Abstract
Homing endonucleases recognize specific long DNA sequences and catalyze double-stranded breaks that significantly stimulate homologous recombination, representing an attractive tool for genome targeting and editing. We previously described a two-plasmid selection system that couples enzymatic DNA cleavage with the survival of host cells, and enables directed evolution of homing endonucleases with altered cleavage sequence specificity. Using this selection system, we successfully evolved mutant I-SceI homing endonucleases with greatly increased cleavage activity towards a new target DNA sequence that differs from the wild-type cleavage sequence by 4 bp. The most highly evolved mutant showed a survival rate ∼100-fold higher than that of wild-type I-SceI enzyme. The degree of selectivity displayed by a mutant isolated from one round of saturation mutagenesis for the new target sequence is comparable to that of wild-type I-SceI for the natural sequence. These results highlight the ability and efficiency of our selection system for engineering homing endonucleases with novel DNA cleavage specificities. The mutant identified from this study can potentially be used in vivo for targeting the new cleavage sequence within genomic DNA.
Keywords: directed evolution, DNA modifying enzymes, gene targeting, homing endonuclease, protein engineering
Introduction
The process of homologous recombination (HR) enables in situ replacement of a disease gene with its therapeutic counterpart, so that the integrated gene can be transcribed from the natural promoter without requiring additional regulatory elements. This process possesses great potential to facilitate treatment of genetic disorders via gene therapy. To date, the most efficient method for inducing HR involves the introduction of a DNA double-stranded break (DSB) in the target gene locus, which results in a 10- to 10 000-fold increased rate of HR (up to 3–5%) at this site (Rouet et al., 1994; Choulika et al., 1995; Smih et al., 1995; Porteus and Baltimore, 2003). It is believed that the introduction of a DSB stimulates the HR machinery of the cell, which then repairs the break by gene exchange with any available homologous template, including an exogenously administered gene-targeting vector (Johnson and Jasin, 2001).
Genomic DSBs can be introduced by either natural or artificial DNA cleavage enzymes. One of the natural endonucleases commonly used for gene targeting is the homing endonuclease I-SceI (Dujon, 1989). This enzyme recognizes an 18 bp asymmetric DNA sequence that is absent in most mammalian genomes (Colleaux et al., 1988; Jasin, 1996) and catalyzes a DSB at the sequence recognition site. However, this system cannot be used for endogenous gene targeting (Jasin, 1996; Cohen-Tannoudji et al., 1998; Donoho et al., 1998). In order to target an endogenous gene, the cleavage sequence of wild-type I-SceI needs to be modified at multiple sites to match the specific target gene. Towards this goal, researchers have attempted to develop in vivo selection systems for engineering homing endonucleases with altered specificity (Gruen et al., 2002; Seligman et al., 2002; Chen and Zhao, 2005a; Doyon et al., 2006). Several independent studies have demonstrated that homing endonuclease mutants with altered specificity towards individual sites could be obtained by using in vivo selection strategy (Sussman et al., 2004; Doyon et al., 2006). However, engineering of homing endonuclease mutants that recognize a new sequence differing at multiple sites remains an overwhelming challenge despite two recent successes (Smith et al., 2006; Arnould et al., 2007). This is partly due to the compact structure of I-SceI where residues for target DNA recognition and cleavage are intricately linked. Thus, to achieve the desired target sequence specificity change, multiple simultaneous mutations might be required, which exceeds the capability of nearly all existing library selection/screening methods.
Another strategy for introducing DSB is through zinc-finger nucleases (ZFNs). ZFNs are constructed by linking a zinc-finger protein, responsible for recognition of a target DNA sequence, and a nonspecific DNA cleavage domain of FokI, a Type IIS restriction enzyme (Kim et al., 1996). The creation of a DSB by a ZFN requires two ZFNs to simultaneously bind the same locus in the genome in a precise orientation and spacing relative to each other (Mani et al., 2005). The requirement for the co-expression of two proteins simultaneously increases the likelihood of eliciting an immunogenic response. In addition, due to the large size of the ZFN gene, the two ZFNs need to be expressed from different plasmids. Thus, the creation of a DSB by ZFNs requires the transfection of both plasmids into the same target cell simultaneously. This is not desirable because gene delivery to the target cell is currently a limiting step in gene therapy (Glover et al., 2005), and the fewer the number of different plasmids required, the higher the efficiency of treatment. In comparison, homing endonuclease I-SceI does not require the co-expression of two proteins for function since it is a single, monomeric protein. Thus, based on this consideration, I-SceI should be less likely to trigger an immunogenic response, and should be much more efficient than ZFNs in catalyzing a desired DSB. More importantly, ZFNs typically recognize a target DNA sequence of 12 bp or fewer, resulting in multiple cleavage events in a mammalian genome. In addition, the FokI domain has nonspecific DNA cleavage activity, which is capable of randomly cleaving or nicking the genomic DNA. Such nonspecific DNA cleavage events are the major sources of cytotoxicity of ZFNs. In contrast, homing endonuclease I-SceI recognizes a unique 18 bp of DNA sequence, guaranteeing a single DNA recognition site on a mammalian genome. It also has an integrated DNA recognition and cleavage domain, ensuring that DNA cleavage is always coupled with DNA recognition. In fact, prior studies have demonstrated that wild-type I-SceI has a lower toxicity than ZFNs, resulting in a higher percentage of permanent integration when used for gene targeting (Steuer et al., 2004).
We previously reported the development of a two-plasmid in vivo selection system that couples enzymatic DNA cleavage with the survival of the host Escherichia coli cell via endonuclease-dependent degradation of a plasmid encoding the toxin CcdB (Chen and Zhao, 2005a). This system is highly sensitive and has a very low background, making it suitable for directed evolution of homing endonucleases with new cleavage sequences. In this study, we report the use of this selection system for the identification of I-SceI variants that selectively cleave a new target sequence that consists of four mismatched base-pairs at the 3′-end of the cleavage sequence. Using a combination of saturation mutagenesis and random mutagenesis, we obtained mutant I-SceI enzymes that cleave the target sequence efficiently. One of the mutants showed a preference for the target sequence over the natural sequence with a comparable selectivity to that of wild-type I-SceI for the natural sequence.
Materials and methods
Materials
Phusion DNA polymerase, T4 DNA ligase and restriction endonucleases were purchased from New England Biolabs (Beverly, MA, USA). QIAprep Spin Plasmid Miniprep Kit, QIAquick Gel Extraction Kit and QIAquick PCR Purification Kit were obtained from Qiagen (Valencia, CA, USA). Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA, USA). All the other reagents unless specified were obtained from Sigma-Aldrich (St Louis, MO, USA).
Plasmid and library construction
The construction of plasmid p11-LacY-wtx1 and pTrc-ISceI has been described previously (Chen and Zhao, 2005b). The new target sequence (tagggataacagggccta) was inserted into p11-LacY-wtx1, replacing the natural I-SceI cleavage sequence, to form p11-LacY-S4. The saturation mutagenized library of I-SceI, where residues 13–16 of I-SceI are randomized, was constructed by megaprimer PCR (Sarkar and Sommer, 1990). Briefly, pTrc-ISceI was used as a PCR template with primers S4_[13–16], aaccaggtaatgaacctg NNS NNS NNS NNS aaactgctgaaagaatac, where N represents a mixture of all four bases and S a mixture of G and C, and KpnI-Isce-2-C: atgccg ggtacc ttattttaaaaaagtttcgg (KpnI recognition site shown in italics). The PCR product was subsequently used as a primer, together with EcoRI-SceI, atcagt gaattc aggaaa ctcgag atgaaa aatat (EcoRI and XhoI sites shown in italics) for amplifying pTrc-ISceI again to form the saturation mutagenized gene library of I-SceI. This PCR product was subsequently digested with KpnI and EcoRI and cloned into the large fragment of pTrc-ISceI digested with the same enzymes to form the saturation mutagenized library pTrc-slib.
The best mutant identified from the saturation library, S4-4, was subjected to a round of random mutagenesis. Error-prone PCR was carried out to amplify mutant S4-4 using primers EcoRI-SceI and XbaI-ISceI-C, atgccg tctaga ttattttaaaaaagtttcgg (XbaI recognition site shown in italics) according to a protocol described elsewhere (Zhao et al., 1999). The PCR product was digested with XhoI and XbaI and cloned into the large fragment of the pUV5-ISceI vector digested with the same enzymes (vector sequence is available upon request) to form pUV5-eplib. Mutant S4.2 selected from the error-prone library was cloned back into the pTrc-ISceI vector as described above to form pTrc-4.2.
For in vitro characterization studies, the wild-type I-SceI and the mutants S4-4 and S4.2 selected from saturation and random mutagenesis experiments, respectively, were amplified with primers NdeI-ISceI-For, atcagt catatg aaaaatattaaaaaaaaccaggt (NdeI recognition site shown in italics) and XhoI-ISce-28-Rev, atgccg ctcgag ttattttaaaaaagtttcggagga (XhoI recognition site shown in italics) and cloned into the NdeI and XhoI recognition sites of pET28a(+) vector (Novagen, WI, USA).
In vivo selection and activity assay
The in vivo selection and activity assay was carried out as described previously (Chen and Zhao, 2005a). Briefly, the selection strain was prepared by transforming E.coli BW25141 (lacIq rrnBT14 ΔlacZWJ16 ΔphoBR580 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 galU95 endABT333 uidA (ΔMluI)::pir+ recA1) (Lessard et al., 1998; Datsenko and Wanner, 2000) with the appropriate reporter plasmid, p11-LacY-wtx1 or p11-LacY-S4, and plating on LB agar plates containing 100 µg/ml ampicillin. The E.coli cells were made electrocompetent following standard protocols (Dower et al., 1988). It was important that cells did not grow to saturation at any stage during competent cell preparation, which would have resulted in loss of the reporter plasmid. Typically, 50 µl of the competent selection strain was transformed with library vector pTrc-slib or pUV5-eplib, or the wild-type pTrc-ISceI or mutant pTrc-S4-4/pTrc-S4.2 plasmids. The culture was immediately recovered in SOC media and shaken at 37°C for 5 min. It was diluted 5-fold with SOC media pre-warmed at 37°C and induced with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). After shaking at 37°C for 70 min and incubating at 30°C for another 1 h, the transformants were plated on agar plates. The selection strain transformed with pUV5 plasmids was plated on agar plates after shaking at 37°C for 40 min. For library selection, the LB agar plates contained 50 µg/ml kanamycin plus 10 mM arabinose (an inducer for the toxin CcdB). An aliquot of cells was spread on plates of LB plus 50 µg/ml kanamycin to estimate the total number of transformants. All plates were incubated at 37°C for 12–24 h until colonies were clearly visible. Colonies were counted manually to estimate the survival rate. The survival rate was calculated by dividing the number of colonies formed on the arabinose-containing plate by the number of colonies on the kanamycin only plate, after accounting for the dilution factor.
Protein purification
Escherichia coli BL21(DE3) cells transformed with the pET28a(+) vector containing the appropriate inserts were grown in LB medium at 37°C to OD600≈0.6, and induced with 0.5 mM IPTG at 18°C overnight. Cells were harvested by centrifugation and resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 15% glycerol, pH 8.0) supplemented with 1 mg/ml lysozyme, frozen at −80°C overnight, thawed and lysed with a sonicator. The soluble proteins were then purified using Ni-NTA resins according to the manufacturer's protocol (QIAgen), followed by gel filtration using HiLoad26/60 Superdex 200 column (GE Healthcare, Piscataway, NJ, USA). The collected fractions containing desired protein were pooled together, concentrated by centrifugation and dialyzed overnight against dialysis buffer (100 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM dithiothreitol and 50% glycerol). Dialyzed proteins were then stored at −80°C until use.
In vitro activity assay
DNA substrates, containing one copy of either the natural I-SceI or TS1 cleavage site, were obtained by PCR using plasmids p11-LacY-wtx1 and p11-LacY-S4 as template, respectively, and primer pair CleaveFor (caagcagatttatcgccagc) and CleaveRev (ccagttaatagtttgcgc). Twelve 15 µl aliquots containing the cleavage buffer (10 mM Tris–HCl pH 8.8, 10 mM MgCl2, 1 mM dithiothreitol, 100 µg/ml BSA) and 100 ng of the appropriate DNA substrate were pre-equilibrated at 4°C in a PTC-200 thermocycler (Bio-Rad). Purified enzyme was added in 2-fold serial dilutions to each of the tubes. The digestion reaction was carried out using the following program: 10 min at 4°C, 2 h at 37°C, 20 min at 80°C, forever at 4°C. All samples were then kept at −20°C until electrophoresis. The digestion reaction (10 µl) was analyzed using agarose gel electrophoresis. The extent of cleavage was quantified by densitometry using a Bio-Rad Gel-Doc with Quantity One software to determine the relative band intensities. The amount of enzyme necessary to cleave 50% of each product was determined from a plot of fraction of cleaved DNA versus enzyme concentrations using Microsoft Excel.
Computational modeling analyses
The homology model of S4-4 mutant bound to its TS1 DNA substrate was created based on the crystal structure of wild-type I-SceI bound to its cognate DNA substrate (PDB accession code: 1R7M) using Insight II Molecular Modeling System (Accelrys Software Inc, San Diego, CA, USA), and was energy minimized using the Molecular Operating Environment (MOE, The Chemical Computing Group, 2008) software package. The figures were generated using the molecular visualization program Chimera (University of California, San Francisco, CA, USA).
Results
Directed evolution of I-SceI variants with altered DNA cleavage specificity
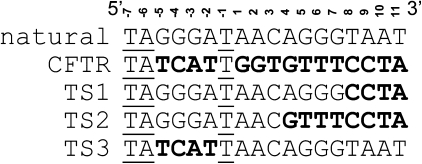
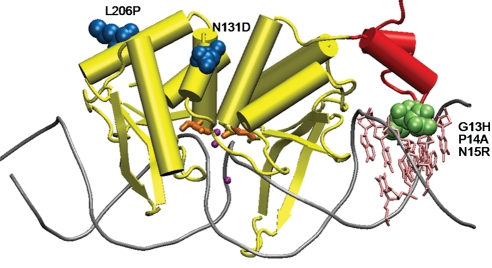
Due to their limited natural repertoire of target sequences, laboratory evolution of homing endonucleases with new sequence specificity that significantly differs from the wild-type enzyme is highly desirable for genome targeting and editing applications. To demonstrate the ability and efficiency of our previously reported in vivo selection system (Chen and Zhao, 2005b) to evolve such mutant enzymes, we designed our target sequence (TS1) based on the sequence surrounding the most common cystic fibrosis mutation ΔF508 in the cystic fibrosis transmembrane conductance regulator (CFTR) gene (Kerem et al., 1989). As shown in Fig. 1, target sequence 1 (TS1, TAGGGATAACAGGGCCTA) contains four mismatched base-pairs compared to the natural I-SceI cleavage sequence at the 3′-end (mismatched bases are in bold, Fig. 1). Transformation of the TS1 selection strain, harboring p11-LacY-S4 plasmid, with pTrc-ISceI, encoding wild-type I-SceI, resulted in a low survival rate of ∼0.9% (Table I). We reasoned that residues directly contacting the substrate would most likely dictate substrate preference (Chockalingam et al., 2005), thus we decided to initiate our directed evolution approach by randomizing residues close to the mismatched base-pairs in the wild-type I-SceI. Based on the crystal structure of I-SceI (Fig. 2), we identified four residues, residue 13 through 16, which form close contacts with the mismatched base-pairs. These four residues were simultaneously randomized with all 20 amino acids. The library we screened contained ∼1.25 × 106 transformants. Transformation of the TS1 selection strain with the saturation mutagenized library, pTrc-slib, resulted in a survival rate of ∼0.007%. Colonies formed on the selection plates were pooled together and grown in LB+50 µg/ml kanamycin media overnight. Plasmids from this pool were isolated, retransformed into the same selection strain and subjected to another round of selection. This process was repeated multiple times in order to enrich positive clones within the population. We reasoned that the presence of a positive clone should confer an increased survival rate after each round of enrichment. After two rounds of enrichment, the survival rate reached a plateau at ∼20%, representing a >20-fold increased survival rate compared to the wild-type enzyme. Individual plasmids isolated from 12 randomly picked colonies from the second round of enrichment were then used to transform the TS1 selection strain and their survival rates were assayed. All selected clones showed higher than 10% survival rates. The best clone, S4-4, exhibited a survival rate of ∼45%, a 50-fold increase compared with wild type I-SceI, and over 6000-fold increase compared with the starting population. DNA sequence analysis revealed that this mutant contained three amino acid substitutions: G13H, P14A and N15R (Table I).
Fig. 1.
Sequence alignment of the natural I-SceI target sequence, CFTR gene sequence surrounding mutation ΔF508 and the target sequences (TS) used in this engineering work. The mismatched base-pairs are shown in boldface. The identical bases are underlined.
Table I.
In vivo activity of wild-type I-SceI and evolved mutants
| Round | Method of mutagenesis | Selected mutants | Survival rate (%)a |
Residuesb | |
|---|---|---|---|---|---|
| Wild-type DNA substrate | TS1 DNA substrate | ||||
| 0 | – | Wild-type | 100 ± 0 | 0.9 ± 0.7 | GPNS |
| 1 | Saturation | S4-4 | <0.05 ± 0.03 | 44.7 ± 14.2 | HARS |
| 2 | Random | S4.2 | 9.1 ± 5.3 | 84.6 ± 13.7 | HARS N131D L206P |
a The data represent the average and standard deviation of three independent experiments.
b Amino acid identity of residues 13–16 is shown with single letters.
Fig. 2.
Cartoon diagram of the crystal structure of homing endonuclease I-SceI in complex with its substrate DNA (PDB accession code: 1R7M). DNA is shown as grey tubes, and base-pairs 8 through 11, which are different in the TS1 sequence compared with the natural cleavage sequence, are indicated in pink. The sub-domain contacting DNA base-pair 8 through 11 is shown in red. Mutations generated from saturation mutagenesis are shown in green space-filled form and mutations from error-prone PCR are in blue space-filled form. The three catalytic Ca2+ ions are shown as purple spheres, and the catalytic residues Asp44 and Asp144 are shown in orange (Moure et al., 2003).
To further increase the cleavage activity of S4-4 toward the TS1 sequence, we performed a second round of directed evolution toward TS1. In order to apply our cell growth-based selection system effectively, the survival rate of S4-4 in the TS1 selection strain needed to be less than 1%. In order to decrease the survival rate, we first reduced the expression level of S4-4 by placing the S4-4 gene under the control of a weaker promoter―UV5. We then modified the assay protocol by shortening the post-transformation recovery time (see Materials and Methods). After these changes, the survival rate of the S4-4 variant in the TS1 selection strain was decreased to less than 0.01%, enabling the system to be used as a selection method to identify variants of S4-4 with improved TS1 cleavage activity.
In the second round of directed evolution, we introduced random mutations into S4-4 using error-prone PCR and screened a library of 104 variants. We identified one variant, S4.2 that showed >5-fold increased survival rate compared with the parental mutant in the new selection system. This variant was then placed under the Trc promoter and assayed for its survival rate using the original assay conditions. Mutant S4.2 showed a survival rate of ∼85% in the TS1 selection strain, >90-fold improvement relative to wild-type I-SceI (Table I). DNA sequencing analysis indicated that mutant S4.2 contained two additional mutations compared to the parent S4-4: N131D and L206P.
In vitro characterization of the evolved homing endonuclease mutants
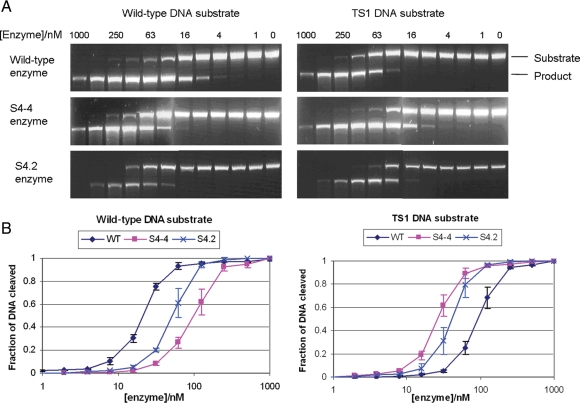
We characterized the in vitro cleavage activity of the mutants S4-4 and S4.2, selected from the saturation and random mutagenesis rounds, respectively, and compared their activity with that of wild-type I-SceI. Enzymes were purified using immobilized nickel affinity chromatography followed by gel filtration, yielding a final purity of >95% estimated by SDS–PAGE (data not shown). The cleavage activity of each enzyme with the natural cleavage sequence and TS1 substrate was assessed using an in vitro digestion assay, as detailed in Materials and Methods. The amount of enzyme required to cleave 50% of each substrate was compared to determine their relative in vitro cleavage efficiency (Fig. 3 and Table I). The mutations identified in S4-4 and S4.2 gave an overall effect of specificity shift and broadening.
Fig. 3.
In vitro cleavage analysis of wild-type and mutant I-SceI enzymes. (A) Representative DNA gel electrophoresis of in vitro cleavage reaction mixtures. Various concentrations of wild-type I-SceI and mutants were incubated with linear DNA substrate, which contains either the wild-type or the TS1 cleavage sequence. Since the recognition sequence is located in the center of the DNA substrate, cleavage of the DNA substrate resulted in a single band product with half the size of the substrate, minimizing the densitometry error. The electrophoresis gel images were captured using different exposure times for a clear contrast between the substrate and the product. (B) Plot of in vitro cleavage assay. The fraction of DNA cleaved was calculated by dividing the product band intensity by the substrate band intensity.
Consistent with in vivo activity assay (Table I), wild-type I-SceI showed strong preference for its natural substrate, while both mutants S4-4 and S4.2 preferred the TS1 substrate. The intermediate mutant S4-4 selected from one round of saturation mutagenesis showed ∼17-fold specificity shift towards TS1 substrate, with little specificity broadening. In contrast, the final mutant, S4.2, which had two additional mutations found in the second round of random mutagenesis, showed more broadened specificity. This resulted in a slightly increased survival rate, from 0 to ∼10%, of the wild-type I-SceI selection strain transformed with mutant S4.2 than that transformed with S4-4 (Table II).
Table II.
In vitro cleavage efficiency and specificity changes of wild-type I-SceI and mutants
| Enzyme | Relative cleavage efficiencya of TS150 to natural sequence50 | Specificity shiftb | Specificity broadeningc |
|---|---|---|---|
| Wild-type | 4.5 | – | – |
| S4-4 | 0.27 | 16.7 | 1.2 |
| S4.2 | 0.77 | 5.8 | 3.4 |
aWild-type I-SceI and mutant S4.2 were incubated with the natural and TS1 recognition sequences individually. The concentration of enzyme necessary to cleave 50% of each site was determined. The relative cleavage efficiency is the ratio of these two concentrations (Sussman et al., 2004).
bThe specificity shift is calculated by dividing the relative cleavage efficiency of the wild-type enzyme by the relative cleavage efficiency of the mutant enzyme.
cThe specificity broadening is the product of the relative cleavage efficiency of the wild-type enzyme and the mutant enzyme. Absence of specificity broadening is represented by the value of 1, indicating no overall change in the breadth of the specificity profile for the mutant enzyme, and broader specificity is indicated by a value greater than 1 (Sussman et al., 2004; Doyon et al., 2006).
Structural basis for altered specificity of I-SceI mutants
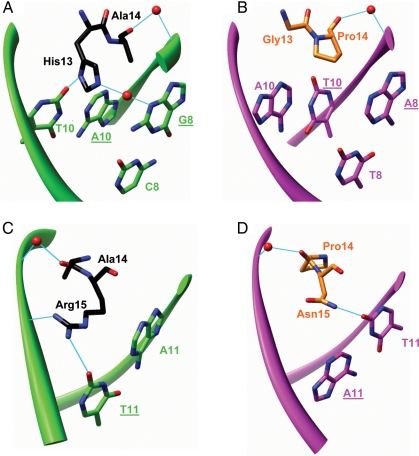
To gain insight into how the introduced amino acid substitutions altered the specificity of I-SceI mutants, a homology model of mutant S4-4 bound to TS1 DNA substrate was built as described in Materials and Methods. As shown in Fig. 4A, mutation G13H forms two new hydrogen bonds with thymine at position 10 (T10) and guanine at positions 8 of the TS1 sequence (G8, underline represents nucleotides from the target sequence) through its δ nitrogen atom and a water molecule, respectively, which may contribute to the specificity alteration at position 8 and 10. P14A in S4-4 mutant interacts with the TS1 backbone through a water-mediated hydrogen bond in a manner similar to P14 in the wild-type complex (Fig. 4A and B), and thus does not contribute directly to the specificity alteration. This mutation might be a result of the stacking effect to accommodate the mutation of glycine to histidine which has a larger side chain. The hydrogen bond between N15 and T11 in the wild-type complex is replaced by a new one formed between R15 and T11 (Fig. 4C and D, underline represents nucleotides from the target sequence), thus mutation N15R maintains the contact with base-pair 11 in the TS1 sequence. In addition, mutation N15R introduces an additional contact with the TS1 backbone through hydrogen-bonding (Fig. 4C). The other two mutations, N131D and L206P, obtained as a result of error-prone PCR mutagenesis, are located on the surface of the protein (Fig. 2). Due to their large distance from the DNA (>15 Å), the molecular basis for their contributions to broadened specificity and reduced activity of S4.2 enzyme towards TS1 sequence is not obvious.
Fig. 4.
Contributions of mutations G13H, P14A and N15R to altered specificity of S4-4 enzyme at base-pairs 8 and 10 (A) and base-pair 11 (C). Base-pair 9 is not contacted by any of the three residues; thus, it is not shown. The corresponding interactions in the wild-type complex are shown in (B) and (D). The backbone of TS1 and wild-type cleavage DNA sequences is shown as green and magenta ribbons, respectively. Nucleotides from the bottom DNA strand are underlined. Water molecules are depicted as red spheres. Hydrogen bonds are shown in cyan.
Discussion
We previously described the development of a two-plasmid selection system that links the DNA cleavage event by a homing endonuclease with E.coli cell survival (Chen and Zhao, 2005a). In this study, we report its application for the identification of mutant I-SceI homing endonucleases that are able to selectively cleave a new target DNA sequence, TS1. The cleavage sequence specificity of the evolved mutants toward TS1 is similar to that of wild-type I-SceI to the natural sequence. The target sequence TS1 differs from the natural cleavage sequence by 4 bp at the 3′-end (Fig. 1). Wild-type I-SceI shows minimal cleavage activity toward TS1 in vivo, with a survival rate of ∼0.9% in the TS1-containing selection strain, according to the E.coli survival-based enzymatic assay (Chen and Zhao, 2005b). After one round of saturation mutagenesis of residues close to the four mismatched base-pairs in TS1, we successfully obtained a mutant, S4-4, which showed a high survival rate (∼45%) in the TS1 selection strain, representing a ∼50-fold increase in survival rate compared with that of wild-type I-SceI. The enzymatic cleavage activities of this mutant and wild-type I-SceI toward TS1 and the natural cleavage sequences were assayed in vitro using purified proteins. Mutant S4-4 exhibited a ∼4-fold preference for the TS1 sequence over the natural sequence, demonstrating ∼17-fold shift in specificity with little specificity broadening (Table II). This degree of specificity is similar to that of wild-type I-SceI for the natural target sequence over TS1. Homing endonuclease I-SceI is known for its high specificity and has been used for genomic engineering (Cohen-Tannoudji et al., 1998; Miller et al., 2003; Storici et al., 2003; Tzfira et al., 2003; Kang et al., 2004). We believe mutant S4-4 may also be used in vivo for gene targeting experiments. The engineering of protein mutants with shifted substrate specificity is rather rare in directed evolution, especially when only positive selection pressure is applied. Most directed evolution experiments result in mutants with broadened substrate specificity (Chen et al., 2004; Chen and Zhao, 2005a). Mutant S4-4 contains three mutations: G13H, P14A and N15R (Fig. 2). Homology modeling analyses showed that G13H and N15R are directly involved in specificity shift by forming new contacts with the new TS1 target sequence, while P14A contributes by providing space to accommodate the bulky side chain of H13.
In an attempt to further improve the cleavage efficiency of S4-4 mutant toward TS1, a second round of random mutagenesis was conducted and yielded a mutant, S4.2, which showed an in vivo survival rate of ∼85% in the TS1 selection strain. This represents a ∼90-fold increase in survival rate compared with that of the wild-type enzyme. Interestingly, although the survival rate of TS1 selection strain transformed with S4-4 was lower than that transformed with S4.2 (Table I), the in vitro enzymatic activity analyses showed that S4-4 cleaved TS1 substrate more efficiently than S4.2 (Fig. 3). This disparity between in vivo and in vitro assays was at least partially, if not all, due to the soluble enzyme expression level difference inside the E.coli cell: S4.2 showed an expression level ∼3–4-fold higher than that of S4-4 (data not shown). These results indicated that the two additional mutations present in S4.2, N131D and L206P might have a role in improving the enzyme folding and/or solubility. In addition, these two mutations were also responsible for reduced cleavage efficiency and broadened specificity of S4.2 (Fig. 3 and Table II), of which the underlying mechanism is not clear from the homology model. To avoid isolation of such mutants with broadened specificity, a negative selection step could be included (Doyon et al., 2006). The broadened specificity may also allow mutant S4.2 to cleave other regions of the reporter plasmid, resulting in its degradation independent of the target sequence cleavage, which increases the in vivo survival rate. Our two-plasmid selection system is readily adaptable to negative selection by incorporating the unwanted cleavage sequence in the homing endonuclease-encoding pTrc plasmid, so that mutants with activity toward the unwanted substrate would cleave the pTrc plasmid, rendering the cell non-viable on kanamycin-containing media.
Although we have achieved a relative large specificity shift of I-SceI (22% mismatch), the ultimate goal of this project is to obtain an I-SceI variant that can efficiently cleave the CFTR sequence (83% mismatch) as shown in Fig. 1. Due to this significant difference, we set out to apply an in vitro coevolution strategy (Chen and Zhao, 2005b) to facilitate the identification of variants with multiple simultaneous mutations. The in vitro coevolution strategy involves stepwise application of directed evolution towards a series of intermediate target DNA sequences that bridge the natural cleavage sequence and the final target cleavage sequence. To date, continued efforts at engineering I-SceI mutants to cleave the TS2 target sequence (Fig. 1), containing an additional four mismatched base-pairs compared with TS1, have not yielded further mutants with shifted specificity. In an effort to continue our engineering of homing endonuclease toward our final target DNA cleavage sequence (CFTR in Fig. 1), we have also tried to introduce fewer mismatched bases, with TS1 plus one or two mutations at the 6 and 7 positions (Fig. 1), as well as starting from the 5′-end (TS3 in Fig. 1). Unfortunately, these endeavors did not yield further specificity-shifted mutants. These results highlight the difficulty in engineering homing endonuclease with altered DNA sequence specificity. Previous efforts on engineering homing endonuclease with a new DNA cleavage sequence containing individual mismatched base-pairs have also yielded limited success (Seligman et al., 2002; Sussman et al., 2004; Doyon et al., 2006; Rosen et al., 2006). Nonetheless, these results are better than those previous results since four nucleotides in the target sequence were changed simultaneously.
The 3′-end of the target TS1 sequence, consisting of base-pairs 8–11, is recognized by a relatively independent sub-domain at the N-terminal of I-SceI that is connected to the central LAGLIDADG helix through a flexible loop (Fig. 2). It is likely that mutations in this region only affect the binding to the target DNA sequence and have little effect on the catalytic activity of the enzyme. Thus, through saturation mutagenesis of residues contacting the DNA directly, we can readily identify mutants with high affinity toward the altered TS1 target sequence. However, the central part of I-SceI is responsible for both DNA recognition and cleavage. Mutations affecting the binding of the substrate can also affect the catalytic efficiency of the enzyme. Our effort at randomizing residues directly contacting DNA base-pairs at the 6 and 7 positions did not yield any mutants with improved cleavage efficiency toward these substrates, possibly due to our inability to accommodate changes in both DNA binding and cleavage simultaneously with a limited set of residues. These results suggested that a drastic specificity alteration (e.g. 83% mismatch of our target CFTR sequence) of homing endonucleases might require an additional library diversity creation strategy, such as domain fusions, to create new protein scaffolds with novel specificity (Chevalier et al., 2002; Epinat et al., 2003; Steuer et al., 2004).
Funding
This work was supported by National Science Foundation CAREER Award (BES-0348107) and National Institutes of Health (R21 HL089418).
Acknowledgements
We thank Karuppiah Chockalingam, Nikhil U. Nair and members of the Zhao laboratory for critical comments of this manuscript.
Footnotes
Edited by Bengt Mannervik
References
- Arnould S., Perez C., Cabaniols J.P., Smith J., Gouble A., Grizot S., Epinat J.C., Duclert A., Duchateau P., Paques F. J. Mol. Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- Chen Z., Zhao H. Nucleic Acids Res. 2005;a 33:e154. doi: 10.1093/nar/gni148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhao H. J. Mol. Biol. 2005;b 348:1273–1282. doi: 10.1016/j.jmb.2005.02.070. [DOI] [PubMed] [Google Scholar]
- Chen Z., Katzenellenbogen B.S., Katzenellenbogen J.A., Zhao H. J. Biol. Chem. 2004;279:33855–33864. doi: 10.1074/jbc.M402118200. [DOI] [PubMed] [Google Scholar]
- Chevalier B.S., Kortemme T., Chadsey M.S., Baker D., Monnat R.J., Stoddard B.L. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- Chockalingam K., Chen Z., Katzenellenbogen J.A., Zhao H. Proc. Natl Acad. Sci. USA. 2005;102:5691–5696. doi: 10.1073/pnas.0409206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulika A., Perrin A., Dujon B., Nicolas J.F. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Tannoudji M., Robine S., Choulika A., Pinto D., El Marjou F., Babinet C., Louvard D., Jaisser F. Mol. Cell. Biol. 1998;18:1444–1448. doi: 10.1128/mcb.18.3.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleaux L., D'Auriol L., Galibert F., Dujon B. Proc. Natl Acad. Sci. USA. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho G., Jasin M., Berg P. Mol. Cell. Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W.J., Miller J.F., Ragsdale C.W. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon J.B., Pattanayak V., Meyer C.B., Liu D.R. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- Dujon B. Gene. 1989;82:91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- Epinat J.C., Arnould S., Chames P., Rochaix P., Desfontaines D., Puzin C., Patin A., Zanghellini A., Paques F., Lacroix E. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D.J., Lipps H.J., Jans D.A. Nat. Rev. Genet. 2005;6:299–310. doi: 10.1038/nrg1577. [DOI] [PubMed] [Google Scholar]
- Gruen M., Chang K., Serbanescu I., Liu D.R. Nucleic Acids Res. 2002;30:e29. doi: 10.1093/nar/30.7.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- Johnson R.D., Jasin M. Biochem. Soc. Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- Kang Y., Durfee T., Glasner J.D., Qiu Y., Frisch D., Winterberg K.M., Blattner F.R. J. Bacteriol. 2004;186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem B., Rommens J.M., Buchanan J.A., Markiewicz D., Cox T.K., Chakravarti A., Buchwald M., Tsui L.C. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- Kim Y.G., Cha J., Chandrasegaran S. Proc. Natl Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard I.A., Pratt S.D., McCafferty D.G., Bussiere D.E., Hutchins C., Wanner B.L., Katz L., Walsh C.T. Chem. Biol. 1998;5:489–504. doi: 10.1016/s1074-5521(98)90005-9. [DOI] [PubMed] [Google Scholar]
- Mani M., Smith J., Kandavelou K., Berg J.M., Chandrasegaran S. Biochem. Biophys. Res. Commun. 2005;334:1191–1197. doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.G., Petek L.M., Russell D.W. Mol. Cell. Biol. 2003;23:3550–3557. doi: 10.1128/MCB.23.10.3550-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moure C.M., Gimble F.S., Quiocho F.A. J. Mol. Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- Porteus M.H., Baltimore D. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Rosen L.E., Morrison H.A., Masri S., Brown M.J., Springstubb B., Sussman D., Stoddard B.L., Seligman L.M. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P., Smih F., Jasin M. Mol. Cell. Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G., Sommer S.S. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- Seligman L.M., Chisholm K.M., Chevalier B.S., Chadsey M.S., Edwards S.T., Savage J.H., Veillet A.L. Nucleic Acids Res. 2002;30:3870–3879. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smih F., Rouet P., Romanienko P.J., Jasin M. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J., et al. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer S., Pingoud V., Pingoud A., Wende W. Chembiochemistry. 2004;5:206–213. doi: 10.1002/cbic.200300718. [DOI] [PubMed] [Google Scholar]
- Storici F., Durham C.L., Gordenin D.A., Resnick M.A. Proc. Natl Acad. Sci. USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman D., Chadsey M., Fauce S., Engel A., Bruett A., Monnat R., Jr, Stoddard B.L., Seligman L.M. J. Mol. Biol. 2004;342:31–41. doi: 10.1016/j.jmb.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Tzfira T., Frankman L.R., Vaidya M., Citovsky V. Plant. Physiol. 2003;133:1011–1023. doi: 10.1104/pp.103.032128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Moore J.C., Volkov A.A., Arnold F.H. Manual of Industrial Microbiology and Biotechnology. 2nd edn. Washington, D.C: American Society for Microbiology Press; 1999. [Google Scholar]