Abstract
The small ubiquitin-like modifier (SUMO) is a ubiquitin-like protein that covalently modifies a large number of cellular proteins. SUMO modification has emerged as an important regulatory mechanism for protein function and localization. SUMOylation is a dynamic process that is mediated by activating (E1), conjugating (E2), and ligating (E3) enzymes and readily reversed by a family of ubiquitin-like protein-specific proteases (Ulp) in yeast and sentrin/SUMO-specific proteases (SENP) in human. This review will focus on the de-SUMOylating enzymes with special attention to their biological function.
Many biochemical pathways are reversible to create an on and off state that is essential for biological regulation. A reversible system allows for quick termination of a biological response that has to be precisely controlled. The SUMO2 modification pathway is an example of a reversible system that is controlled by a series of on and off enzymes (Fig. 1) (1). In contrast to the much more complex ubiquitin pathway (2), SUMOylation utilizes only a single conjugating enzyme, Ubc9 (3), and a limited number of ligases (4–6). This simplicity also manifests in the off step because there are only two SUMO-deconjugating enzymes in yeast and six in human. One may assume that these limited numbers of on and off enzymes would sufficient to regulate only be a small number of substrates and biological pathways. However, the number of SUMO substrates continues to expand, and the varieties of systems that are known to be regulated by SUMO also proliferate quickly. Here, I will review only the enzymes that are involved in the de-SUMOylation pathways to provide insights into how these limited numbers of proteases are able to regulate a diverse array of biological responses.
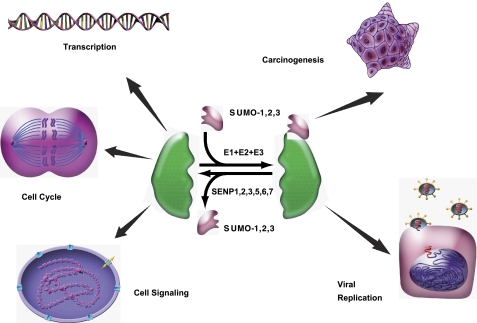
FIGURE 1.
SUMOylation and de-SUMOylation. SUMOylation is a dynamic process that is mediated by activating (E1), conjugating (E2), and ligating (E3) enzymes and readily reversed by the SENP family in human. SUMOylation and de-SUMOylation regulate a diverse spectrum of biological responses, from transcription, cell division, and signal transduction to carcinogenesis and viral replication.
Localization and Enzymatic Activity of SUMO-specific Proteases
SUMO-specific proteases are C48 cysteine proteases that possess a conserved catalytic domain characterized by the catalytic triad (histidine, aspartate, and cysteine) and a conserved glutamine residue required for the formation of the oxyanion hole in the active site (7). Members of the C48 cysteine protease family have N- and C-terminal sequences that differ from each other. Homologs of these proteases are present in plant, yeast, and mammalian cells. In this review, I will focus on the two yeast ubiquitin-like protein-specific proteases (Ulp) and the six human sentrin/SUMO-specific proteases (SENP) (Table 1).
TABLE 1.
Human SENPs
| Human SENPs (1) | Other names | Primary location | Specificity | Enzymatic activity |
|---|---|---|---|---|
| SENP1 | SuPr-2 | Nucleoplasm (11) | SUMO1/2/3 (11) | C-terminal hydrolase, isopeptidase (11) |
| SENP2 | SuPr-1, AXAM2, SMT3IP2 | Nuclear pore, nuclear speckle (16-18) | SUMO1/2/3 (16, 17) | C-terminal hydrolase, isopeptidase (16, 17) |
| SENP3 | SSP3, SMT3IP1 | Nucleolus (19-21) | SUMO2/3 (20, 21) | Isopeptidase (19-21) |
| SENP5 | Nucleolus (20, 21) | SUMO2/3 (20, 21) | Isopeptidase (20, 21) | |
| SENP6 | SUSP1, SSP1 | Nucleoplasm (18, 23) | SUMO2/3 (23) | Chain editing (23) |
| SENP7 | Nucleoplasm (18) | ?SUMO2/3 | ?Chain editing |
Yeast has a single SUMO-like modifier, Smt3, and two Smt3-specific proteases, Ulp1 and Ulp2. Both Smt3 and Ulp2 (Smt4) were identified from the same screen as suppressors of the Mif2 (a centromeric protein) mutation (8). Ulp1 is a protein of 621 amino acids that contains the catalytic domain at the C terminus and an N-terminal domain that attaches this protease to the nuclear pore (7). Ulp1 possesses the C-terminal hydrolase activity required for removing C-terminal amino acids from Smt3 to reveal the diglycine residues important for conjugation to Smt3 substrates. Ulp1 also has the isopeptidase activity that is essential for removing Smt3 from conjugated substrates. Ulp2 (Smt4) is a 1034-amino acid protease that possesses only isopeptidase activity (9). It is localized in the nucleoplasm. Yeast deficient in Ulp2 accumulates Smt3 polymers, suggesting that Ulp2 is also involved in the processing of the Smt3 chains (10).
The SENPs can be divided into three families. The first family consists of SENP1 and SENP2, which have broad specificity for the three mammalian SUMOs (SUMO1–3). The second family includes SENP3 and SENP5, which favor SUMO2/3 as substrates and are localized in the nucleolus. The third family contains SENP6 and SENP7, which have an additional loop inserted in the catalytic domain and also appear to prefer SUMO2/3. From an evolutionary standpoint, SENP1–3 and SENP5 are more closely related to Ulp1, whereas SENP6 and SENP7 are related to Ulp2.
SENP1 is localized in the nucleoplasm but not in the nucleolus (11). It contains a nuclear localization signal in the N terminus (12) and a nuclear export sequence near the C terminus (13). In the SENP1–/– embryo, the SUMO1 precursor cannot be processed, suggesting that SENP1 is the main SUMO1 C-terminal hydrolase (14). However, SENP1 is an efficient isopeptidase for all SUMOs. SENP2 also possesses nuclear localization and export signals (15). When exported from the nucleus, it is quickly ubiquitinated and degraded. SENP2 was reported to be tethered to the nuclear pore through binding to Nup153 nucleoporin (16, 17). Furthermore, SENP2 is also localized in a yet undefined nuclear speckle that is distinct from the nuclear body (18). SENP2 has isopeptidase activity for all SUMOs. SENP3 and SENP5 are both localized predominately in the nucleolus (19–21). The nucleolar localization signals are positioned in their N termini. Both SENP3 and SENP5 show preference for SUMO2/3. SENP6 has a distinct split of its catalytic domain by an insertion (22). It was originally shown to be localized to the cytosol in NIH3T3 and HeLa cells (22). However, studies from other laboratories suggest that SENP6 is localized in the nucleoplasm (18, 23). SENP6 also appears to prefer SUMO2/3 as substrates (23). SENP7 is the least characterized SENP that is localized in the nucleoplasm (18). However, its catalytic activity has not been convincingly demonstrated.
Structural Studies on Ulp1, SENP1, and SENP2
Analysis of the crystal structure containing Ulp1-(403–621) and Smt3-(13–98) confirms that Ulp1 belongs to the cysteine protease superfamily (24). The secondary structure of Ulp1 possesses seven α-helices and seven β-strands and shares similarity with other cysteine proteases in the active site that includes the central α-helix and three β-strands and the catalytic triad. The Ulp1 and Smt3 interface is extensive and includes the exposed β-sheet of Smt3 and an entire face of the protease. The Gly-Gly-X motif of Smt3 apparently passes through a hydrophobic tunnel above the active site during cleavage. The Ulp1 active site resembles other cysteine protease active sites in that the nucleophilic Cys-580 is coordinated by His-514 and stabilized by Asp-531 and the oxyanion hole is created by Gln-574 and Cys-580.
The crystal structure of SENP2-(364–589) shows that its active site comprises Cys-548, His-478, and Asp-495, resembling other cysteine proteases (25). Furthermore, the crystal of SENP2 and SUMO1-(18–97) reveals that SENP2 undergoes local structural rearrangements after binding to SUMO1. His-478 undergoes a 180° rotation in the SENP2-SUMO1 complex such that the imidazole ring now points directly toward Asp-495 and away from Cys-478. In addition, Trp-410, Trp-479, and His-474 all rotate about their C-β atoms to adapt to interactions with the SUMO1 Gly-Gly motif. The SUMO1/SENP2 interface is similar to that of Smt3/Ulp1. Comparative proteolysis assays shows that SENP2 hydrolyzes SUMO2 better than SUMO1 or SUMO3 precursors. This C-terminal hydrolase activity is apparently dependent on the respective C-terminal tails of these SUMOs. However, SENP2 efficiently hydrolyzes SUMO1, SUMO2, or SUMO3 from RanGAP1. Furthermore, the isopeptidase activity of SENP2 is ∼20-fold stronger than its C-terminal hydrolase activity, suggesting that SENP2 interacts more readily with SUMO conjugates than with SUMO precursors.
Further insight into the structural basis of isopeptidase activity comes from the crystal of catalytically inactive SENP1-(415–644) in complex with a SUMO1-modified fragment of RanGAP1-(418–587) (26). It shows that there is a minimally specific and biologically relevant recognition of RanGAP1 by SENP1. The binding between the conserved C-terminal strands of SUMO1 and the main chain of SENP1 provides the major interaction surface. Furthermore, the side chain of Lys-524 forms a right angle between RanGAP1 and SUMO1. When the SUMO1 precursor bound to SENP1 is analyzed, the scissile peptide bond has the cis-arrangement of the amide nitrogen atoms similar to the isopeptide bond in SUMO1-conjugated RanGAP1. Based on these studies, it was proposed that an initial association between SUMO conjugates or SUMO precursors and SENP1 stimulates the opening of the tryptophan tunnel. Closing of the tunnel causes trans,cis-isomerization of the amide nitrogens of the scissile bond of the substrate. Chemical catalysis then proceeds.
Structural analysis of catalytically inactive SENP2 in complex with RanGAP1-SUMO1, RanGAP1-SUMO2, or SUMO precursors also reveals a similar 90° kink proximal to the scissile bond that forces C-terminal amino acid residues or the lysine side chain toward the SENP2 surface (27). This geometry appears to be unfavorable for processing of SUMO precursors. The structural analysis described above is based on the interaction between the catalytic domain of either SENP1 or SENP2 and SUMO-RanGAP1 or SUMO precursors. The structural studies provide insights regarding substrate binding and the potential catalytic mechanism. However, these studies do not account for the contribution of the N termini of the SENPs to substrate specificity.
Function of Smt3-specific Proteases
Ulp1—Ulp1 performs a dual function as both the C-terminal hydrolase and isopeptidase for Smt3 (7). The isopeptidase activity of Ulp1 is essential for growth of yeast cells, particularly at the G2/M transition. The catalytic domain of Ulp1 is sufficient to rescue the growth defect of Ulp1 deletion in yeast (28). In Schizosaccharomyces pombe, Ulp1 is not essential for cell viability, but cells lacking Ulp1 display severe nuclear abnormality and are more sensitive to ultraviolet radiation (29). This nuclear defect is dependent on the isopeptidase activity because it is not rescued by the mature form of Pmt3 (homolog of Smt3). Furthermore, Ulp1 exhibits a cell cycle-specific pattern of localization. During the S and G2 phases, Ulp1 is localized at the nuclear periphery. However, during mitosis, Ulp1 is localized in the nucleus. The bulk of cellular Ulp1 is not associated with nucleoporins but with three karyopherins (Pse1, Kap95, and Kap60) (30). Expression of the catalytic domain of Ulp1 in the nucleus reduces cell viability; however, the mechanism of this dominant lethality is unknown. In addition, two myosin-like proteins, Mlp1 and Mlp2, which form filaments attached to the nucleoplasmic side of the nuclear pore complex, are required to localize Ulp1 to the nuclear envelope (31). Deletion of the Mlp proteins results in DNA damage sensitivity and clonal lethality. In a recent study, Esc1, a nuclear envelope protein not associated with the nuclear pore complex, was shown to be required for proper assembly of the nuclear basket and for normal nuclear pore complex localization of Ulp1 (32). Both Esc1 and Ulp1 help to retain unspliced pre-mRNAs in the nucleus.
Ulp2—Ulp2-null cells exhibit temperature-sensitive growth, chromosome instability, and hypersensitivity to DNA-damaging agents (9). Ulp2 was shown to control chromosome cohesion at centromeric regions and through modulation of the function of DNA topoisomerase II (Top2) (33). Top2 mutants resistant to Smt3 modification suppress the Ulp2 cohesion defect, suggesting that Top2 is a major substrate regulating a component of chromatin structure required for centromeric cohesion. More recently, it was shown that Ulp2 is required for resumption of cell division following DNA damage-induced arrest but is not required for DNA double-strand break repair (34).
Function of SUMO-specific Proteases
SENP1—The initial report of SENP1 showed that it is active against both SUMO-modified RanGAP1 and PML in vitro (11). However, SENP1 deconjugates only SUMO-PML but not SUMO-RanGAP1 in vivo. The differential effect of SENP1 on these two substrates in vivo was attributed to the nuclear localization of SENP1. Thus, SENP1 is not able to regulate SUMOylated RanGAP1, which is localized to the cytoplasmic fibrils of the nuclear pore complex. This is probably an oversimplification because SENP1 can potentially shuttle between the nucleus and cytosol through a nuclear localization signal and nuclear export sequence (12, 13).
Because the androgen receptor and its co-regulators are SUMOylated (35, 36), the androgen/AR system is an ideal model to study the role of SENPs in the regulation of a complex biological pathway. In a functional screen, we found that SENP1 is the most potent regulator of androgen receptor-dependent transcription (37). SENP1 deconjugates SUMOylated HDAC1 (histone deacetylase 1) to reduce its deacetylase activity, allowing transcription to increase manyfold over background levels. This is biologically relevant because SENP1 is highly expressed in an early prostate cancer lesion called prostatic intraepithelial neoplasia and prostate cancer tissues but not in normal prostate tissues (18). Furthermore, the androgen receptor directly regulates SENP1 transcription by binding to an androgen-response element in the SENP1 promoter (38). To further define the role of SENP1 in the development of prostate cancer, we generated several SENP1 transgenic mouse lines that were driven by an androgen receptor-dependent probasin promoter (18). Preliminary studies showed that androgen receptor-driven overexpression of SENP1 in the prostate gland indeed leads to development of a prostatic intraepithelial neoplasia-like lesion at 4 months, which progressively increases in severity. Thus, SENP1 is the first SENP shown to play a role in disease pathogenesis in human.
In addition to regulating HDAC1, SENP1 also regulates SIRT1, a class III histone deacetylase. Similar to HDAC1, SUMOylation of SIRT1 increases its deacetylase activity, and SENP1 reduces the deacetylase activity of SIRT1 (39). Furthermore, stress-inducing agents, such as UV radiation and hydrogen peroxide, promote the association of SIRT1 with SENP1. Knocking down SENP1 by siRNA reduces stress-induced apoptosis. Thus, SENP1 plays a critical role in stress-induced apoptosis through de-SUMOylation of SIRT1, which regulates members of the p53 family. In another study, it was shown that SENP1 can be trapped in the cytosol by thioredoxin and that TNF induces the release of SENP1 from thioredoxin through a reactive oxygen species-dependent mechanism (40). TNF-induced nuclear transport of SENP1 correlates with de-SUMOylation of HIPK1 (homeodomain-interacting protein kinase 1) and cytoplasmic translocation of HIPK1, leading to an increase in ASK-1 (apoptosis signal-regulating kinase 1)-dependent apoptosis. Thus, it is clear that SENP1 can enhance apoptosis through multiple mechanisms.
SENP1 also regulates c-Jun transcription by de-SUMOylating CRD1 (cell cycle regulator domain 1) of p300, thereby releasing the cis-repression of CRD1 on p300 (41). This activity may also add to the pro-carcinogenic profile of SENP1. Many of the functional studies reported above were based on overexpression of SENP1 in cell lines or knocking down SENP1 by siRNA. An important tool to reveal the true biology of SENP1 is through the gene deletion approach in mice. A random retroviral insertion within the first intron the mouse SENP1 gene leads to a marked reduction in the expression of SENP1 and embryonic lethality at embryonic day 13.5 (42). It was shown that this mutation causes an increase in SUMO1 conjugates but not SUMO2/3 conjugates. The embryonic lethality was attributed to placental abnormalities that are incompatible with embryonic development.
My laboratory has generated SENP1 knock-out embryos using a gene trapping strategy. We showed that inactivation of the SENP1 gene causes embryonic lethality in mid-gestation in mice as a result of severe fetal anemia stemming from deficient Epo production (14). SENP1 controls Epo production by regulating the stability of HIF1α (hypoxia-inducible factor 1α) under hypoxic conditions. During normoxia, HIF1α is hydroxylated at two critical proline residues by a family of oxygen-sensing enzymes called PHD/EGLN (prolyl hydrolase domain/egg laying-defective nine). Proline hydroxylation is important for HIF1α to binds to its ubiquitin ligase complex, the VHL-elongin B/C complex, leading to ubiquitination and proteasomal degradation. It was assumed that under hypoxic conditions, HIF1α is inefficiently proline-hydroxylated and thus failed to bind to the VHL E3 complex and to become stabilized. We showed that hypoxia induces rapid SUMOylation of HIF1α, which allows it to bind to the VHL protein in a hydroxyproline-independent manner, also leading to ubiquitination and proteasomal degradation. SENP1 reverses SUMOylation of HIF1α, reduces binding to the VHL protein, and consequently stabilizes HIF1α. These results reveal that SUMO can also serve as a signal for ubiquitin-mediated protein degradation. Thus, SENP1 plays a critical role in regulating HIF1α stability during hypoxia. Because HIF1α regulates multiple, critical downstream genes, such as Epo, vascular endothelial growth factor, and GLUT-1, it has the potential to regulate erythropoiesis, angiogenesis, glycolysis, and the entire hypoxic response.
SENP2—SuPr-1 is an N-terminally truncated form of murine SENP2 that has been shown to localize to the PML-containing nuclear body (43). SuPr-1 induces c-Jun-dependent transcription that does not require its isopeptidase activity; however, mutations that affects SuPr-1 binding to PML impair transcriptional activity. Thus, this particular transcriptional regulation is not related to de-SUMOylation.
Another group reported an Axin-binding protein called Axam, which is identical to SuPr-1 (44). The Axin-binding domain is located in the central region of Axam, distant from the catalytic domain. Axam was shown to decrease the level of β-catenin dependent in its catalytic activity. Axam strongly inhibits axis formation and the Wnt signal in Xenopus embryos. Thus, de-SUMOylation through Axam plays a role in regulation of the Wnt signaling pathway.
We have generated SENP2–/– embryos through a gene trapping technique (14). SENP2–/– embryos died at embryonic day 9.5, much earlier than SENP1–/– embryos. Although not fully characterized, SENP2 knock-out mice provide several important insights. First, SENP1 and SENP2 perform non-redundant functions in cells. Thus, they are not able to compensate for each other in specific knock-out embryos. Comparing the SENP1–/– and SENP2–/– mouse embryonic fibroblast cells, we observed that hypoxia-regulated HIF1α stability was affected only in the SENP1–/– mouse embryonic fibroblast cells. Thus, SENP1 and SENP2 clearly have different substrate specificity. This is important because most of the in vitro overexpression systems will not be able to distinguish the activity between these two closely related SENPs.
SENP3—The nucleolar localization of SENP3 suggests that it may regulate certain aspects of nucleolar function (19–21). In addition, SENP3 prefers SUMO2/3 as substrates (20, 21). These two observations have narrowed down the number of SENP3 substrates. Indeed, SENP3 has recently been shown to associate with nucleophosmin NPM1, a crucial factor in ribosome biogenesis (45). SENP3 deconjugates NPM1-SUMO2 conjugates in vitro and counteracts ADP-ribosylation factor-induced modification of NPM1 by SUMO2 in vivo. Depletion of SENP3 by siRNA interferes with nucleolar rRNA processing and inhibits the conversion of the 32 S rRNA species to the 28 S form, a phenotype similar to knockdown of NPM1. These results define SENP3 as an essential factor for ribosome biogenesis.
SENP5—Knockdown of SENP5 by siRNA inhibits cell proliferation, exhibiting defects in nuclear morphology with appearance of binucleate cells. These findings suggest that SENP5 may play a role in mitosis and/or cytokinesis (21). Another potential target for SENP5 is the mitochondrial fission GTPase DRP1 (46). Overexpression of SENP5 deconjugates SUMO1 from a number of mitochondrial substrates and rescues SUMO1-induced mitochondrial fragmentation. Knocking down SENP5 by siRNA leads to development of mitochondrial abnormality. The reduction of SENP5 levels also results in an increase in free radicals. Thus, SENP5 may play a role in the regulation of mitochondrial morphology and metabolism. It is unclear whether mitochondria are a true target for SENP5 because SENP5 is mainly a nucleolar protease and prefers SUMO2/3 as substrates.
SENP6—There is a discrepancy in terms of SENP6 localization in the literature. The initial study showed that SENP6 is in the cytosol (22), but our laboratory and others have clearly demonstrated that SENP6 is in the nucleus (18, 23). Silencing SENP6 causes redistribution of enhanced green fluorescent protein-SUMO2 and -SUMO3 into PML bodies (23). It is not clear whether SENP6 is involved in the regulation of PML bodies in vivo.
Perspectives
Here, I would like to return to the issues raised at the opening of this review. How can a limited number of SENPs regulate such a large universe of SUMOylated substrates in mammalian cells?
Each SENP contains an N-terminal sequence that is involved in cellular localization. There are also nuclear localization signals and nuclear export signals in different regions of the SENPs, so the SENPs should not be viewed as stationary proteases that are limited to a single cellular localization. In fact, the SENPs can be dynamically regulated by their import and export signals through environmental influences. A good example is the regulation of cytosolic and nuclear localization of SENP1 by TNF (40).
Another way to regulate the SENPs is through transcriptional control and post-translational regulation. We have shown that SENP1 transcription is induced by androgen and interleukin-6 (18, 38). Similar results were also reported for interleukin-6 induction of SENP1, which leads to de-SUMOylation of PML (47). We also have evidence that SENP1 is regulated by ubiquitin-mediated degradation and has a short half-life. This can be regulated by androgen and hypoxia. Thus, the level of each SENP can be regulated by environmental factors.
The third way is to regulate the binding of SENPs to their substrates. An example is that stress-inducing agents, such as UV radiation and hydrogen peroxide, promote the binding of SENP1 to SIRT1 (39). Another example is KLF5 (Kruppel-like transcription f–actor 5), a crucial regulator of energy metabolism. Under basal conditions, KLF5 is modified by SUMO and is associated with transcriptionally repressive regulatory complexes (48). Upon agonist stimulation of peroxisome proliferator-activated receptor δ, KLF5 is de-SUMOylated by SENP1 and becomes associated with transcriptional activation complexes.
The fourth way is to regulate the activity of the SENPs. It has been shown that at low concentrations, reactive oxygen species result in the rapid disappearance of many SUMO conjugates, including key transcription factors. This is due to direct and reversible inhibition of SUMO-conjugating enzymes through the formation of disulfide bonds involving the catalytic cysteines of the SUMO E1 subunit Uba2 and the E2-conjugating enzyme Ubc9 (49). Similarly, H2O2 has been shown to induce formation of an intermolecular disulfide linkage of SENP1 via the active-site Cys-603 and a unique Cys-613 (50). This reversible modification is also observed in Ulp1 but not in SENP2.
Through a combination of these mechanisms, SENPs should be able to regulate a large number of biological systems in a precise manner. In the coming years, conditional knock-out of individual SENP genes will reveal interesting biology that will further broaden our knowledge of this emerging field.
Supplementary Material
Acknowledgments
I thank H. M. Chang, J. Cheng, T. Bawa, S. Zhang, X. Kang, T. Kamitani, and L. Gong for contributions to works cited in this review.
This is the third of three articles in the Thematic Minireview Series on Regulation of Signaling by Non-degradative Ubiquitination. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
Footnotes
The abbreviations used are: SUMO, small ubiquitin-like modifier; SENP, sentrin/SUMO-specific protease; siRNA, small interfering RNA; TNF, tumor necrosis factor; Epo, erythropoietin; VHL, von Hippel-Lindau.
References
- 1.Yeh, E. T., Gong, L., and Kamitani, T. (2000) Gene (Amst.) 248 1–14 [DOI] [PubMed] [Google Scholar]
- 2.Hershko, A., Ciechanover, A., and Varshavsky, A. (2000) Nat. Med. 6 1073–1081 [DOI] [PubMed] [Google Scholar]
- 3.Gong, L., Kamitani, T., Fujise, K., Caskey, L. S., and Yeh, E. T. (1997) J. Biol. Chem. 272 28198–28201 [DOI] [PubMed] [Google Scholar]
- 4.Johnson, E. S., and Gupta, A. A. (2001) Cell 106 735–744 [DOI] [PubMed] [Google Scholar]
- 5.Pichler, A., Gast, A., Seeler, J. S., Dejean, A., and Melchior, F. (2002) Cell 108 109–120 [DOI] [PubMed] [Google Scholar]
- 6.Kahyo, T., Nishida, T., and Yasuda, H. (2001) Mol. Cell 8 713–718 [DOI] [PubMed] [Google Scholar]
- 7.Li, S. J., and Hochstrasser, M. (1999) Nature 398 246–251 [DOI] [PubMed] [Google Scholar]
- 8.Meluh, P. B., and Koshland, D. (1995) Mol. Biol. Cell 6 793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, S. J., and Hochstrasser, M. (2000) Mol. Cell. Biol. 20 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bylebyl, G. R., Belichenko, I., and Johnson, E. S. (2003) J. Biol. Chem. 278 44113–44120 [DOI] [PubMed] [Google Scholar]
- 11.Gong, L., Millas, S., Maul, G. G., and Yeh, E. T. (2000) J. Biol. Chem. 275 3355–3359 [DOI] [PubMed] [Google Scholar]
- 12.Bailey, D., and O'Hare, P. (2004) J. Biol. Chem. 279 692–703 [DOI] [PubMed] [Google Scholar]
- 13.Kim, Y. H., Sung, K. S., Lee, S. J., Kim, Y. O., Choi, C. Y., and Kim, Y. (2005) FEBS Lett. 579 6272–6278 [DOI] [PubMed] [Google Scholar]
- 14.Cheng, J., Kang, X., Zhang, S., and Yeh, E. T. (2007) Cell 131 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itahana, Y., Yeh, E. T., and Zhang, Y. (2006) Mol. Cell. Biol. 26 4675–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hang, J., and Dasso, M. (2002) J. Biol. Chem. 277 19961–19966 [DOI] [PubMed] [Google Scholar]
- 17.Zhang, H., Saitoh, H., and Matunis, M. J. (2002) Mol. Cell. Biol. 22 6498–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng, J., Bawa, T., Lee, P., Gong, L., and Yeh, E. T. (2006) Neoplasia (N. Y.) 8 667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida, T., Tanaka, H., and Yasuda, H. (2000) Eur. J. Biochem. 267 6423–6427 [DOI] [PubMed] [Google Scholar]
- 20.Gong, L., and Yeh, E. T. (2006) J. Biol. Chem. 281 15869–15877 [DOI] [PubMed] [Google Scholar]
- 21.Di Bacco, A., Ouyang, J., Lee, H. Y., Catic, A., Ploegh, H., and Gill, G. (2006) Mol. Cell. Biol. 26 4489–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, K. I., Baek, S. H., Jeon, Y. J., Nishimori, S., Suzuki, T., Uchida, S., Shimbara, N., Saitoh, H., Tanaka, K., and Chung, C. H. (2000) J. Biol. Chem. 275 14102–14106 [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay, D., Ayaydin, F., Kolli, N., Tan, S. H., Anan, T., Kametaka, A., Azuma, Y., Wilkinson, K. D., and Dasso, M. (2006) J. Cell Biol. 174 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mossessova, E., and Lima, C. D. (2000) Mol. Cell 5 865–876 [DOI] [PubMed] [Google Scholar]
- 25.Reverter, D., and Lima, C. D. (2004) Structure (Camb.) 12 1519–1531 [DOI] [PubMed] [Google Scholar]
- 26.Shen, L., Tatham, M. H., Dong, C., Zagorska, A., Naismith, J. H., and Hay, R. T. (2006) Nat. Struct. Mol. Biol. 13 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reverter, D., and Lima, C. D. (2006) Nat. Struct. Mol. Biol. 13 1060–1068 [DOI] [PubMed] [Google Scholar]
- 28.Li, S. J., and Hochstrasser, M. (2003) J. Cell Biol. 160 1069–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor, D. L., Ho, J. C., Oliver, A., and Watts, F. Z. (2002) J. Cell Sci. 115 1113–1122 [DOI] [PubMed] [Google Scholar]
- 30.Panse, V. G., Kuster, B., Gerstberger, T., and Hurt, E. (2003) Nat. Cell Biol. 5 21–27 [DOI] [PubMed] [Google Scholar]
- 31.Zhao, X., Wu, C. Y., and Blobel, G. (2004) J. Cell Biol. 167 605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis, A., Felberbaum, R., and Hochstrasser, M. (2007) J. Cell Biol. 178 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachant, J., Alcasabas, A., Blat, Y., Kleckner, N., and Elledge, S. J. (2002) Mol. Cell 9 1169–1182 [DOI] [PubMed] [Google Scholar]
- 34.Schwartz, D. C., Felberbaum, R., and Hochstrasser, M. (2007) Mol. Cell. Biol. 27 6948–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poukka, H., Karvonen, U., Janne, O. A., and Palvimo, J. J. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 14145–14150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinlein, C. A., and Chang, C. (2002) Endocr. Rev. 23 175–200 [DOI] [PubMed] [Google Scholar]
- 37.Cheng, J., Wang, D., Wang, Z., and Yeh, E. T. (2004) Mol. Cell. Biol. 24 6021–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bawa-Khalfe, T., Cheng, J., Wang, Z., and Yeh, E. T. (2007) J. Biol. Chem. 282 37341–37349 [DOI] [PubMed] [Google Scholar]
- 39.Yang, Y., Fu, W., Chen, J., Olashaw, N., Zhang, X., Nicosia, S. V., Bhalla, K., and Bai, W. (2007) Nat. Cell Biol. 9 1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, X., Luo, Y., Yu, L., Lin, Y., Luo, D., Zhang, H., He, Y., Kim, Y. O., Kim, Y., Tang, S., and Min, W. (2008) Cell Death Differ. 15 739–750 [DOI] [PubMed] [Google Scholar]
- 41.Cheng, J., Perkins, N. D., and Yeh, E. T. (2005) J. Biol. Chem. 280 14492–14498 [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi, T., Sharma, P., Athanasiou, M., Kumar, A., Yamada, S., and Kuehn, M. R. (2005) Mol. Cell. Biol. 25 5171–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Best, J. L., Ganiatsas, S., Agarwal, S., Changou, A., Salomoni, P., Shirihai, O., Meluh, P. B., Pandolfi, P. P., and Zon, L. I. (2002) Mol. Cell 10 843–855 [DOI] [PubMed] [Google Scholar]
- 44.Kadoya, T., Yamamoto, H., Suzuki, T., Yukita, A., Fukui, A., Michiue, T., Asahara, T., Tanaka, K., Asashima, M., and Kikuchi, A. (2002) Mol. Cell. Biol. 22 3803–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haindl, M., Harasim, T., Eick, D., and Muller, S. (2008) EMBO Rep. 9 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zunino, R., Schauss, A., Rippstein, P., Andrade-Navarro, M., and McBride, H. M. (2007) J. Cell Sci. 120 1178–1188 [DOI] [PubMed] [Google Scholar]
- 47.Ohbayashi, N., Kawakami, S., Muromoto, R., Togi, S., Ikeda, O., Kamitani, S., Sekine, Y., Honjoh, T., and Matsuda, T. (2008) Biochem. Biophys. Res. Commun. 371 823–828 [DOI] [PubMed] [Google Scholar]
- 48.Oishi, Y., Manabe, I., Tobe, K., Ohsugi, M., Kubota, T., Fujiu, K., Maemura, K., Kubota, N., Kadowaki, T., and Nagai, R. (2008) Nat. Med. 14 656–666 [DOI] [PubMed] [Google Scholar]
- 49.Bossis, G., and Melchior, F. (2006) Mol. Cell 21 349–357 [DOI] [PubMed] [Google Scholar]
- 50.Xu, Z., Lam, L. S., Lam, L. H., Chau, S. F., Ng, T. B., and Au, S. W. (2008) FASEB J. 22 127–137 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.