of 6-TGMP into DNA is not directly toxic to cells, it prevents subsequent replication by halting the cell cycle in G2 phase after one round of replication probably as a result of one or more of the many effects of 6-TG described above (Bohon and de los Santos, 2003). Needless to say, no single event has been associated with 6-TG toxicity.
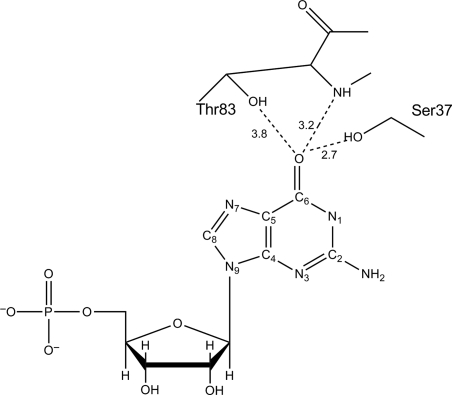
Despite the wide application of 6-TG as an anti-leukemic agent, one major drawback with its use as a chemotherapeutic drug is the development of drug resistance. The majority of 6-TG resistance can be attributed to the increased activity/production of TPMT, or to a lack of or reduced HGPRT activity (Lennard, 1992; Karran, 2006). To a lesser extent, an inactive MMR system or 6-TG transport impairment has also been reported to contribute to drug resistance (Swann et al., 1996; Fotoohi et al., 2006). However, not all 6-TG drug resistance can be accounted for by these mechanisms. Evidence indicates the presence of unknown and random inter-individual variations in 6-TG resistant cells of phenotypically normal individuals (Yamanaka et al., 1985; Aubrecht et al., 1997). There is also considerable inter-individual variability in mutation frequency that gives rise to 6-TG resistance in normal, nontransformed cells from patients with cancer prone disease such as Werner's syndrome (Fukuchi et al., 1990; Davies et al., 1992; Aubrecht et al., 1997). Because there appears to be a subset of 6-TG resistance that cannot be attributed to mutations affecting HGPRT, MMR, TPMT or transporter activities, we hypothesize that mutations could arise in the enzyme responsible for the second and rate-limiting step in the 6-TG activation pathway, i.e. the phosphorylation of 6-TGMP to 6-TGDP by GMK. Such mutants would need to retain essential GMP phosphorylation activity but lack the ability to phosphorylate 6-TGMP. Our initial studies presented here focus on evaluating the potential that such mutation(s) in GMK exist. Toward that end, mouse GMK (MGMK) serine 37 (S37) was targeted for mutagenesis based on structural information that suggests that activation occurs when a bond is formed between S37 and the oxygen on carbon six (C6) of guanine or at the sulfur at C6 in 6-TG (Fig. 1). Site-directed mutagenesis was used to introduce the larger polar residue, tyrosine, medium-size polar residue, threonine, or a small non-polar residue, alanine. These residues were selected to evaluate the influence of steric strain or a hydroxyl group on substrate binding and catalysis especially with regard to the 6-TG substrate. This study explores the role of MGMK residue S37 in enzyme/substrate recognition with respect to the natural substrate GMP and the important anti-metabolite, 6-TGMP.
Fig. 1.
Schematic of active site with the location of S37 and T83 with respect to guanine in MGMK based on the analogous residues (S34 and S80) in yeast GMK. (Adapted from Sekulic et al. (2002) and Stehle and Schulz (1992)).
Materials and methods
Materials
Restriction endonucleases and T4 DNA ligase were purchased from Gibco BRL (Rockville, MD, USA) or New England Biolabs (Beverly, MA, USA). Oligonucleotides used for site-directed mutagenesis and DNA sequencing were obtained from Operon (San Pablo, CA, USA). Nickel columns (Ni-NTA Spin Kit) used to purify wild-type and mutant GMKs were purchased from Qiagen (Valencia, CA, USA). 6-TGMP used in enzyme assays was custom synthesized by Moravek Biochemical (Brea, CA, USA). Enzyme assay reagents and other chemicals were purchased from Sigma (St. Louis, MO, USA) unless otherwise specified.
Bacterial strains
Escherichia coli strain NM522 [F′ lacIqΔ(lacZ)-M15proA+B+/supE thiΔ(lac-proAB)Δ(hsdMS-mcrB)5(rk−mk−McrBC−)] was used as a recipient for certain cloning procedures. E.coli strain CJ236 (F+ LAM−, ung-1, relA1, dut-1, spoT1, thi-1) was used to produce single-stranded DNA for site-directed mutagenesis (Kunkel, 1985). E.coli strain TS202A(DE3) (LAM−, tdk-1, IN(rrnD-rrnE)1, ilv−276 kanR ara−), a conditional GMK deficient strain, was used in genetic complementation experiments to assess GMK activity and sensitivity to 6-TG. Growth conditions were as described previously (Stolworthy et al., 2003).
Construction of GMK mutants
The bacterial expression vector, pETHT:mgmk, described previously, was used as a template for the introduction of amino acid substitutions at S37 by site-directed mutagenesis using the Kunkel method (Brady et al., 1996; Kunkel, 1985; Stolworthy and Black, 2001). The mutagenic oligonucleotides used were designed to also introduce a new restriction site and were as follows: S37A, 5′-CAGTGTCGCGCATACTACAAGG-3′, BstUI; S37T, 5′-CAGTGTGACTCATACTACAAGG-3′, HinfI and; S37Y, 5′-CAGTGTGTACCATACTACAAGG-3′, RsaI. The resulting mutations were confirmed by DNA sequencing and the plasmids designated pETHT:mgmk-S37A, pETHT:mgmk-S37T, and pETHT:mgmk-S37Y. Sequencing was performed at the core sequencing facility at Washington State University.
Genetic complementation
For complementation studies, E.coli strain TS202A(DE3) harboring pETHT, pETHT:mgmk, pETHT:mgmk-S37A, pETHT:mgmk-S37T or pETHT:mgmk-S37Y was grown at 37oC on selection media as described previously (Stolworthy et al., 2003). Briefly, all transformants were grown under nonselective conditions on minimal medium [for 1 l—15 g Bacto Agar, 100 μl of 1 M CaCl2, 1 mM MgSO4, 11 mM glucose, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 47 mM Na2HPO4, 3 mM l-isoleucine, 3 mM l-valine] containing carbenicillin (carb) and kanamycin (kan) each at 50 µg/ml and supplemented with 0.2% l-arabinose (+ARA) to provide expression of an endogenous GMK from an arabinose promoter. For selective conditions (genetic complementation by GMK), arabinose was omitted from the minimal medium (−ARA). For drug sensitivity assays, the −ARA medium was supplemented with 300 µg/ml of 6-TG (−ARA+6-TG).
Growth curves
E.coli strain TS202A(DE3) harboring pETHT, pETHT:mgmk, pETHT:mgmk-S37A, pETHT:mgmk-S37T or pETHT:mgmk-S37Y was grown at 37°C on +ARA media. A single colony from each sample was grown overnight. Five milliliters of overnight cultures (+ARA) of TS202A(DE3) cells harboring pETHT, pETHT:mgmk, pETHT:mgmk-S37A, pETHT:mgmk-S37T or pETHT:mgmk-S37Y were used to inoculate 50 ml of +ARA, −ARA, or –ARA+6TG media to an initial OD600 of ∼0.05–0.07. Cultures were then grown at 37°C with vigorous shaking and monitored for growth by reading the OD600 every 30 min using a Bio-Rad Smart Spec 3000 (Hercules, CA, USA). The growth experiments were repeated at least three times, and similar results were obtained.
Protein overexpression and purification
E.coli TS202A(DE3) harboring pETHT:mgmk, pETHT:mgmk-S37A, pETHT:mgmk-S37T or pETHT:mgmk-S37Y was grown at 30°C in M9ZB [For 1 l—10 g Bacto tryptone, 5 g of NaCl, 100 µl of CaCl2, 1 mM MgSO4, 11 mM glucose, 22 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 47 mM Na2HPO4] containing carbenicillin (carb) and kanamycin (kan) at 50 µg/ml and supplemented with 0.2% l-arabinose (M9ZB+ARA). Five milliliters of overnight culture was transferred to 500 ml of M9ZB+ARA and the cultures grown at 30°C until an OD600 of 0.2–0.25 was reached. Protein overexpression was induced by the addition of 0.4 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG).
Proteins were purified using a Qiagen Ni-NTA Spin Kit (Valencia, CA, USA) under native conditions as specified by the manufacturer. Three hours following IPTG induction, cells were cooled on ice for 15–30 min and then centrifuged at 4000 rpm for 10 min at 4°C (Beckman JLA-10.500). Cells were lysed by adding 17.5 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole at pH 8.0) and lysozyme at 1 mg/ml, incubated on ice for 30 min and subjected to sonication with 6–10 s bursts with a 10 s cooling period between bursts. To pellet cellular debris, the lysate was centrifuged at 9000 rpm (Beckman JA-17) for 20–30 min at 4°C. Cleared lysate was produced by filtering the lysate through 0.2 µM Supor® Membrane Low Protein Binding (PALL-Life Sciences, East Hills, NY, USA). For affinity purification, 1 ml of Ni-NTA slurry was added to 4 ml cleared lysate and gently mixed by shaking at 4°C for 60 min. The Ni-NTA resin/cleared lysate mixture was added into a column and flow through fractions were collected. The resin was subjected to washing by adding 8 ml of wash buffer (50 mM NaH2PO4, 600 mM NaCl and 20 mM imidazole at pH 8.0) three times. Bound protein was eluted by adding four aliquots of 500 µl of elution buffer (50 mM NaH2PO4, 300 mM NaCl and 250 mM imidazole at pH 8.0). The elution fractions were dialyzed against dialysis buffer (50 mM NaCl, 50 mM Tris at pH 7.5) at 4°C. After dialysis, the samples were collected and stored at −20°C. A small sample (1 µg) of each purified protein was subjected to electrophoresis in a 12% polyacrylamide–SDS-containing gel. Protein concentrations were quantified by the Bradford method using reagents supplied by Bio-Rad. Known concentrations of bovine serum albumin were used as standards and used to generate calibration curves according to the manufacturer's instructions.
Spectrophotometric assay
Enzyme activities toward the substrates GMP and 6-thioguanine-monophosphate (6-TGMP) were measured using a lactate dehydrogenase–pyruvate kinase coupled assay (Agarwal et al., 1978). Briefly, 1 µg or 7 µg of enzyme, for GMP or 6-TGMP assays, respectively, was mixed with 500 µl GMK assay cocktail reaction (1 M Tris, pH 7.5, 2.5 M KCl, 1 M MgCl2, 1 M NaPEP, 10 mM NADH, 100 mM ATP, 8.5 µl pyruvate kinase, 8.5 µl lactate dehydrogenase in 5 ml). The reaction was started by the addition of either various concentrations of GMP or 6-TGMP. The A340 was measured at 2 s intervals over 1800 s using an HP™ 8452 Olis SpectralWorks diode array spectrophotometer. For kinetic analysis studies, the A340 was measured at 10 s intervals over 900 s (GMP) or at 60 s intervals over 1800 s (6-TGMP) using a Shimadzu UV-1700 PharmaSpec UV-VIS Spectrophotometer. Each enzyme assay experiment was repeated at least three times.
Results
Construction of mutants and genetic complementation
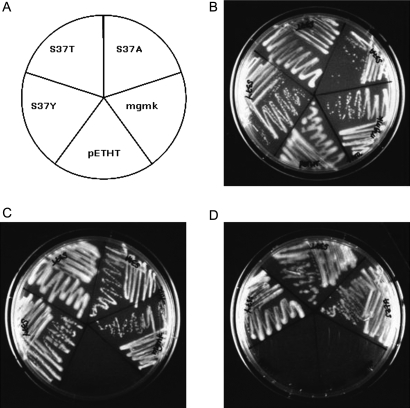
Using site-directed mutagenesis, single amino acid substitutions were introduced at S37 to create the MGMK mutants S37A, S37T and S37Y as described in Materials and Methods. To ascertain whether these mutants displayed GMK activity, pETHT, pETHT:mgmk, pETHT:mgmk-S37A, pETHT:mgmk-S37Y and pETHT:mgmk-S37T were used to transform a previously established conditional GMK-deficient E.coli strain, TS202A(DE3), and plated onto selection medium in the presence or absence of arabinose (Fig. 2). E.coli strain TS202A(DE3) was created by integration of the mouse gmk gene downstream of the bacterial chromosomal arabinose promoter and disruption of the endogenous bacterial gmk gene by insertion of a kanamycin resistance gene into the bacterial gmk locus. Because GMK is an essential enzyme, the growth of E.coli TS202A(DE3) requires the presence of arabinose to induce expression of the integrated mgmk gene. In the absence of arabinose, the cells are not viable on selective medium unless a functional GMK is expressed from an introduced gmk-encoded plasmid (Stolworthy et al., 2003).
Fig. 2.
Functional complementation of the conditional guanylate kinase (GMK) strain E.coli TS202A(DE3) by wild-type and mutant mouse GMKs. (A) E.coli TS202A(DE3) strains harboring the expression vector (pETHT) or the vector containing wild-type mgmk (pETHT:mgmk) or mgmk mutants (pETHT:S37A, pETHT:S37T, pETHT:S37Y) was grown at 37°C for 36–48 h on (B) mgmk selection medium with arabinose (+ARA), (C) without (−ARA), or (D) in the absence of arabinose with the addition of 300 µg/ml 6-TG (−ARA+6-TG).
Growth of individual transformants was assessed visually following streaking onto minimal selection plates containing arabinose (+ARA) or without arabinose (−ARA) and incubation at 37°C for 36–48 h (Fig. 2). All transformants grew on the +ARA plates as expected since no selection is imposed (Fig. 2B). The ability of transformants to grow on selection media lacking arabinose (−ARA) indicates that the transformants express sufficient functional GMK activity to complement the conditionally GMK-deficient E.coli. Figure 2C shows that on −ARA media, pETHT (empty vector), was unable to complement the conditional GMK-deficient E.coli, whereas cells expressing the wild-type and the three mutant MGMKs, S37A, S37T and S37Y, were viable. This indicates that the S37 substitutions retain activity toward GMP.
To assess whether these substitutions impact the ability of GMK to phosphorylate 6-TGMP and thereby restrict cell growth, transformants were streaked onto –ARA plates containing a concentration of 6-TG (300 µg/ml) that prevents growth of cells expressing wild-type GMK (pETHT:mgmk). As shown in Fig. 2D, cells harboring wild-type mgmk were not able to grow on –ARA+6-TG plates. Surprisingly, all three S37 mutants were viable in the presence of 6-TG and display a resistant phenotype.
Growth in selective media
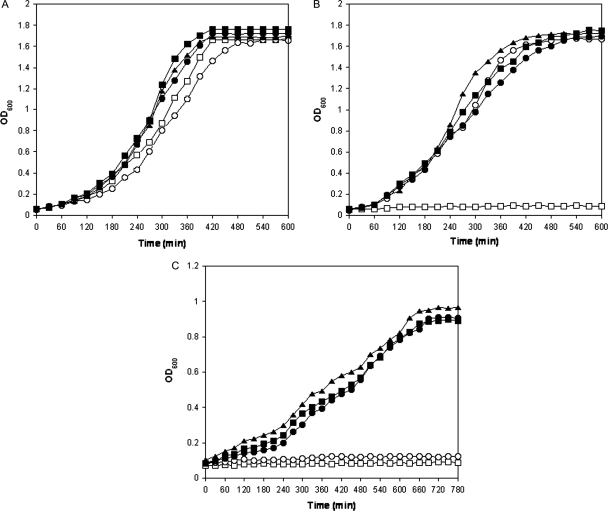
As a means to better understand the impact of the different S37 substitutions on substrate activity and specificity, we evaluated the ability of wild-type and mutant mgmk-containing strains on TS202A(DE3) for growth in non-selective, selective and 6-TG-containing media. Results of representative growth curves in these media are shown in Fig. 3. Doubling times of each sample in different media were calculated and are shown in Table I. When +ARA medium was used, all MGMK expressing transformants grew approximately at the same rate as shown in Fig. 3A. Cells harboring pETHT grew somewhat slower than the S37 mutants with a doubling time of 125.6 min. We speculate that this reduced growth rate may be due to the reliance of arabinose induction of the chromosomal-integrated GMK gene. Doubling times for cells harboring wild-type MGMK and MGMK mutants S37A and S37T were in the range of 100–108 min in +ARA media. Cells harboring MGMK mutant S37Y, however, showed the highest doubling time of 118.2 min (P > 0.05). In the absence of arabinose (−ARA), no growth is observed with pETHT (TS202A(DE3)). However, cells harboring wild-type MGMK and MGMK mutants initially grew at comparable rates as shown in Fig. 3B but at later time points diverged slightly. It is unclear why this occurred but accumulation of some enzymes with time may be responsible. Doubling times were 104.5 min (wild type), 114.4 min (S37A), 106.5 min (S37T) and 155.7 min (S37Y) with a P-value of >0.05, indicating the difference in growth is not statistically significant. Figure 3C shows a representative graph of cells incubated in −ARA medium supplemented with 300 µg/ml 6-TG (−ARA+6TG). As demonstrated in Fig. 2D, pETHT:mgmk (TS202A(DE3)) is unable to grow under these conditions and demonstrates sensitivity to 6-TG. Doubling times for cells harboring mutant MGMKs were fairly comparable with each other for S37A and S37Y (211.6 and 204.6 min, respectively) and slightly higher for S37T (242.6 min) although the growth rate is somewhat impaired (2–3-fold) in −ARA+6-TG compared with their growth in the absence of 6-TG (−ARA). The results shown in Fig. 3 provide further evidence that the substitutions at S37 not only yield enzymes with activity toward GMP but that this activity has shifted away from 6-TGMP.
Fig. 3.
Growth of conditional GMK-deficient strain TS202A(DE3) harboring vector (open squares, pETHT), wild-type mgmk (open circles, pETHT:mgmk) and mgmk mutants (filled squares, S37A; filled triangles, S37T; filled circles, S37Y) at 37°C in mgmk selection medium (A) with arabinose (+ARA), (B) in the absence of arabinose (−ARA) and (C) in the absence of arabinose with the addition of 300 µg/ml 6-TG (−ARA+6TG) was monitored over 10–13 h by OD600 readings. This experiment was repeated at least three times with similar results. Representative plots are shown.
Table I.
Growth pattern profile in E.coli
| Doubling time (min) |
|||
|---|---|---|---|
| +ARA | −ARA | −ARA +300 µg/ml 6-TG | |
| pETHT | 125.62 (23.05)a | ND | ND |
| pETHT.mgmk | 104.42 (8.50) | 104.51 (9.11) | ND |
| pETHT:S37A | 108.28 (13.22) | 114.41 (5.31) | 211.6 (7.56) |
| pETHT:S37T | 100.51 (2.39) | 106.53 (3.22) | 242.56 (7.46) |
| pETHT:S37Y | 118.23 (11.51) | 155.71 (16.26) | 204.6 (18.42) |
ND, not detected.
aStandard error mean.
Enzyme assays
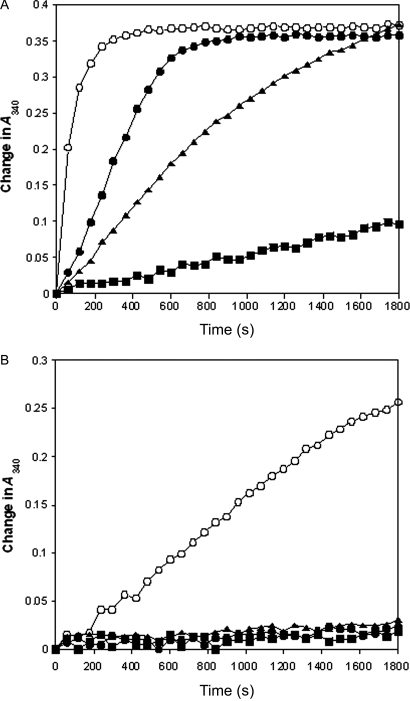
Induction of the pETHT:mgmk constructs with IPTG led to high levels of MGMK protein expression. All proteins were purified to near homogeneity (data not shown) with concentrations obtained at 609 mg/l (wild-type MGMK), 114.2 mg/l (S37A), 746.6 mg/l (S37T) and 386 mg/l (S37Y). To extend the complementation studies and growth curve results, we performed enzyme assays using purified enzymes and the substrates, GMP and 6-TGMP. All assays were performed at least three times and similar results were obtained. As shown in Fig. 4A, the ability of mutant GMKs (S37A, S37T and S37Y) to phosphorylate GMP was altered with respect to the wild-type GMK activity. S37A activity was the most dramatically altered and retained only very low to modest activity. Two mutants, S37T and S37Y, displayed impaired but significant activities toward GMP. Compared with wild-type GMK activity at 420 s (set at 100%), S37Y had the highest GMK activity at ∼70%, S37T at ∼35% and S37A at ∼7%. Interestingly, this difference in activities is not apparent in the biological systems examined (plate and growth assays).
Fig. 4.
Guanylate kinase enzyme assays of purified wild-type (open circles, MGMK) and mutant GMKs (filled squares, S37A; filled triangles, S37T; filled circles, S37Y). (A) One microgram of each purified enzyme was used with 30 µM GMP in a lactate dehydrogenase–pyruvate kinase coupled enzyme assay described in Agarwal et al. (1978). (B) Seven micrograms of each purified enzyme were subjected to coupled enzyme assays in the presence of 60 µM 6-TGMP. The enzyme assays were repeated at least three times with similar results observed. Representative plots are shown.
To better understand the influence of the different substitutions at S37, kinetic analyses were performed using purified enzymes. These studies show that wild-type MGMK displays a Km value toward GMP of 59.0 µM (Table II). Mutant S37Y has a slightly better Km value at 48.7 µM. As suggested by the impaired growth rate, both mutants S37A and S37T display reduced GMP-binding ability, as indicated by a 6.5–9.5-fold increase in Km as compared with wild-type MGMK. All three mutant MGMKs, however, have reduced turnover rates (kcat) relative to wild-type MGMK, ranging from 32.5-fold lower for S37A to 12- and 9-fold lower for S37T and S37Y, respectively (Table II). Overall, wild-type MGMK has the highest relative catalytic efficiency (kcat/Km) toward GMP. Corresponding to enzyme assay results, mutant S37A shows the greatest reduction in relative catalytic activity at about 318-fold lower than that of the wild-type enzyme. Mutant S37T also shows significant reduction in catalytic efficiency (∼81-fold less efficient) whereas mutant S37Y displays a modest reduction in catalytic efficiency (7-fold) (Table II).
Table II.
Kinetic parameters of wild-type MGMK and S37 mutants
| GMP |
6-TGMP |
|||||
|---|---|---|---|---|---|---|
| Km (μM) | kcata (s−1) | kcat/Km (s−1/μM) | Km (μM) | kcat (s−1) | kcat/Km (s−1/μM) | |
| MGMK | 59.02 (0.23)b | 20.8 (2.1) | 0.35 (0.033) | 171.5 (11.65) | 0.0074 (0.00055) | 4.3 × 10−5 (8.22 × 10−7) |
| S37A | 558.6 (18.37) | 0.64 (0.05) | 0.0011 (5 × 10−5) | ND | ND | ND |
| S37T | 381.7 (3.64) | 1.7 (0.09) | 0.0043 (0.00019) | ND | ND | ND |
| S37Y | 48.7 (0.26) | 2.35 (0.36) | 0.048 (0.0065) | ND | ND | ND |
ND, not detected.
aThe Kcat values were calculated using the equation Vmax = Kcat/[E] where [E] = total enzyme concentration and is based on one active site per monomer. Assay conditions are described in Materials and methods.
bStandard error mean.
When 6-TGMP is used as the substrate in crude lysate assays (Fig. 4B), detectable phosphorylation was observed only with wild-type MGMK. Wild-type MGMK displays a clear preference for GMP as 6-TGMP activity is greatly reduced even when 7-fold more enzyme and twice as much substrate (6-TGMP) are used. When 6-TGMP was used as a substrate with wild-type MGMK, only 15% of the activity observed with GMP as the substrate was detected. Kinetic analyses lend further support that only wild-type enzyme retains the ability to phosphorylate 6-TGMP. When the overall wild-type enzyme catalytic efficiencies (kcat/Km) between 6-TGMP and GMP are compared, an 8140-fold reduction in catalytic efficiency toward 6-TGMP is observed (2% kcat and 2.9-fold higher Km). This is in line with earlier reports that human GMK is the rate-limiting enzyme in the phosphorylation pathway of 6-TG and displays a Km over 2 mM for 6-TGMP and a maximal velocity of 3% of that with GMP (Miller et al., 1977; Sekulic et al., 2002). When 6-TGMP was used as the substrate in kinetic studies, none of the three S37 mutants displayed any detectable activity toward 6-TGMP even at high enzyme (35 µg or 5-fold higher than the amount of wild-type MGMK used) and high substrate concentrations [up to 2000 µM 6-TGMP or 12-fold higher than the wild-type Km for 6-TGMP (171.5 µM)]. All assays were performed at least three times, and similar results were observed (Table II). These results lend further support to the data shown in Figs 2 and 3, which indicate that wild-type MGMK is capable of phosphorylating GMP and, to a lesser extent 6-TGMP, whereas the S37 mutants are capable of phosphorylating GMP but lack detectable activity toward 6-TGMP, i.e. are resistant to 6-TGMP.
Discussion
Guanylate kinase is an essential enzyme in the purine nucleotide biosynthesis pathway and plays a key role in generating GDP and dGDP precursors for RNA and DNA metabolism, respectively. In addition, GMK is also responsible for the activation of several antiviral and anti-neoplastic drugs, including the chemotherapeutic drug, 6-TG, which has been used as the first line of treatment for leukemia for five decades (Karran and Attard, 2008). Unfortunately, treatment failure due to the development of 6-TG resistance has widely been reported (Aubrecht et al., 1997). Loss of activity of the key enzyme, HGPRT, is the best characterized resistance mechanism and, more recently, inactive MMR, increased TPMT activity and impaired 6-TG transport systems have also been reported to be responsible for 6-TG resistance in cancer cells (Lennard, 1992; Morgan et al., 1994; Fotoohi et al., 2006; Karran, 2006). Because not all 6-TG resistance can be accounted for by these mechanisms, we sought to evaluate whether mutations in GMK, the rate-limiting step in the activation of 6-TG, could lead to drug resistance.
We have focused this study on the mouse rather than the human GMK primarily because, in our hands, the mouse enzyme is soluble in E.coli whereas the human enzyme is not (Brady et al., 1996). Since the mouse enzyme shares high sequence similarity to human GMK (88% identity, 93% homology, information learned about MGMK is likely to be directly transferable to the human enzyme (Brady et al., 1996). Furthermore, the crystal structure of mouse GMK was solved in 2002 (no structure of the human enzyme has been forthcoming) and facilitates visualization of important molecular interactions (Sekulic et al., 2002). Indeed, structural studies by Sekulic et al. (2002) suggest substrate discrimination between the two purine nucleotides, AMP and GMP, in GMK occurs in part at the interactions between S37 and the C6 position on guanine with additional hydrogen bonding with threonine 83 (T83). From their report of the yeast GMK structure, Stehle and Schulz (1992) suggest that the S37 analogous serine (S34) and the T83 analogous residue (S80) in yeast are important in substrate specificity. Furthermore, modeling of the interactions between MGMK and 6-TG indicates that the formation of bonds with S37 and T83 occurs in a similar fashion to when GMP is bound but that steric clashes between these groups and the substrate might occur and diminish enzyme activity (Sekulic et al., 2002).
In this study, we sought to evaluate the role of S37 to discriminate between the oxygen or sulfur in GMP or 6-TGMP substrate interactions, respectively, as well as to provide evidence that this proposed discrimination could result in drug resistance in a biological setting. Three site-directed mutants of MGMK were created at S37 (S37A, S37T and S37Y) and their activities toward the native substrate or the anti-leukemic drug 6-TG were assessed in a conditional GMK-deficient E.coli strain using genetic complementation in both plate assays and growth curve experiments (Figs 2 and 3). While the growth of wild-type and mutant MGMK expressing E.coli under non-permissive conditions (−ARA) was robust, it contrasted sharply with the growth pattern exhibited in the presence of 6-TG. On drug-containing plates, wild-type mgmk expressing cells were non-viable whereas all three S37 mutants grew well (Fig. 2D). In 6-TG containing broth cultures, a similar pattern emerged; all S37 mutants grew whereas no growth was observed with the wild-type mgmk-expressing cells (Fig. 3C). While the growth rate under selective pressure and in the presence of 6-TG (−ARA+6-TG) of all mutants is diminished compared with that in the absence of 6-TG (−ARA), this may be partly attributable to reduced production of GMP as a result of competition between 6-TG and guanine with HGPRT. These data indicate that mutations at S37 display sufficient GMP activity for growth but result in a resistance phenotype to 6-TG.
Initial enzyme assays using GMP as the substrate also confirm that all three mutants display GMK activity although activity was reduced compared with that of wild-type MGMK. Kinetic analyses of the three MGMK mutants demonstrate that the overall enzyme catalytic efficiency (kcat/Km) toward GMP is reduced varying degrees from the wild-type value and ranges from 7- to 318-fold lower. For S37T and S37A with GMP as the substrate both Km and kcat values are significantly impaired (6.5- to 9.5-fold and 12- to 32.5-fold, respectively). However, for S37Y, the Km is slightly improved while the kcat is reduced 8.9-fold compared with wild-type values. This improved Km may be a result of the aromatic ring base stacking with the guanine. The lower kcat values of all mutants suggest an appropriate positioning of a hydroxyl group, such as with the wild-type serine, to the oxygen moiety at the C6 position of guanine may be important for facilitating efficient catalytic turnover (kcat) with GMP. S37A displays the worst binding and catalysis and this may be a result of a very loose/sloppy active site afforded by the presence of a small side chain.
Only wild-type GMK displays detectable activity toward 6-TGMP, although the Km is almost 3-fold higher and the kcat is 2810-fold lower than when GMP is the substrate. Even when 5-fold more enzyme (35 µg) and 12-times the wild-type Km concentration (2000 µM) are used with the S37 mutants, no 6-TGMP activity is detected. The kinetic data support the suggestion by Sekulic et al. (2002) that steric hindrance between T83, GMP or 6-TGMP and the presence of certain residues at position 37 (i.e. S37, S37T and S37Y) plays a role in substrate-binding and catalysis. With S37A, the substrate may not be able to be positioned appropriately due to inefficient side-chain interactions. These data suggest that there is a level of discrimination between the side chain at S37 and the ability of the enzyme to interact with the substrate and that the rules that govern this distinction differ when there is oxygen or sulfur at the C6 position. Crystal structure determinations of the S37 variants with 6-TGMP and GMP in future studies would likely provide further insight to the nature of the impact of these substitutions on the kinetic parameters described here.
The emergence of resistance to chemotherapeutic agents is a key cause of treatment failure. A better understanding of the molecular factors that result in the loss of drug potency may lead to the development of improved dosing regimens and treatment options for patients. A complete list of the potential mechanism(s) of drug resistance to 6-TG remains elusive because not all cases of resistance can be explained by mutational events in known targets. In this report, we sought to demonstrate that mutations in GMK could reveal a previously undescribed 6-TG resistance mechanism. Our results demonstrate that MGMK containing amino acid substitutions at S37 (S37A, S37T and S37Y) retain varying degrees of essential GMP activity but do not display detectable activity toward 6-TGMP. Furthermore, the S37 mutants provide sufficient GMK activity in a genetic complementation system for robust cell viability and also confer resistance to otherwise lethal doses of 6-TG. Taken together, these data suggest that the substitutions examined at S37 in MGMK are biologically relevant and display critical and appropriate discrimination in substrate specificity/recognition that results in drug resistance.
Funding
This work was supported by the National Institutes of Health [CA85939 to M.E.B.].
Acknowledgements
We thank Shannon Winn for technical assistance on the initial aspects of this study.
Footnotes
Edited by Paul Carter, Board Member for PEDS
References
- Agarwal K.C., Miech R.P., Parks R.E. Methods Enzymol. 1978;51:483–490. doi: 10.1016/s0076-6879(78)51066-5. [DOI] [PubMed] [Google Scholar]
- Aubrecht J., Goad M.E., Schiestl R.H. J. Pharmacol. Exp. Ther. 1997;282:1102–1108. [PubMed] [Google Scholar]
- Boehme R.R. J. Biol. Chem. 1984;269:12346–12349. [PubMed] [Google Scholar]
- Bohon J. Nucl Acids Res. 2003;31:1331–1338. doi: 10.1093/nar/gkg203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady W.A., Kokoris M.S., Fitzgibbon M., Black M.E. J. Biol. Chem. 1996;271:16734–16740. doi: 10.1074/jbc.271.28.16734. [DOI] [PubMed] [Google Scholar]
- Davies M.J., Lovell D.P., Anderson D. Mutation Res. 1992;265:165–171. doi: 10.1016/0027-5107(92)90045-4. [DOI] [PubMed] [Google Scholar]
- Elion G.B. Science. 1989;244:41–47. doi: 10.1126/science.2649979. [DOI] [PubMed] [Google Scholar]
- Fotoohi A.K., Lindqvist M., Peterson C., Albertioni F. Biochem. Biophys. Res. Commun. 2006;343:208–215. doi: 10.1016/j.bbrc.2006.02.134. [DOI] [PubMed] [Google Scholar]
- Fukuchi K., Tanaka K., Kumahara Y., Marumo K., Pride M.B., Martin G.M., Monnat R.J. Hum. Genet. 1990;84:249–252. doi: 10.1007/BF00200569. [DOI] [PubMed] [Google Scholar]
- Gaidarov I.O., Suslov O.N., Abdulev N.G. FEBS Lett. 1993;335:81–84. doi: 10.1016/0014-5793(93)80444-y. [DOI] [PubMed] [Google Scholar]
- Karran P. Br. Med. Bull. 2006;79–80:153–170. doi: 10.1093/bmb/ldl020. [DOI] [PubMed] [Google Scholar]
- Karran P., Attard N. Nat. Rev. Cancer. 2008;8:24–36. doi: 10.1038/nrc2292. [DOI] [PubMed] [Google Scholar]
- Kunkel T. Proc. Natl Acad. Sci. USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennard L. Eur. J. Clin. Pharmacol. 1992;43:329–339. doi: 10.1007/BF02220605. [DOI] [PubMed] [Google Scholar]
- Ling Y.H., Chan J., Beattie K.L., Nelson J.A. Mol. Pharmacol. 1992;42:802–807. [PubMed] [Google Scholar]
- Miller W.H., Miller R.L. J. Biol. Chem. 1980;255:7204–7207. [PubMed] [Google Scholar]
- Miller R.L., Adamczyk D.L., Spector T. Biochem. Pharmacol. 1977;26:1573–1576. doi: 10.1016/0006-2952(77)90071-5. [DOI] [PubMed] [Google Scholar]
- Morgan C.J., Chawdry R.N., Smith A.R., Siravo-Sagraves G., Trewyn R.W. Cancer Res. 1994;54:5387–5393. [PubMed] [Google Scholar]
- Sekulic N., Shuvalova L., Spangenberg O., Konrad M., Lavie A. J. Biol. Chem. 2002;277:30236–30243. doi: 10.1074/jbc.M204668200. [DOI] [PubMed] [Google Scholar]
- Stehle T., Schulz G.E. J. Mol. Biol. 1992;224:1127–1141. doi: 10.1016/0022-2836(92)90474-x. [DOI] [PubMed] [Google Scholar]
- Stolworthy T.S., Black M.E. Protein Eng. Des. Sel. 2001;14:903–909. doi: 10.1093/protein/14.11.903. [DOI] [PubMed] [Google Scholar]
- Stolworthy T.S., Krabbenhoft E., Black M.E. Anal. Biochem. 2003;322:40–47. doi: 10.1016/j.ab.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Swann P.F., Waters T.R., Moulton D.C., Xu Y.Z., Zheng Q., Edwards M., Mace R. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- Woods D.F., Bryant P.J. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- Yamanaka H., Kamatani N., Nishoka K., Kobayashi M., Wada Y., Ohtani T., Mikanagi K. Hum.Hered. 1985;35:358–363. doi: 10.1159/000153580. [DOI] [PubMed] [Google Scholar]
- Yan T., Berry S.E., Desai A.B., Kinsella T.J. Clin. Cancer Res. 2003;9:2327–2334. [PubMed] [Google Scholar]