The induction of an appropriate immune response upon infection of an invading pathogen is controlled by host PRRs2 that specifically recognize essential structural components of infectious agents termed PAMPs (1–3). Detection of PAMPs by PRRs triggers intracellular signaling pathways leading to the expression of immune mediators such as cytokines, chemokines, and type I IFNs. TLRs represent a now well defined PRR family because much has been discovered recently in terms of the PAMPs they interact with, the downstream signaling pathways triggered, and the complex yet specific immune responses elicited.
TLRs are expressed on both immune and nonimmune cells (1) and are part of the IL-1R/TLR superfamily, a family of proteins defined by the presence of a common cytoplasmic signaling domain (the TIR domain), which also includes the IL-1, IL-18, and IL-33 receptors (4). The TLRs can be divided into two subgroups based on their subcellular location: one comprising TLR1, TLR2, and TLR4–6, which signal from the plasma membrane, and a second containing TLR3 and TLR7–9, which are localized to endosomal compartments. TLR2 functions as a heterodimer with either TLR1 or TLR6 to respond to either bacterial triacyl or diacyl lipopeptides, respectively. TLR4 forms homodimers in response to LPS from Gram-negative bacteria. TLR3 binds viral double-stranded RNA, whereas TLR7 and TLR8 detect viral single-stranded RNA. Finally, TLR9 responds to viral and bacterial DNA in endosomes in a process likely to be dependent on the sugar backbone of DNA rather than on the presence of unmethylated CpG dinucleotides as was previously thought (5).
Among the transcription factors activated by TLR signaling are three classes known to be important in the innate immune response, namely NF-κB, IRFs, and AP-1 family members. Signaling pathways shared with IL-1R are used to mediate NF-κB and AP-1 activation, leading to the induction of cytokines and chemokines. A subset of TLRs (TLR3, TLR4, and TLR7–9) can also activate IRF3 and IRF7, leading to the induction of IFN-α and/or IFN-β.
Similar to most known signaling pathways, the role of phosphorylation in regulating TLR signaling is well established. However, it is now appreciated that ubiquitination plays at least as important a role in the control and regulation of TLR signaling as phosphorylation (6). Historically, the primary function of ubiquitination identified in these pathways was to target proteins for proteasomal degradation. It is now clear, however, that the role of ubiquitin is far more diverse. For example, monoubiquitination of a target protein can act as a signal for receptor internalization, vesicle sorting, DNA repair, or gene silencing (7). Furthermore, polyubiquitin chains of different topologies direct distinct outcomes for target proteins. Hence, the formation of Lys48-linked polyubiquitin chains of at least four ubiquitins is required to signal a protein for degradation via the 26 S proteasome, whereas polyubiquitin chains linked through Lys63 do not mediate protein degradation but rather direct protein interactions that function in DNA repair, signal transduction, endocytosis, and ribosomal protein synthesis. Although the role of Lys63-linked polyubiquitination in signaling pathways leading to NF-κB activation is well established, it is also emerging as a potentially key event in IRF activation. This minireview focuses on the role of these non-degradative Lys63-linked polyubiquitination events in regulating signal transduction by TLRs to NF-κB. The importance of Lys63-linked polyubiquitination in the regulation of TLR signaling pathways to NF-κB is underscored by the fact that DUBs such as CYLD and A20 function as key silencers of NF-κB activation by these innate immune receptors (8). The removal of Lys63-linked polyubiquitin chains from TLR signaling molecules such as RIP1, TRAF6 (TNF receptor-associated factor 6), TAK1 (transforming growth factor-β-activated kinase 1), and NEMO by these DUBs is a vital control mechanism preventing excessive induction of pro-inflammatory responses upon TLR stimulation.
Polyubiquitination as an Activation Signal in TLR Signaling to NF-κB
Subsequent to ligand engagement and receptor dimerization, different TLRs engage one or a subset of TIR domain-containing adaptors to initiate signaling to NF-κB (9). For example, TLR4 recruits four TIR adaptors (MyD88, Mal, TRAM, and TRIF), whereas TLR3 engages only TRIF, and TLR2 heterodimers signal via both Mal and MyD88 (9). TLRs largely use the “classical” pathway to NF-κB activation, involving the IKK complex, which contains the kinases IKKα and IKKβ associated with NEMO, a scaffold protein (10). Phosphorylation of IκB proteins by the IKK complex targets them for Lys48-linked ubiquitination and subsequent degradation by the proteasome, which releases NF-κB dimers in the cytoplasm to translocate into the nucleus and activate target genes (10). The IKK complex also phosphorylates the NF-κB subunit p65, which is necessary for transactivation of genes.
Role of TRAF6 E3 Ligase Activity in NF-κB Activation by TLRs
The current understanding of TLR-mediated NF-κB activation proposes that the majority (and potentially all) of TLR signal transduction pathways to NF-κB converge at TRAF6, at a point upstream of IKK activation, irrespective of the TIR adaptor used (Fig. 1). Hence, the identification of TRAF6 as a RING (really interesting new gene) domain E3 ubiquitin ligase (11, 12) represented a seminal discovery in understanding these pathways. TRAF6 works in combination with a heterodimeric E2 enzyme complex known as TRIKA1 (TRAF6-regulated IKK activator 1), consisting of the E2 enzyme Ubc13 (ubiquitin-conjugating enzyme 13) and the Ubc-like protein Uev1a (ubiquitin E2 variant 1a) (11). Together, TRAF6 and TRIKA1 catalyze the synthesis of Lys63-linked polyubiquitin chains on target components of TLR signaling pathways, including on TRAF6 itself. This polyubiquitination of TRAF6 leads to recruitment and activation of a downstream trimeric complex comprising TAK1, TAB1 (TAK1-binding protein 1), and TAB2 or TAB3 (12). TAB2 and TAB3 contain a novel conserved zinc finger domain that recognizes Lys63-linked polyubiquitin chains conjugated to TRAF6, thus facilitating recruitment of the TAK1-TAB1-TAB2/3 complex to TRAF6 (13). Mutations of the TAB2 and TAB3 ubiquitin-binding domains that abolished their ability to bind TRAF6 polyubiquitin chains showed that this association is essential for turning on TAK1 kinase activity and hence the subsequent TAK1-mediated phosphorylation and activation of IKKβ (13). MKK (MAPK kinase) family members are also targets of TAK1 phosphorylation, and these in turn phosphorylate and activate JNK (Jun N-terminal kinase) and p38 MAPKs (12, 14). Within the IKK complex, NEMO acts as both a target of polyubiquitination (15–18) and, like TAB2/TAB3, as a ubiquitin receptor preferentially binding Lys63-linked polyubiquitin chains on other signaling molecules (19). It would appear that both functions of NEMO are integral to TLR signaling. TRAF6-mediated polyubiquitination of NEMO at Lys285 was found to be required for optimal NF-κB activation by TLR4 (20). Furthermore, a mutation in NEMO that results in a severe form of incontinentia pigmenti, a genetic disease that primarily manifests as a severe inflammation of the skin, was found to impede NF-κB activation via TNF, IL-1, LPS, and phorbol 12-myristate 13-acetate due to impaired NEMO polyubiquitination by TRAF6 (21). The recognition of Lys63-linked polyubiquitin chains on IRAK-1 (IL-1R-associated kinase 1) by NEMO is also essential for the activation of signaling cascades downstream of IL-1R/TLRs (see below). Thus, NEMO undergoes polyubiquitination at numerous Lys residues upon IL-1R/TLR stimulation and is also recruited to key signaling complexes through the recognition of Lys63-linked polyubiquitination. However, the relative contribution of the ubiquitin-binding function and the polyubiquitination of NEMO in different TLR signaling cascades merits further investigation, as it is as yet unclear how these ubiquitin interactions activate signal transduction.
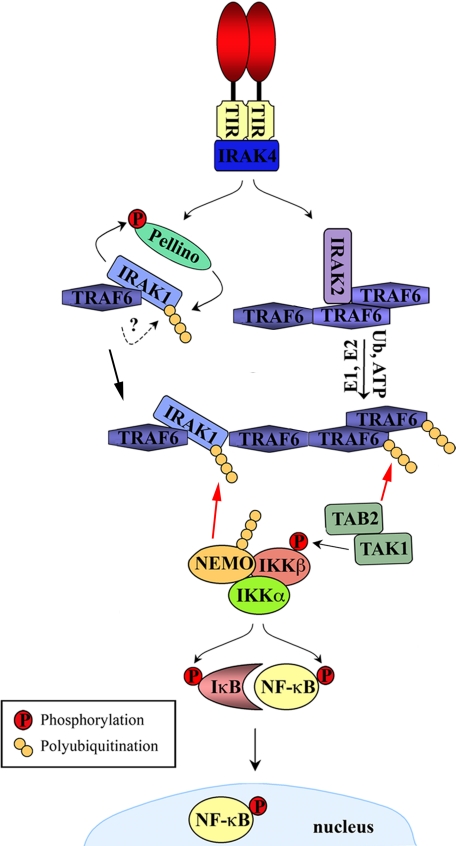
FIGURE 1.
Role of Lys63-linked polyubiquitination in IL-1R/TLR signaling to NF-κB. IL-1R/TLR stimulation leads to receptor dimerization, allowing recruitment of the appropriate downstream TIR adaptor via TIR/TIR domain associations. This is followed by activation of IRAK-1 and IRAK-2. IRAK-1 activation involves phosphorylation by IRAK-4, but this has yet to be confirmed for IRAK-2. IRAK-2 triggers the E3 ligase activity of TRAF6, potentially by promoting its oligomerization, leading to TRAF6 autoubiquitination and the synthesis of Lys63-linked polyubiquitin chains on target molecules such as NEMO. TAB2 (or TAB3) specifically recognizes Lys63-linked polyubiquitin chains on TRAF6, thus recruiting TAK1 to the TRAF6 complex, resulting in TAK1 activation. In parallel, Pellino proteins are phosphorylated by IRAK-1, a process that results in their degradation. However, the Pellino proteins also mediate Lys63-linked polyubiquitination of IRAK-1, facilitating the recruitment of NEMO and therefore the IKK complex through the specific recognition of these ubiquitin (Ub) modifications. Activated TAK1, now in close proximity to the IKK complex, phosphorylates and activates this complex. Red arrows indicate recruitment via binding to polyubiquitin chains.
Role of Ubc13 as an E2 Ubiquitin-conjugating Enzyme in TLR Signaling
Akira and co-workers (22) tested the requirement for Ubc13 in TLR signaling in vivo by conditional knock-out of ubc13 in mice. Although previously assumed to be central for NF-κB activation due to its role in TRA6 E3 ligase activity (see above), surprisingly, in cells lacking ubc13, NF-κB activation by numerous IL-1R/TLR agonists in different cell types was normal, whereas only MAPK activation was grossly impaired (22). IL-1-dependent TRAF6 polyubiquitination proceeded normally in the absence of Ubc13, indicating that TRAF6 E3 ligase activity can be coupled to E2 enzymes other than Ubc13 to mediate its autoubiquitination. However, NEMO polyubiquitination was severely inhibited and was shown to be required at least in part for IL-1-induced MAPK activation (22). Another study using cells from mice heterozygous for the ubc13 gene found that LPS-induced IκBα degradation was significantly blocked in both macrophages and splenocytes (23). Strikingly, in that study, the induction of TRAF6 polyubiquitination following LPS stimulation was prevented in the absence of Ubc13. Thus, it may be that the requirement for Ubc13 in catalyzing TRAF6 polyubiquitination and mediating downstream signal transduction is cell type- and signal-dependent.
Function of IRAK-2 in TRAF6 Ubiquitination and NF-κB Activation
Although the importance of TRAF6 ubiquitination in TLR signaling has been established, how exactly receptor-adaptor oligomerization enhances or turns on TRAF6 E3 ligase activity is still somewhat unclear. Originally, IRAK-1 was shown to be required for certain IL-1R/TLR pathways to NF-κB and to interact with TRAF6, and as it could also interact with receptor complexes, it was placed “upstream” of TRAF6 in signaling to NF-κB. IRAK-4 is also recruited to activated receptor complexes and can phosphorylate and activate IRAK-1. Certainly, IRAK-1 and/or IRAK-4 seems to be involved to some degree in almost all IL-1R/TLR signaling pathways (but not necessarily in NF-κB activation). Thus, when TRAF6 was found to function as an E3 ligase, it was assumed that IRAK-1 must somehow turn on this activity. However, this was never proven, and it has been shown that IRAK-1 is dispensable for TLR3, TLR4, and TLR9 pathways to NF-κB (24–26). Furthermore, a study of the Epstein-Barr virus-encoded oncogene LMP1, a known viral homolog of the TNF receptor superfamily members, was able to trigger TRAF6 polyubiquitination and subsequent IKKβ activation in IRAK-1-deficient cells (27).
In keeping with this, we have recently proposed that IRAK-2, rather than IRAK-1, is required for turning on TRAF6 E3 ligase activity in IL-1R/TLR signaling pathways to NF-κB (28). IRAK-2 was discovered in 1997 (29), but its particular role in TLR signaling has remained unclear until recently. In our hands, exogenous IRAK-1 could not induce the polyubiquitination of TRAF6, whereas IRAK-2 could (28), even in IRAK-1-deficient cells.3 Furthermore, point mutation of a TRAF6-binding motif within IRAK-2 gave rise to a nonfunctional molecule that could no longer activate NF-κB and MAPKs and could no longer drive TRAF6 polyubiquitination. The importance of IRAK-2 in the activation of NF-κB by TLR3, TLR4, and TLR8 in human cell lines was confirmed by IRAK-2 small interfering RNA, whereas knockdown of IRAK-2 expression also inhibited TLR4/LPS-mediated IL-8 production in primary human cells (28).
An essential requirement for IRAK-2 in NFκB-dependent signaling by TLRs has now also been shown in mice. Kawagoe et al. (30) generated Irak-2–/– mice and showed that IRAK-2 was critical for the induction of NF-κB-dependent cytokines by a range of TLRs tested. Mice lacking IRAK-2 displayed greatly enhanced survival in response to either LPS or CpG challenge, thus further verifying the potent pro-inflammatory function of IRAK-2. Furthermore, in the case of TLR2, although early NF-κB and MAPK activation was normal in cells lacking IRAK-2, sustained or late NF-κB activation was seriously impaired (30). How this fits with a role for IRAK-2 in triggering TRAF6 autoubiquitination has yet to be ascertained given that this is often an early and transient event, at least for IL-1 signaling (11). However, the kinetics of TRAF6 polyubiquitination for TLR ligands is less clear, and it will be of interest to measure this in both normal and IRAK-2-deficient cells.
The mechanism whereby IRAK-2 activates TRAF6 E3 ligase activity also remains to be deciphered, but one possibility is that IRAK-2 may direct TRAF6 oligomerization. The coupling of TRAF6 oligomerization to ubiquitination has been suggested as a central feature of IKK activation following antigen binding to T cell receptors (16). Only high molecular weight oligomeric forms of both BCL10 and MALT1, two essential T cell receptor signaling mediators, could activate IKK and NF-κB in vitro. MALT1 oligomers (and not the monomeric protein) bound TRAF6 directly, triggering its subsequent oligomerization and simultaneously switching on its E3 ligase activity such that NEMO polyubiquitination was observed. Thus, for TLR signaling, IRAK-2 may provide a TRAF6 oligomerization trigger, similar to MALT1 in T cells.
Lys63-linked Polyubiquitination of IRAK-1 in the NF-κB Activation Pathway
Although IRAK-1 may not have a role in stimulating TRAF6 E3 ligase activity, it is itself ubiquitinated. Previously, it was considered that following its phosphorylation and concomitant activation, IRAK-1 was rapidly ubiquitinated and targeted for proteasomal degradation (31). Somewhat surprisingly, it has now emerged that IRAK-1 undergoes Lys63-linked polyubiquitination and that, in fact, this modification is critical for signal transduction rather than marking a degradation event (32–34). The attachment of Lys63-linked polyubiquitin chains to IRAK-1 has now been demonstrated after both IL-1 and LPS stimulation (33, 34).
Both TRAF6 and the Pellino proteins have been put forward as candidate E3 ligases involved in assembling polyubiquitin chains on IRAK-1 (34, 35). The presence of recombinant glutathione S-transferase-TRAF6 appeared to enhance the polyubiquitination of IRAK-1 in combination with all other components necessary to catalyze a ubiquitination reaction in an in vitro reconstitution system (34). However, IL-1-stimulated polyubiquitination of IRAK-1 was not impaired in IL-1-stimulated TRAF6–/– fibroblasts (33). The presence of a RING-like domain within the secondary structure of the Pellino proteins initially led to the proposal that these proteins may act as E3 ligases, and indeed, overexpression of Pellino1 and Pellino2 was found to promote non-degradative polyubiquitination of IRAK-1 (36). The Pellino proteins have been shown to directly catalyze the elongation of polyubiquitin chains in vitro, thus solidly confirming the role of these proteins as E3 ubiquitin ligases (32, 35). The topology of the polyubiquitin chains constructed by the Pellino proteins appears, in fact, to be governed by the particular E2 ubiquitin ligase engaged (32). Thus, when in combination with the Ubc13-Uev1a E2 complex, Pellino1 specifically triggered the formation of Lys63-linked polyubiquitin chains, whereas when working in conjunction with UbcH3, polyubiquitin chains linked via Lys48 were preferentially assembled. Both IRAK-1 and IRAK-4 can directly phosphorylate the Pellino proteins in vitro, leading to their enhanced E3 ligase activity (32). Interestingly, this also leads to degradative (Lys48) ubiquitination of Pellino proteins, and hence, not only can Pellino proteins regulate IRAK-1 by catalyzing its Lys63-linked ubiquitination, but IRAK-1 can also cause Pellino degradation (35). The exact role of Pellino proteins in the complex regulation of TLR signaling is still being elucidated, with some family members likely having a positive role and others negatively regulating these pathways (35, 37).
What then is the role of IRAK-1 Lys63 polyubiquitination in TLR signaling? Windheim et al. (33) showed that Lys63 polyubiquitination of IRAK-1 caused its association with NEMO in a ligand-dependent manner, whereas unmodified IRAK-1 failed to interact with NEMO, and this was recently confirmed by another group (34). Furthermore, a NEMO point mutant unable to interact with Lys63-polyubiquitinated IRAK-1 failed to restore IL-1-stimulated NF-κB activation in NEMO-deficient cells (33). Thus, ubiquitinated IRAK-1 likely has a role in recruiting NEMO to a post-receptor complex containing TAK1, which would contribute to NF-κB activation by bringing the IKK complex into proximity to TAK1.
Model for the Role of Polyubiquitination in TLR Signaling to NF-κB
Taken together, the data described above lead to the following current model for how Lys63-linked ubiquitination contributes to activation of the IL-1R/TLR/NF-κB axis (Fig. 1). After ligand stimulation, IRAK-2 induces TRAF6 E3 ligase activity, leading to the Lys63-linked polyubiquitination of TRAF6 itself and other substrates, including NEMO. TAK1 is then recruited to ubiquitinated TRAF6 via TAB2 and TAB3. Concurrently, Pellino proteins polyubiquitinate IRAK-1, which allows recruitment of the IKK complex (via NEMO) to IRAK-1. Upstream of these events, IRAK-4 may “turn on” both the IRAK-2/TRAF6/TAK1 and Pellino/IRAK-1/NEMO pathways. At some early time point post-stimulation, ubiquitinated TRAF6 and ubiquitinated IRAK-1 may act in close proximity or be part of the same TRAF6 complex such that TAK1 recruited by TRAF6 can phosphorylate and hence activate the IKK complex recruited via IRAK-1. For TLR2 at least, it has now been proven that both IRAK-1 and IRAK-2 are required for maximal and sustained NF-κB activation in mice (30). However, in other cases, the role of IRAK-1 described in this model would be stimulus-specific because at least some TLRs such as TLR3 and TLR9 have been shown to activate NF-κB without a requirement for IRAK-1. However, the IRAK-2/TRAF6/TAK1 axis may be utilized by all TLRs.
Future Perspectives
The importance of the regulation of signaling by ubiquitin is reflected in the fact that it is so highly conserved in nature. For example, activation of Drosophila TAK1 and the IKK complex in response to stimulation of the IMD signaling pathway by Gram-negative bacteria has been shown to be dependent on the Drosophila Ubc13 and Uev1A homologs (38). Thus, a signal activation function for ubiquitination in innate immunity is evolutionarily conserved from Drosophila to humans. A further indication of the importance of ubiquitin in signaling pathways sensing microorganisms is the fact that pathogens target this host process to shut down immune responses. For example, the Yersinia pestis virulence factor YopJ acts as a deubiquitinating protease that specifically targets TRAF6 and TRAF3 to effectively block the activation of NF-κB and MAPKs as well as IFN induction by TLRs (39). Further investigation of the means whereby pathogens manipulate E3 ligases such as TRAF6 and TRAF3 will be invaluable in uncovering the relevance of (and detailed mechanism whereby) Lys63-linked polyubiquitination leads to activation of signaling mechanisms.
Whereas ubiquitination involving TRAF6 is essential in linking receptor proximal events to the IKK complex for NF-κB activation, there is now evidence to suggest that polyubiquitination by TRAF3 may play an equivalent role in IRF activation (40, 41), and the targeting of this activity by Y. pestis to block TLR-mediated IFN induction further supports this. Overexpressed TRAF3 was modified by the addition of Lys63-linked polyubiquitin chains, and using small interfering RNA targeted to DUBA (a DUB found to promote the removal of Lys63-linked polyubiquitin chains from TRAF3) enhanced the IFN response induced by TLR3, TLR4, TLR7, and RIG-I-like receptors (42). The downstream targets of TRAF3 polyubiquitination have yet to be characterized, but one may be TANK (43).
TRAF6 is also used by TLR7–9 to activate IRFs via MyD88 (24, 44), thus raising the question as to whether its E3 ligase activity is equally important here. Ectopically expressed TRAF6 associated with and stimulated IRF7 ubiquitination in a Ubc13-dependent manner (44). However, the relative contribution of TRAF3 and TRAF6 to IFN-α induction via TLRs remains to be assessed. IRAK-1 has similarly been implicated in the stimulation of IFN-α release by TLR7–9 while being dispensable for NF-κB activation in this pathway (24). It will be interesting to see whether IRAK-1 needs to be polyubiquitinated via a Lys63 ubiquitin linkage for this function. If so, deciphering the role of the Pellino proteins (or indeed TRAF3 or TRAF6) as E3 ligases of IRAK-1 in IRF activation will be required.
Although we have discussed the mechanisms whereby Lys63-linked polyubiquitination contributes to IL-1R/TLR signaling for the family as a whole and the role of TRAF6 and TRAF3 in NF-κB and IRF activation, respectively, it is almost inevitable that receptor-specific differences will exist, and some of these differences are already apparent. For example, the protein kinase RIP1, which is required for TLR3 and TLR4 signaling to NF-κB, but not IRF3 activation, undergoes polyubiquitination upon TLR3 stimulation (45). RIP1 is used only by TLR3 and TLR4 for TRIF-dependent activation of NF-κB (45, 46), so this polyubiquitination step is likely not required for TLR signaling to NF-κB in any other context. As well as receptor-specific differences, the observation that NF-κB activation was largely normal in certain cell types lacking Ubc13 but not in others (22, 23) indicates that the ubiquitinating enzymes used by a given TLR pathway may differ between different cell types. These issues highlight the need for further exploration of the regulation of TLR signaling by Lys63-linked polyubiquitination, and this will no doubt reveal further complexities inherent in these pathways.
Supplementary Material
This work was supported by Science Foundation Ireland, Enterprise Ireland, and the Irish Health Research Board. This is the first of three articles in the Thematic Minireview Series on Regulation of Signaling by Non-degradative Ubiquitination. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: PRR, pattern recognition receptor; PAMP, pathogen-associated molecular pattern; IFN, interferon; TLR, Toll-like receptor; IL, interleukin; IL-1R, IL-1 receptor; TIR, Toll/IL-1 receptor; LPS, lipopolysaccharide; NF-κB, nuclear factor-κB; IRF, IFN regulatory factor; DUB, deubiquitinating enzyme; TNF, tumor necrosis factor; NEMO, NF-κB essential modifier; IKK, IκB kinase; MAPK, mitogen-activated protein kinase.
S. E. Keating and A. G. Bowie, unpublished data.
References
- 1.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783–801 [DOI] [PubMed] [Google Scholar]
- 2.Meylan, E., and Tschopp, J. (2006) Mol. Cell 22 561–569 [DOI] [PubMed] [Google Scholar]
- 3.Unterholzner, L., and Bowie, A. G. (2008) Biochem. Pharmacol. 75 589–602 [DOI] [PubMed] [Google Scholar]
- 4.Thompson, A. J., and Locarnini, S. A. (2007) Immunol. Cell Biol. 85 435–445 [DOI] [PubMed] [Google Scholar]
- 5.Haas, T., Metzger, J., Schmitz, F., Heit, A., Muller, T., Latz, E., and Wagner, H. (2008) Immunity 28 315–323 [DOI] [PubMed] [Google Scholar]
- 6.Chen, Z. J. (2005) Nat. Cell Biol. 7 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickart, C. M., and Fushman, D. (2004) Curr. Opin. Chem. Biol. 8 610–616 [DOI] [PubMed] [Google Scholar]
- 8.Sun, S.-C. (2008) Nat. Rev. Immunol. 8 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Neill, L. A., and Bowie, A. G. (2007) Nat. Rev. Immunol. 7 353–364 [DOI] [PubMed] [Google Scholar]
- 10.Chau, T. L., Gioia, R., Gatot, J. S., Patrascu, F., Carpentier, I., Chapelle, J. P., O'Neill, L., Beyaert, R., Piette, J., and Chariot, A. (2008) Trends Biochem. Sci 33 171–180 [DOI] [PubMed] [Google Scholar]
- 11.Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C., and Chen, Z. J. (2000) Cell 103 351–361 [DOI] [PubMed] [Google Scholar]
- 12.Wang, C., Deng, L., Hong, M., Akkaraju, G. R., Inoue, J., and Chen, Z. J. (2001) Nature 412 346–351 [DOI] [PubMed] [Google Scholar]
- 13.Kanayama, A., Seth, R. B., Sun, L., Ea, C. K., Hong, M., Shaito, A., Chiu, Y. H., Deng, L., and Chen, Z. J. (2004) Mol. Cell 15 535–548 [DOI] [PubMed] [Google Scholar]
- 14.Ninomiya-Tsuji, J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z., and Matsumoto, K. (1999) Nature 398 252–256 [DOI] [PubMed] [Google Scholar]
- 15.Tang, E. D., Wang, C. Y., Xiong, Y., and Guan, K. L. (2003) J. Biol. Chem. 278 37297–37305 [DOI] [PubMed] [Google Scholar]
- 16.Sun, L., Deng, L., Ea, C. K., Xia, Z. P., and Chen, Z. J. (2004) Mol. Cell 14 289–301 [DOI] [PubMed] [Google Scholar]
- 17.Zhou, H., Wertz, I., O'Rourke, K., Ultsch, M., Seshagiri, S., Eby, M., Xiao, W., and Dixit, V. M. (2004) Nature 427 167–171 [DOI] [PubMed] [Google Scholar]
- 18.Abbott, D. W., Wilkins, A., Asara, J. M., and Cantley, L. C. (2004) Curr. Biol. 14 2217–2227 [DOI] [PubMed] [Google Scholar]
- 19.Wu, C. J., Conze, D. B., Li, T., Srinivasula, S. M., and Ashwell, J. D. (2006) Nat. Cell Biol. 8 398–406 [DOI] [PubMed] [Google Scholar]
- 20.Abbott, D. W., Yang, Y., Hutti, J. E., Madhavarapu, S., Kelliher, M. A., and Cantley, L. C. (2007) Mol. Cell. Biol. 27 6012–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sebban-Benin, H., Pescatore, A., Fusco, F., Pascuale, V., Gautheron, J., Yamaoka, S., Moncla, A., Ursini, M. V., and Courtois, G. (2007) Hum. Mol. Genet. 16 2805–2815 [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto, M., Okamoto, T., Takeda, K., Sato, S., Sanjo, H., Uematsu, S., Saitoh, T., Yamamoto, N., Sakurai, H., Ishii, K. J., Yamaoka, S., Kawai, T., Matsuura, Y., Takeuchi, O., and Akira, S. (2006) Nat. Immunol. 7 962–970 [DOI] [PubMed] [Google Scholar]
- 23.Fukushima, T., Matsuzawa, S., Kress, C. L., Bruey, J. M., Krajewska, M., Lefebvre, S., Zapata, J. M., Ronai, Z., and Reed, J. C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 6371–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uematsu, S., Sato, S., Yamamoto, M., Hirotani, T., Kato, H., Takeshita, F., Matsuda, M., Coban, C., Ishii, K. J., Kawai, T., Takeuchi, O., and Akira, S. (2005) J. Exp. Med. 201 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, Y., Li, T., Sane, D. C., and Li, L. (2004) J. Biol. Chem. 279 51697–51703 [DOI] [PubMed] [Google Scholar]
- 26.Jiang, Z., Zamanian-Daryoush, M., Nie, H., Silva, A. M., Williams, B. R., and Li, X. (2003) J. Biol. Chem. 278 16713–16719 [DOI] [PubMed] [Google Scholar]
- 27.Song, Y. J., Jen, K. Y., Soni, V., Kieff, E., and Cahir-McFarland, E. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2689–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keating, S. E., Maloney, G. M., Moran, E. M., and Bowie, A. G. (2007) J. Biol. Chem. 282 33435–33443 [DOI] [PubMed] [Google Scholar]
- 29.Muzio, M., Ni, J., Feng, P., and Dixit, V. M. (1997) Science 278 1612–1615 [DOI] [PubMed] [Google Scholar]
- 30.Kawagoe, T., Sato, S., Matsushita, K., Kato, H., Matsui, K., Kumagai, Y., Saitoh, T., Kawai, T., Takeuchi, O., and Akira, S. (2008) Nat. Immunol. 9 684–691 [DOI] [PubMed] [Google Scholar]
- 31.Yamin, T. T., and Miller, D. K. (1997) J. Biol. Chem. 272 21540–21547 [DOI] [PubMed] [Google Scholar]
- 32.Ordureau, A., Smith, H., Windheim, M., Peggie, M., Carrick, E., Morrice, N., and Cohen, P. (2008) Biochem. J. 409 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Windheim, M., Stafford, M., Peggie, M., and Cohen, P. (2008) Mol. Cell. Biol. 28 1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conze, D. B., Wu, C. J., Thomas, J. A., Landstrom, A., and Ashwell, J. D. (2008) Mol. Cell. Biol. 28 3538–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler, M. P., Hanly, J. A., and Moynagh, P. N. (2007) J. Biol. Chem. 282 29729–29737 [DOI] [PubMed] [Google Scholar]
- 36.Schauvliege, R., Janssens, S., and Beyaert, R. (2006) FEBS Lett. 580 4697–4702 [DOI] [PubMed] [Google Scholar]
- 37.Xiao, H., Qian, W., Staschke, K., Qian, Y., Cui, G., Deng, L., Ehsani, M., Wang, X., Qian, Y.-W., Chen, Z. J., Gilmour, R., Jiang, Z., and Li, X. (2008) J. Biol. Chem. 283 14654–14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou, R., Silverman, N., Hong, M., Liao, D. S., Chung, Y., Chen, Z. J., and Maniatis, T. (2005) J. Biol. Chem. 280 34048–34055 [DOI] [PubMed] [Google Scholar]
- 39.Sweet, C. R., Conlon, J., Golenbock, D. T., Goguen, J., and Silverman, N. (2007) Cell. Microbiol. 9 2700–2715 [DOI] [PubMed] [Google Scholar]
- 40.Hacker, H., Redecke, V., Blagoev, B., Kratchmarova, I., Hsu, L. C., Wang, G. G., Kamps, M. P., Raz, E., Wagner, H., Hacker, G., Mann, M., and Karin, M. (2006) Nature 439 204–207 [DOI] [PubMed] [Google Scholar]
- 41.Oganesyan, G., Saha, S. K., Guo, B., He, J. Q., Shahangian, A., Zarnegar, B., Perry, A., and Cheng, G. (2006) Nature 439 208–211 [DOI] [PubMed] [Google Scholar]
- 42.Kayagaki, N., Phung, Q., Chan, S., Chaudhari, R., Quan, C., O'Rourke, K. M., Eby, M., Pietras, E., Cheng, G., Bazan, J. F., Zhang, Z., Arnott, D., and Dixit, V. M. (2007) Science 318 1628–1632 [DOI] [PubMed] [Google Scholar]
- 43.Gatot, J. S., Gioia, R., Chau, T. L., Patrascu, F., Warnier, M., Close, P., Chapelle, J. P., Muraille, E., Brown, K., Siebenlist, U., Piette, J., Dejardin, E., and Chariot, A. (2007) J. Biol. Chem. 282 31131–31146 [DOI] [PubMed] [Google Scholar]
- 44.Kawai, T., Sato, S., Ishii, K. J., Coban, C., Hemmi, H., Yamamoto, M., Terai, K., Matsuda, M., Inoue, J., Uematsu, S., Takeuchi, O., and Akira, S. (2004) Nat. Immunol. 5 1061–1068 [DOI] [PubMed] [Google Scholar]
- 45.Cusson-Hermance, N., Khurana, S., Lee, T. H., Fitzgerald, K. A., and Kelliher, M. A. (2005) J. Biol. Chem. 280 36560–36566 [DOI] [PubMed] [Google Scholar]
- 46.Meylan, E., Burns, K., Hofmann, K., Blancheteau, V., Martinon, F., Kelliher, M., and Tschopp, J. (2004) Nat. Immunol. 5 503–507 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.