Abstract
Ribonucleotide reductase (RNR) catalyzes the rate-limiting step in deoxyribonucleotide synthesis essential for DNA replication and repair. RNR in S phase mammalian cells comprises a weak cytosolic complex of the catalytic R1 protein containing redox active cysteine residues and the R2 protein harboring the tyrosine free radical. Each enzyme turnover generates a disulfide in the active site of R1, which is reduced by C-terminally located shuttle dithiols leaving a disulfide to be reduced. Electrons for reduction come ultimately from NADPH via thioredoxin reductase and thioredoxin (Trx) or glutathione reductase, glutathione, and glutaredoxin (Grx), but the mechanism has not been clarified for mammalian RNR. Using recombinant mouse RNR, we found that Trx1 and Grx1 had similar catalytic efficiency (kcat/Km). With 4 mm GSH, Grx1 showed a higher affinity (apparent Km value, 0.18 μm) compared with Trx1 which displayed a higher apparent kcat, suggesting its major role in S phase DNA replication. Surprisingly, Grx activity was strongly dependent on GSH concentrations (apparent Km value, 3 mm) and a Grx2 C40S mutant was active despite only one cysteine residue in the active site. This demonstrates a GSH-mixed disulfide mechanism for glutaredoxin catalysis in contrast to the dithiol mechanism for thioredoxin. This may be an advantage with the low levels of RNR for DNA repair or in tumor cells with high RNR and no or low Trx expression. Our results demonstrate mechanistic differences between the mammalian and canonical Escherichia coli RNR enzymes, which may offer an explanation for the nonconserved shuttle dithiol sequences in the C terminus of the R1.
Ribonucleotide reductase (RNR),3 the essential enzyme generating the four deoxyribonucleoside triphosphates (dNTPs) required for DNA synthesis (1), plays a critical role in the high fidelity DNA replication and repair. The class Ia enzyme, present in all eukaryotes (from yeast to mammals), some bacteria, and some viruses, is composed of two subunits: R1 and R2 (2). The R2 protein (β) is a homodimer (2 × 45 kDa), and each polypeptide harbors a tyrosyl free radical that is generated and stabilized by an iron center (1, 2).
The R1 protein (α) contains the substrate-binding site, the active site including three cysteine residues, two redox active shuttle cysteines, and allosteric sites. The allosteric sites control substrate specificity and enzyme activity via binding of nucleotide triphosphates (3). In the absence of nucleotide effectors, the mammalian R1 is a 90-kDa monomer. Binding of effectors to the specificity site induces the formation of R1 dimers that interact with the R2 dimer, forming an active α2β2 complex. However, the R1 hexamer induced by ATP/dATP (4) is suggested to form the major complex of RNR with the R2 dimer (5).
In enzyme turnover, a disulfide is formed in the active site, from the catalytically active SH group of the cysteines; consequently the 2′-OH of the ribonucleoside, is replaced with a hydrogen atom (6). As shown in Escherichia coli RNR, which is the best characterized class I enzyme, the narrow active site cleft in R1 excludes direct reduction of the active site disulfide by external dithiol-dependent redoxins, but the mobile tail of R1 transfers reducing equivalents from C-terminally located cysteine residues (shuttle) to the active site by a thiol-disulfide exchange reaction (7). The resulting C-terminal disulfide has to be reduced by an external thiol-dependent reductase system (8).
Both thioredoxin (Trx) (9) and glutaredoxin (Grx) (10) were discovered as dithiol electron donors of E. coli ribonucleotide reductase and are today well known multifunctional thiol-dependent redoxins in cells with catalytically active cysteine thiols in a CXXC active site (11, 12). The oxidized Trx is reduced by NADPH via the flavoprotein thioredoxin reductase (TrxR), which is a selenoenzyme in mammalian cells. In contrast, the disulfide in oxidized Grx is reduced by two molecules of the monothiol GSH. The resulted GSSG in turn is reduced by NADPH and glutathione reductase (11, 12). In E. coli the class Ia enzyme, NrdAB, which is essential for aerobic growth, requires the dithiol form of at least one of Grx1, Trx1, or Trx2 to be viable (13–15). A 10-fold lower Km value of Grx1 (0.13 μm) than that for Trx1, with similar Vmax of both dithiol redoxins, makes Grx1 the most efficient electron donor for the E. coli enzyme.
Despite intense research on mammalian RNR (4, 16, 17), virtually nothing is known about the electron donor systems, and in enzyme assays, 10 mm DTT has been used as a strong artificial dithiol reductant. The cells have to critically balance dNTP pools and avoid excessive or too low concentrations of dNTP because this will increase the error rates of DNA polymerases (18). In E. coli, a balanced supply of deoxyribonucleotides is obtained by a regulatory mechanism that up-regulates the level of RNR in response to the lack of any of its two main hydrogen donors, Trx1 or GSH plus Grx1 (13). It is not known whether the electron donor system is rate-limiting for mammalian RNR activity. However, the requirements for DNA replication consuming large amounts of dNTPs during S phase and house-keeping DNA repair and mitochondrial DNA synthesis in postmitotic cells may be widely different; because the enzyme concentration and composition differs during cell cycle, and only the R1 subunit is the same (19, 20). The R2 subunit of RNR is induced during S phase and degraded via a KEN box when the cell enters mitosis (21). In postmitotic cells instead, the p53R2 protein is present in complex with R1, both at low concentrations (16, 22).
In mammalian cells Trx1 and Grx1 are present in the cytosol or nucleus, whereas Trx2 is localized in the mitochondria (12). Grx2 has two isoforms that are located in the mitochondria and the cytosol/nucleus (23–25). Previous studies did not reveal a clear co-localization of RNR with Trx1 or Grx1 in cells using immunohistochemistry (26, 27). Furthermore in rabbit bone marrow it has been reported that Trx is not active as a hydrogen donor for the homologous ribonucleotide reductase (28).
Luthman et al. (29, 30) used purified calf thymus RNR and observed activity with Trx and GSH plus Grx. Here we have used recombinant RNR to evaluate the kinetics of the electron donors. Our results show marked differences in mechanisms for mammalian RNR compared with the well known E. coli enzyme. Considering the amount of R1 in the assays and using R2 subunit, our system represents the S phase RNR. These data are also of general interest for understanding tumor cell growth.
EXPERIMENTAL PROCEDURES
Materials and Enzymes—The [3H]CDP was from Amersham Biosciences. The cation exchanger resin AG® 50W was purchased from Bio-Rad. Yeast glutathione reductase, GSH, DTT, NADPH, and insulin were from Sigma. Human Trx1 and Grx1 were prepared as described (31, 32). Human Grx2 and the Grx2C40S were prepared by Dr. Catrine Johansson (24, 32). Rat recombinant TrxR was a kind gift from Dr. Olle Rengby (33). Human recombinant R1 was kindly provided by Prof. Pär Nordlund from the Department of Medical Biochemistry and Biophysics (Karolinska Institute).
Expression and Purification of Mouse R1 Subunit—The E. coli strain expressing R1 was kindly provided by Dr. Lars Thelander (Umeå University, Umeå, Sweden), and purification was carried out essentially as described by Davis et al. (34). Induction of R1 expression with 50 μm isopropyl β-d-thiogalactopyranoside was done at an A600 of 1 at a temperature of 15 °C in 20 liters of fermenter. The bacterial pellet (30 g) was dissolved in 90 ml of buffer A (50 mm HEPES, pH 7.3, 1 mm EDTA, 1 mm DTT, and 25 mm KCl) and lysed with 0.1% lysozyme. After shock freezing the bacteria in liquid nitrogen, the cell debris were pelleted. Solid ammonium sulfate to obtain 50% final saturation was added to the supernatant, and the pellet obtained after centrifugation was dissolved in 10 ml of buffer A.
After desalting the solution on a Sephadex G-25 column (100 ml) with buffer A, the sample was applied to a 20-ml dATP-Sepharose column (34) equilibrated with the same buffer. The column was washed with 60 ml of high salt buffer (50 mm HEPES, pH 7.3, 1 mm EDTA, 1 mm DTT, and 500 mm KCl) followed by 30 ml of 50 mm HEPES, pH 7.3. The R1 protein was eluted with 50 mm HEPES, pH 7.3, and 50 mm ATP. Protein was concentrated with Centriprep YM-30 (Amicon), and the buffer was changed to 50 mm HEPES, pH 7.3, l00 mm KCl on a Sephadex G-25 column. The purity of R1 was finally analyzed by SDS-PAGE, and the protein concentration was determined with a Bio-Rad protein assay kit.
Expression and Purification of Mouse R2 Subunit—The E. coli strain expressing R2 was a kind gift from Dr. Lars Thelander, and the R2 protein was expressed and purified as described before (35). After homogenizing 30 g of frozen bacteria in 90 ml of buffer B (50 mm Tris-HCl, pH 7.5, 1 mm phenylmethylsulfonyl fluoride, and 1 mm EDTA), 2% streptomycin sulfate, and 0.1% lysozyme were added to the extract; the bacterial suspension was frozen in liquid nitrogen and thawed once, and cell debris and nucleic acids were pelleted. Solid ammonium sulfate to 40% saturation was then added to the supernatant fraction, and the precipitate was collected.
After dissolving the pellet in 6 ml of buffer B, the sample was desalted on a G-25 column. The solution was then subjected to anion exchange chromatography using a 30-ml DEAE-cellulose column (DE 52) (35). Thereafter the protein was precipitated using 70% solid ammonium sulfate and dissolved in 50 mm Tris-HCl, pH 7.5, followed by G-25 chromatography. Reactivation of the R2 was done by regenerating of the iron center and tyrosyl radical as described by Thelander and co-workers (35).
Determination of Ribonucleotide Reductase Activity—RNR was reconstituted by mixing recombinant R1 and R2 proteins. Activity was assayed following the conversion of [3H]CDP into [3H]dCDP. The reaction was initiated by adding reaction mixture containing 40 mm Tris-HCl buffer, pH 7.6, 2 mm ATP, 10 mm MgCl2, 200 mm KCl, 200 μm FeCl3, and 0.5 mm [3H]CDP (∼20,000 cpm/nmol) in a final volume of 50 μl. Incubation was carried out at 37 °C for 30 min. The reaction was terminated by the addition of 1 m HClO4 and hydrolysis to dCMP. The amount of [3H]dCMP radioactivity was quantified by liquid scintillation counting after ion exchange chromatography on Dowex-50 columns (36).
As reductant, the indicated concentrations of DTT or human Trx1 and 0.1 μm TrxR plus 1 mm NADPH were used in separate experiments. When the Grx system was used together with RNR, the samples contained 0.1 μm GR, 1 mm NADPH, and varying amounts of GSH generally 4 mm. To avoid oxidation of GSH to GSSG, each time a new tube of frozen stock of GSH titrated to pH 7.0 was used. The activity was calculated as nmol of dCDP produced per time of incubation. Modifications to the protocol are indicated in individual table and figure legends.
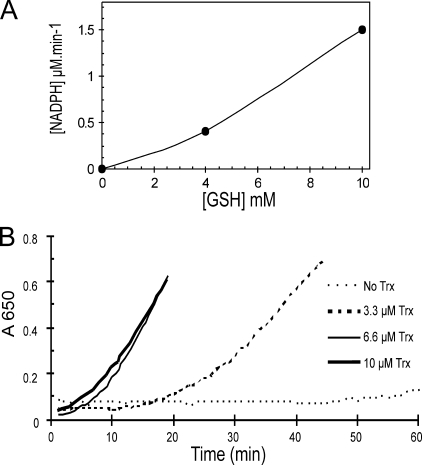
Insulin Disulfide Bond Reduction Activity of Thioredoxin1—Two assays were utilized to study the reduction of Trx1 with GSH. First, in a final volume of 150 μl, 10 μm Trx1 was mixed with 0.16 mm insulin, 0.5 mm NADPH, 0.2 mm EDTA, and 100 mm phosphate buffer, pH 7.0. Two samples with 4 and 10 mm GSH were monitored at A340 after adding 0.1 μm GR. A molar extinction coefficient of 6200 m–1 cm–1 was used in calculations (37, 38).
Insulin precipitation assay was also used by recording the increase in turbidity at A650. Different concentrations of Trx1 were checked in the same buffer using 10 mm GSH. The mixture without Trx1 and GSH was set as a blank, and the basic level of NADPH consumption with adding GR was measured in the sample without Trx1.
Expression and Purification of Mouse Glutaredoxin1—Oligonucleotide primers containing NdeI and BamHI restriction sites were designed to amplify the open reading frame of mouse Grx1 with polymerase chain reaction. The DNA fragment was cloned into the NdeI-BamHI sites of the pGEM-T vector (Promega). The construct was verified by DNA sequence determination. The insert was subcloned into the same restriction sites of the pET-15b vector (Novagen) to express the polyhistidine tag at the N terminus.
Expression and purification was done as previously described (24). The reductase activity was checked using a hydroxyethyl disulfide assay (24).
Cell Cultures and Preparation of Cell Extracts—HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in an humidified atmosphere of 5% CO2 at 37 °C.
The cells were trypsinized, spun down, and resuspended in the same volume of packed cell volume of 20 mm HEPES, pH 7.4, 2 mm MgCl2 with freshly added protease inhibitor (Roche Applied Science). The lysate was passed through five cycles of freeze-thaw; debris was removed by centrifugation (13,000 rpm, 10 min, 4 °C). The supernatant was heated to 75 °C in a boiling water bath. Precipitated proteins were spun down, and the supernatant was assayed. The protein concentration was quantified with Bio-Rad protein assay. Cell extracts were added to a series of tubes containing constant amount of recombinant R1 and R2. The samples were supplemented with TrxR and NADPH to couple the Trx; or GSH, NADPH, and GR to couple Grx in the extract. To compensate the oxidative environment produced by cell lysis, the amount of NADPH was raised to 3 mm. DTT served as a control.
RESULTS
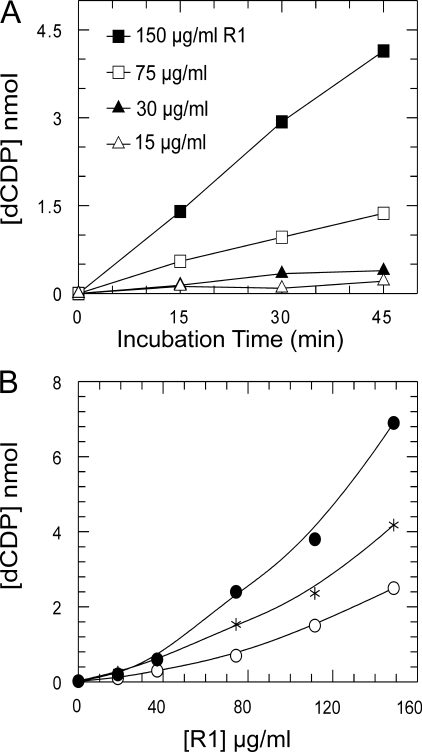
Characterization of the Mammalian RNR—We expressed and purified the recombinant mouse R1 and R2 proteins to apparent homogeneity reconstituting the tyrosyl radical in R2 (34, 35). By mixing R1 and R2 proteins in the presence of Mg2+ ions, active RNR was formed, and CDP was reduced to dCDP in a linear reaction with DTT as electron donor during 45-min incubations (Fig. 1A). As known from previous experiments, R1 and R2 formed a weak complex, and only at high concentrations (75–150 μg/ml) of R1, activity was recorded. Increasing R1 with a fixed amount of R2 resulted in a marked cooperative-like increase in activity; this behavior was identical for all three electron donors (Fig. 1B).
FIGURE 1.
Initial rate of RNR assay with different concentrations of R1 depending on time and electron donor. A, four series of samples with different concentrations of R1 and constant 36 μg/ml R2 were assayed using 10 mm DTT as reductant. The reaction was stopped after the desired time. B, a range of 0–150 μg/ml R1 was assayed with 36 μg/ml R2 within 45 min of incubation time. Each concentration was checked with the Trx system: 4 μm Trx1, 1 mm NADPH, and 0.1 μm TrxR (•); or the Grx system: 1.76 μm Grx1, 1 mm NADPH, 10 mm GSH, and 0.1 μm GR (○); and 10 mm DTT (*).
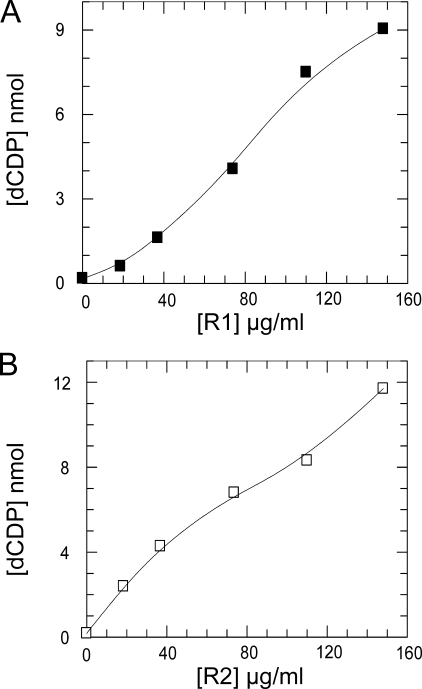
The specific activity for several different preparations was typically 60 and 250 nmol/min/mg for R1 and R2, respectively, measured in the presence of an excess of the other subunit, which are values similar to previously reported mammalian recombinant preparations (34, 35). Note that R1 has the lowest specific activity and is the apparent rate-limiting subunit consistent with its content of the shuttle disulfide and catalytic center. The weak complex of the two subunits forming active RNR makes it difficult to determine kinetic parameters and also true kcat values. As shown in Fig. 2B with a low amount of R2, which is not enough to saturate R1, the shape of the activity curve is not sigmoidal because R1 disulfides were reduced by the electron donor irrespective of whether it was free or bound to R2. However, in contrast the curve for constant R2 (Fig. 2A) is sigmoidal because R1 is the limiting subunit in the weak complex.
FIGURE 2.
Characterization of mouse RNR subunits. Different concentrations of R1 with 74 μg/ml R2 (A) or different concentrations of R2 with 110 μg/ml R1 (B) were assayed for 30 min incubation time in a total volume of 50 μl. As electron donor a Trx system containing 3 μm Trx1, 1 mm NADPH, and 0.1 μm TrxR was used.
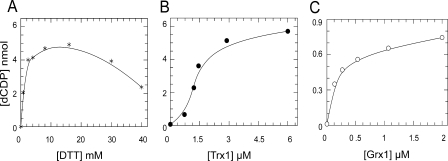
Effect of DTT on RNR—To investigate the optimal amount of DTT for RNR activity, experiments were carried out with R1 quite similar to concentrations calculated from S phase fibroblasts of 48 μg/ml (20). The highest activity was obtained with 4–10 mm DTT (Km = 2 mm). Inhibition by higher concentrations of DTT was observed (Fig. 3A). No increase in activity was obtained by R1 prereduced with DTT.
FIGURE 3.
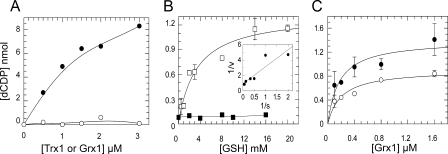
Efficiency of DTT, Trx1, and Grx1 for reduction of mouse RNR. A, enzyme activity of 74 μg/ml R1 plus 36 μg/ml R2 was measured in the presence of increasing amounts of DTT with standard assay conditions. B and C, samples with 120 μg/ml R1 and 40 μg/ml R2 were assayed with varying concentrations of Trx1, 1 mm NADPH, and 0.1 μm TrxR as the Trx system (B) or increasing amounts of Grx1, 1 mm NADPH, 4 mm GSH, and 0.1 μm GR as the Grx system (C).
Trx1 and Grx1 as Hydrogen Donors for S Phase RNR—Enzyme activity was measured with varying concentrations of human Trx1 (0.7–6 μm) using excess TrxR and NADPH or different amounts of Grx1 (0.1–2 μm) in combination with 4 mm GSH, NADPH, and excess GR (Fig. 3, B and C). Typical apparent Km values of 1.9 and 0.18 μm were obtained for Trx1 and Grx1, respectively. With the higher concentrations of R1, the Trx activity curve showed a tendency to sigmoidal behavior probably reflecting rate-limiting reduction by low Trx. In contrast Grx activity showed no such tendency, but remarkably the apparent Vmax and corresponding kcat was much lower for Grx than for Trx. However, the catalytic efficiency (kcat/Km) was similar or 2 × 105 m–1 s–1.
Because the reaction of Grx with RNR may be species specific like for E. coli RNR (29), mouse Grx1 was expressed to determine whether that would be more active with the homologous RNR. Because there was no difference between the reaction rates with human and mouse Grx1 (data not shown), we used the human Grx1 in further experiments.
Preincubation of R1 with DTT did not change the activity with either the Trx or Grx systems. This finding was in contrast to results with the E. coli RNR enzyme, which has to be freshly prereduced with DTT before assay for a Grx system to show activity (39). We also tested preincubation of R1 plus R2 with excess DTT followed by a Sephadex G-25 desalting step before assays. This treatment resulted in no difference in enzyme activity; nor did higher ATP (5 mm) as an allosteric effector (4). All of several R1 and R2 preparations showed the low kcat with the Grx system. The same relative activity of Trx and Grx was also detected with a human R1 preparation (Table 1).
TABLE 1.
Characterization of human R1 in reduction with Trx1 and Grx1
Enzyme activity of human R1 plus 50 μg/ml mouse R2 was measured in the presence of 10 μm Trx1, 0.5 mm NADPH, and 0.3 μm TrxR for the Trx system and 10 μm of Grx1, 5 mm GSH, 0.1 μm GR, and 0.5 mm NADPH for the Grx system. The samples were incubated at 37 °C for 30 min in a total volume of 50 μl.
|
R1
|
Enzyme
activitya
|
||
|---|---|---|---|
| Trx system | Grx system | ||
| nmol of dCDP formed | |||
| 80 μg/ml | 1.18 | 0.12 | |
| 160 μg/ml | 3.40 | 0.24 | |
Enzyme activity of 100 μg/ml human R1 with 10 mm DTT was 3.1 nmol.
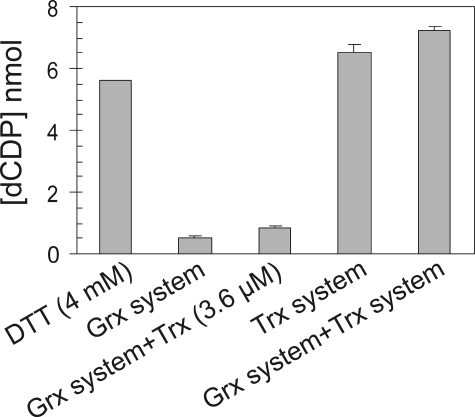
Combination of Trx with the Grx System for RNR Activity—To investigate whether the Trx system may activate RNR by reducing any critical disulfide bond to enable the Grx system to express higher turnover, both systems were added to the reaction mixture; no positive effect was observed, and the activity with combination of both systems was equal to the sum of their individual activities (Fig. 4).
FIGURE 4.
Contribution of Trx with Grx system in RNR assay. Samples with 120 μg/ml R1 and 40 μg/ml R2 were assayed with different reductants as indicated in each column. The Grx system contained 1 μm Grx1, 4 mm GSH plus NADPH, and GR. As the Trx system 3.6 μm Trx1 with NADPH and TrxR were used. Combination of 3.6 μm Trx1 or the whole Trx system to the Grx system was checked. The results are expressed as the means ± S.E. of two independent experiments performed in duplicates.
Because Trx strengthens the R1-R2 complex of E. coli RNR (40), we wondered whether Trx could stimulate Grx activity by making more active enzyme; therefore just Trx was added to the Grx system in an RNR assay. The data showed higher activity in the sample, which contained Grx system plus Trx (without TrxR) compared with the Grx system alone (Fig. 4). However, with insulin reduction assays, we showed that human Trx1 could catalyze the reduction of insulin disulfide bonds with high GSH and NADPH plus GR (Fig. 5). This is a consequence of a lower redox potential of human Trx (–230 mV) (41) compared with E. coli Trx (–270 mV) (42). Therefore, Trx did not increase Grx activity in RNR reduction per se, but it acted directly as a dithiol by reduction with high GSH and GR. This result may explain how mammalian cells can survive without TrxR activity.
FIGURE 5.
Trx1 can reduce disulfide bonds with high amount of GSH and GR. A, using insulin as the substrate for Trx1, the effect of 4 and 10 mm GSH on the reduction of disulfides with 10 μm Trx1 was investigated after adding 0.1 μm GR. The consumption of NADPH was measured at A340. The rate of oxidation without Trx1 was subtracted from each point. B, this experiment was performed with 10 mm GSH and different concentrations of Trx1; precipitation of insulin at A650 was monitored after adding 0.1 μm GR.
Grx2 and Its Monothiol Mutant Protein Are as Active as Grx1—Mammalian Grx2 (24, 25) has a splice form Grx2b that has been suggested to be a possible electron donor for RNR in DNA repair (24). Experiments were carried out with human Grx2 (0.15–2.4 μm) and 120 μg/ml R1 plus 40 μg/ml R2 using standard glutaredoxin assay conditions (4 mm GSH, 0.1 μm GR, and 1 mm NADPH). Grx2 had the same activity as Grx1 (Table 2) except for a marginally higher Km value. Similar experiments with a C40S mutant of Grx2 unexpectedly showed full activity with RNR. This demonstrates that Grx operates with mouse RNR by a monothiol mixed disulfide mechanism in sharp contrast to Grx1 and E. coli RNR (43). There is no possibility for Trx to be active as a monothiol mutant, because this is not a substrate for TrxR, nor does it work as a disulfide reductase and consequently cannot be active as an electron donor for RNR (41, 42).
TABLE 2.
Kinetic constants of Trx1, Grx1, wild type, and mutant Grx2 for reduction of mouse RNR
The values derived from fitting data in Fig. 3 (B and C).
Activity of 120 μg/ml R1 and 40 μg/ml R2 was measured with varying concentrations of wild type or mutant Grx2 in the presence of 4 mm GSH, 0.1 μm GR, and 1 mm NADPH. The samples were incubated at 37 °C for 30 min in a total volume of 50 μl.
Because Grx2 is a substrate for TrxR (32), we asked whether it can reduce RNR via a TrxR and NADPH pathway. Using 0.1 μm TrxR with Grx2 the activity was about 10-fold lower than that with GR and 4 mm GSH (data not shown), showing that dithiol Grx is an inefficient electron donor for mouse RNR.
Role of GSH in Mammalian RNR Catalytic Mechanism—DTT at 1 mm can reduce E. coli Trx1 and Grx1, which then both give the same Vmax with E. coli RNR (39); E. coli RNR has a 30 mm Km with DTT (44). Using this experiment with mouse RNR full activity with Trx1 was obtained, but Grx1 was not giving any activity above DTT, suggesting that dithiol Grx was not an electron donor (Fig. 6A).
FIGURE 6.
Requirement of GSH for reduction of RNR with Grx. A, assay mixture contained 120 μg/ml R1, 40 μg/ml R2, and 0.4 mm DTT as ultimate reducing power. RNR activity with different concentrations of Trx1 (•) or Grx1 (○) was measured. The background activity of RNR with this amount of DTT was subtracted from each point. B, two series of samples with 74 μg/ml R1 and 36 μg/ml R2, and increasing concentrations of GSH were prepared; the reaction was started by adding reaction mixture supplemented with 1 μm Grx1, 1 mm NADPH, and 0.1 μm GR (□). Each point represents the mean value of two independent experiments with duplicate samples. The inset shows Lineweaver-Burk plot. In the second series (▪) standard reaction mixture was used to detect the background activity with different amounts of GSH. C, enzyme activity of 74 μg/ml R1 and 36 μg/ml R2 versus different concentrations of Grx1 is shown. Utilizing 4 mm (○) or 10 mm (•) GSH plus 1 mm NADPH and 0.1 μm GR were compared.
The effect of GSH was further investigated in RNR assays using different concentrations of GSH (0.5–20 mm). Samples contained 1 μm Grx1 plus excess GR and 1 mm NADPH. Results revealed that the apparent Km value for GSH was 3 mm. Thus GSH was necessary for RNR reduction with Grx; experiments performed with only high GSH as the only reducing source for RNR confirmed that GSH itself was not active (Fig. 6B). Furthermore we measured the kinetics of Grx1 using 4 and 10 mm GSH (Fig. 6C), and 10 mm GSH resulted in a higher apparent kcat.
Activity of Trx and Grx Systems in Cell Extracts—The assay of RNR activity in the crude cell extracts is complicated by the presence of competing activities that rapidly deplete the substrate; either hydrolyzing enzymes that dephosphorylate or kinases that phosphorylate the ribonucleoside diphosphate substrate (45). Using a noncleavable ATP analog, which was effective at minimizing substrate diversion (45) or conducting assays in permeable cells (46, 47) have been solutions so far used. Because Trx and Grx are heat stable proteins (10), we instead applied heating to 75 °C treatment and used cell extracts as a source of redox proteins. As described under “Experimental Procedures,” the heated extract gave no evidence of an unknown system. Furthermore the results showed that Trx was the dominant electron donor in the extract (Fig. 7).
FIGURE 7.
Cell extract as source of heat stable electron donors for RNR. The columns show the activity of 120 μg/ml R1 plus 40 μg/ml R2 with 5 μg heat-treated cell extract. The enzyme activity with the addition of 0.1 μm TrxR and 3 mm NADPH (black bar) or 4 mm GSH, 3 mm NADPH, and 0.1 μm GR (white bar) were compared. Each column represents the average of two independent assays with duplication. The count with 1 mm DTT (gray bar) was plotted after subtracting the background activity of recombinant RNR with 1 mm DTT as a control.
DISCUSSION
The dNTP pools increase several-fold as cells enter S phase, and this is accompanied by increased synthesis of R1 and induction of R2 (1). The mechanism of the electron donors with mammalian RNR has not previously been characterized in detail. This study provides some unexpected differences compared with the well known E. coli system (39).
The thioredoxin system showed a Vmax value with RNR similar to that of DTT and a Km value in the range of 1.5–2.0 μm. This is in contrast to yeast, which has lower activity with the Trx system compared with DTT (48). Our results showed that the glutaredoxin system with 4 mm GSH had a 10-fold lower apparent Km value but also a 10-fold lower Vmax. The Vmax for Grx was strongly dependent on the GSH concentration with an apparent Km value of 3 mm. With E. coli RNR, E. coli Grx1 has a low Km value but the same Vmax as thioredoxin, and the apparent Km for GSH is 0.4 mm (39).
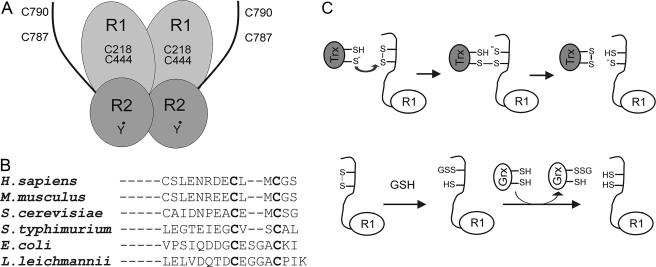
Another unexpected result was that the monothiol mutant C40S of Grx2 was fully active. The data for E. coli RNR shows clearly that the bacterial Grx1 mutant C14S is completely inactive (43) compatible with a dithiol-disulfide mechanism. Subsequent results by Bushweller and co-workers (49) demonstrates that binding of Grx1 to RNR involved a complex formation. The C-terminal sequence of the R1 subunit is not conserved between E. coli and mammalian cells. In fact the CXXXXC shuttle sequence in E. coli R1 is an exception, whereas yeast, human, or mouse R1 have a CXXC sequence (Fig. 8B). Because Grx2C40S cannot catalyze reduction of disulfides, this evidence strongly suggests a monothiol mechanism involving a glutathionylation reaction (Fig. 8C) compatible with the high Km of GSH.
FIGURE 8.
A, schematic model of mouse RNR with two pairs of redox active cysteines on the active site and C-terminal tail of R1, and the tyrosyl radical on R2 subunit. B, sequence alignment of the C-terminal region of the R1 subunit from different species. C, a model showing the dithiol and monothiol reduction pathways of mammalian RNR.
One advantage of a glutathionylation mechanism may be with very low levels of R1 involved in repair and production of dNTPs for mitochondrial DNA synthesis. In many resting postmitotic cells, thioredoxin is present at very low levels. The sigmoidal curve of Trx activity (Fig. 3B) showed that reduced Trx could not be efficient with a low concentration of R1 in postmitotic cells. The high concentration of GSH (5–20 mm in most mammalian cells) (50) would ensure that there is glutathionylated R1, and then any Grx should be able to catalyze reduction of the C-terminal disulfide (Fig. 8C). The cell contains glutaredoxins with a single Cys residue belonging to the GST family of proteins like the ω-GSTs (51). A study showing no variation in dNTP pools by down-regulating TrxR in malignant mouse cells (52) supports the conclusion that the Trx system is not the only pathway used by RNR in tumor cells and highlights the activity by GSH and Grx.
In recent studies it is shown that yeast with deletion of both cytoplasmic thioredoxins has a longer S phase because of the loss of the high rate of dNTP synthesis (53). Analysis of the in vivo redox state of RNR established Trx as the major reductant in yeast (54). Our data has also revealed the advantage of Trx as electron donor because of its high kcat.
RNR is a slow enzyme, and the kcat calculated for the Trx system (12 × min–1) will not be enough for DNA replication. Considering that 3 billion base pairs (6 × 109 nucleotides equal to 10–14 mol) are made in a mammalian cell during an S phase of 6 h and that the average volume of a fibroblast is 3.4 pl (20), the rate of nucleotide production can be calculated to be at least 140 nm s–1 or an estimated turnover of R1 of ∼300 × min–1. This is far away from the RNR activity that we measured with the Trx system as electron donor. Although the Trx and Grx concentrations (55, 56) seem high enough in the cell to predict that they can drive RNR activity, the activity of the enzyme in vivo must have higher turnover particularly with the Grx system. The highly active enzyme in vivo remains to be isolated and characterized, as is also true for E. coli (6, 17, 57).
The concentration of R1 protein in logarithmically growing and S phase of mouse fibroblast Balb/3T3 cells has been calculated to be around 48 μg/ml (0.5 μm), which reduces to 5.3 μg/ml in resting cells (20). We tested this low amount of R1 but could hardly detect any activity; the sigmoidal curve of the activity of the subunits also prompted us to choose a higher level of R1 in our experiments.
Our results are representative of the S phase in the cell because we have only used the R2 subunit, which is implicated in DNA replication (58). The recently discovered p53R2 has been shown to be continuously present in resting cells in low concentration (16, 22) in contrast to the R2 protein, which is degraded (58). Resting cells expressing low levels of R1+p53R2 in the cytosol are assumed to supply dNTPs for DNA repair and mitochondrial DNA synthesis (19). Upon DNA damage, p53R2 would be used with R1 to supply dNTPs for repair. The human R1 and R2 as well as the p53R2 have been suggested to relocate from cytoplasm to nucleus after genotoxic stress by UV irradiation (59). However, recent data demonstrate that ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage (60).
Previous experiments showed no change in growth rate, DNA synthesis, or the size of the dNTP pools in mouse 3T6 fibroblasts with depleted GSH after treatment with buthionine sulfoximine (61). This result implies that in the cultured cells, Grx is not the only hydrogen donor for RNR and DNA synthesis. On the other hand the level of dNTP needed for DNA repair is low in mammalian cells, because in mouse fibroblasts there is no increase in the level of dNTP pools after DNA damage (20). Furthermore, DNA repair in cells has a dependence on GSH, because there is evidence of accumulation of DNA damage in the organs of mice with a defect in GSH metabolism inducing low levels of GSH (62).
Our results show that Trx1 and Grx1 have similar catalytic efficiency (kcat/Km) with RNR and favor Trx1 as an S phase electron donor. Glutaredoxin is implicated in DNA repair and mitochondrial DNA synthesis via a glutathionylation mechanism. It remains to be determined whether and how the difference in spacing between the two C-terminal cysteine residues in E. coli and mammalian R1 dictates the difference in the mechanism of the Trx and Grx systems to disulfide reduction.
Acknowledgments
We gratefully thank Rolf Eliasson for excellent technical assistance and Jacek Andrzejewski for large scale cultivation of bacteria. We thank Prof. Lars Thelander for providing the plasmids encoding the RNR subunits and Prof. Pär Nordlund for human R1. The gift of human Grx2 and Grx2C40S by Dr. Catrine Johansson and TrxR by Dr. Olle Rengby are also acknowledged.
This work was supported in part by Swedish Research Council Medicine Grant 3529, Swedish Cancer Society Grant 961, and funds from the K. A. Wallenberg Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RNR, ribonucleotide reductase; dNTP, deoxyribonucleoside triphosphate; DTT, dithiothreitol; GR, glutathione reductase; Grx, glutaredoxin; Trx, thioredoxin; TrxR, thioredoxin reductase; GST, glutathione S-transferase.
References
- 1.Nordlund, P., and Reichard, P. (2006) Annu. Rev. Biochem. 75 681–706 [DOI] [PubMed] [Google Scholar]
- 2.Kolberg, M., Strand, K. R., Graff, P., and Andersson, K. K. (2004) Biochim. Biophys. Acta 1699 1–34 [DOI] [PubMed] [Google Scholar]
- 3.Reichard, P. (2002) Arch. Biochem. Biophys. 397 149–155 [DOI] [PubMed] [Google Scholar]
- 4.Kashlan, O. B., Scott, C. P., Lear, J. D., and Cooperman, B. S. (2002) Biochemistry 41 462–474 [DOI] [PubMed] [Google Scholar]
- 5.Rofougaran, R., Vodnala, M., and Hofer, A. (2006) J. Biol. Chem. 281 27705–27711 [DOI] [PubMed] [Google Scholar]
- 6.Thelander, L., and Reichard, P. (1979) Annu. Rev. Biochem. 48 133–158 [DOI] [PubMed] [Google Scholar]
- 7.Uhlin, U., and Eklund, H. (1994) Nature 370 533–539 [DOI] [PubMed] [Google Scholar]
- 8.Sjoberg, B. M., and Sahlin, M. (2002) Methods Enzymol. 348 1–21 [DOI] [PubMed] [Google Scholar]
- 9.Laurent, T. C., Moore, E. C., and Reichard, P. (1964) J. Biol. Chem. 239 3436–3444 [PubMed] [Google Scholar]
- 10.Holmgren, A. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 2275–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lillig, C. H., Berndt, C., and Holmgren, A. (2008) Biochim. Biophys. Acta 1780 1304–1317 [DOI] [PubMed] [Google Scholar]
- 12.Lillig, C. H., and Holmgren, A. (2007) Antioxid. Redox Signal. 9 25–47 [DOI] [PubMed] [Google Scholar]
- 13.Gallardo-Madueno, R., Leal, J. F., Dorado, G., Holmgren, A., Lopez-Barea, J., and Pueyo, C. (1998) J. Biol. Chem. 273 18382–18388 [DOI] [PubMed] [Google Scholar]
- 14.Prieto-Alamo, M. J., Jurado, J., Gallardo-Madueno, R., Monje-Casas, F., Holmgren, A., and Pueyo, C. (2000) J. Biol. Chem. 275 13398–13405 [DOI] [PubMed] [Google Scholar]
- 15.Gon, S., Faulkner, M. J., and Beckwith, J. (2006) Antioxid. Redox Signal. 8 735–742 [DOI] [PubMed] [Google Scholar]
- 16.Guittet, O., Hakansson, P., Voevodskaya, N., Fridd, S., Graslund, A., Arakawa, H., Nakamura, Y., and Thelander, L. (2001) J. Biol. Chem. 276 40647–40651 [DOI] [PubMed] [Google Scholar]
- 17.Wang, J., Lohman, G. J., and Stubbe, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14324–14329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinberg, G., Ullman, B., and Martin, D. W., Jr. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 2447–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pontarin, G., Ferraro, P., Hakansson, P., Thelander, L., Reichard, P., and Bianchi, V. (2007) J. Biol. Chem. 282 16820–16828 [DOI] [PubMed] [Google Scholar]
- 20.Hakansson, P., Hofer, A., and Thelander, L. (2006) J. Biol. Chem. 281 7834–7841 [DOI] [PubMed] [Google Scholar]
- 21.Nasmyth, K. (2001) Annu. Rev. Genet. 35 673–745 [DOI] [PubMed] [Google Scholar]
- 22.Tanaka, H., Arakawa, H., Yamaguchi, T., Shiraishi, K., Fukuda, S., Matsui, K., Takei, Y., and Nakamura, Y. (2000) Nature 404 42–49 [DOI] [PubMed] [Google Scholar]
- 23.Lonn, M. E., Hudemann, C., Berndt, C., Cherkasov, V., Capani, F., Holmgren, A., and Lillig, C. H. (2008) Antioxid. Redox Signal. 10 547–557 [DOI] [PubMed] [Google Scholar]
- 24.Lundberg, M., Johansson, C., Chandra, J., Enoksson, M., Jacobsson, G., Ljung, J., Johansson, M., and Holmgren, A. (2001) J. Biol. Chem. 276 26269–26275 [DOI] [PubMed] [Google Scholar]
- 25.Gladyshev, V. N., Liu, A., Novoselov, S. V., Krysan, K., Sun, Q. A., Kryukov, V. M., Kryukov, G. V., and Lou, M. F. (2001) J. Biol. Chem. 276 30374–30380 [DOI] [PubMed] [Google Scholar]
- 26.Hansson, H. A., Rozell, B., Stemme, S., Engstrom, Y., Thelander, L., and Holmgren, A. (1986) Exp. Cell Res. 163 363–369 [DOI] [PubMed] [Google Scholar]
- 27.Rozell, B., Barcena, J. A., Martinez-Galisteo, E., Padilla, C. A., and Holmgren, A. (1993) Eur. J. Cell Biol. 62 314–323 [PubMed] [Google Scholar]
- 28.Hopper, S., and Iurlano, D. (1983) J. Biol. Chem. 258 13453–13457 [PubMed] [Google Scholar]
- 29.Luthman, M., Eriksson, S., Holmgren, A., and Thelander, L. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 2158–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luthman, M., and Holmgren, A. (1982) J. Biol. Chem. 257 6686–6690 [PubMed] [Google Scholar]
- 31.Ren, X., Bjornstedt, M., Shen, B., Ericson, M. L., and Holmgren, A. (1993) Biochemistry 32 9701–9708 [DOI] [PubMed] [Google Scholar]
- 32.Johansson, C., Lillig, C. H., and Holmgren, A. (2004) J. Biol. Chem. 279 7537–7543 [DOI] [PubMed] [Google Scholar]
- 33.Rengby, O., Johansson, L., Carlson, L. A., Serini, E., Vlamis-Gardikas, A., Karsnas, P., and Arner, E. S. (2004) Appl. Environ. Microbiol. 70 5159–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis, R., Thelander, M., Mann, G. J., Behravan, G., Soucy, F., Beaulieu, P., Lavallee, P., Graslund, A., and Thelander, L. (1994) J. Biol. Chem. 269 23171–23176 [PubMed] [Google Scholar]
- 35.Mann, G. J., Graslund, A., Ochiai, E., Ingemarson, R., and Thelander, L. (1991) Biochemistry 30 1939–1947 [DOI] [PubMed] [Google Scholar]
- 36.Engstrom, Y., Eriksson, S., Thelander, L., and Akerman, M. (1979) Biochemistry 18 2941–2948 [DOI] [PubMed] [Google Scholar]
- 37.Holmgren, A. (1979) J. Biol. Chem. 254 9627–9632 [PubMed] [Google Scholar]
- 38.Luthman, M., and Holmgren, A. (1982) Biochemistry 21 6628–6633 [DOI] [PubMed] [Google Scholar]
- 39.Holmgren, A. (1979) J. Biol. Chem. 254 3672–3678 [PubMed] [Google Scholar]
- 40.Kasrayan, A., Birgander, P. L., Pappalardo, L., Regnstrom, K., Westman, M., Slaby, A., Gordon, E., and Sjoberg, B. M. (2004) J. Biol. Chem. 279 31050–31057 [DOI] [PubMed] [Google Scholar]
- 41.Watson, W. H., Pohl, J., Montfort, W. R., Stuchlik, O., Reed, M. S., Powis, G., and Jones, D. P. (2003) J. Biol. Chem. 278 33408–33415 [DOI] [PubMed] [Google Scholar]
- 42.Holmgren, A. (1985) Annu. Rev. Biochem. 54 237–271 [DOI] [PubMed] [Google Scholar]
- 43.Bushweller, J. H., Aslund, F., Wuthrich, K., and Holmgren, A. (1992) Biochemistry 31 9288–9293 [DOI] [PubMed] [Google Scholar]
- 44.Aberg, A., Hahne, S., Karlsson, M., Larsson, A., Ormo, M., Ahgren, A., and Sjoberg, B. M. (1989) J. Biol. Chem. 264 12249–12252 [PubMed] [Google Scholar]
- 45.Slabaugh, M. B., and Mathews, C. K. (1984) J. Virol. 52 501–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kucera, R., and Paulus, H. (1982) Arch. Biochem. Biophys. 214 102–113 [DOI] [PubMed] [Google Scholar]
- 47.veer Reddy, G. P., and Pardee, A. B. (1982) J. Biol. Chem. 257 12526–12531 [PubMed] [Google Scholar]
- 48.Ge, J., Perlstein, D. L., Nguyen, H. H., Bar, G., Griffin, R. G., and Stubbe, J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10067–10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berardi, M. J., Pendred, C. L., and Bushweller, J. H. (1998) Biochemistry 37 5849–5857 [DOI] [PubMed] [Google Scholar]
- 50.Meister, A. (1988) J. Biol. Chem. 263 17205–17208 [PubMed] [Google Scholar]
- 51.Whitbread, A. K., Masoumi, A., Tetlow, N., Schmuck, E., Coggan, M., and Board, P. G. (2005) Methods Enzymol. 401 78–99 [DOI] [PubMed] [Google Scholar]
- 52.Yoo, M. H., Xu, X. M., Carlson, B. A., Patterson, A. D., Gladyshev, V. N., and Hatfield, D. L. (2007) PLoS ONE 2 e1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koc, A., Mathews, C. K., Wheeler, L. J., Gross, M. K., and Merrill, G. F. (2006) J. Biol. Chem. 281 15058–15063 [DOI] [PubMed] [Google Scholar]
- 54.Camier, S., Ma, E., Leroy, C., Pruvost, A., Toledano, M., and Marsolier-Kergoat, M. C. (2007) Free Radic. Biol. Med. 42 1008–1016 [DOI] [PubMed] [Google Scholar]
- 55.Lundberg, M., Fernandes, A. P., Kumar, S., and Holmgren, A. (2004) Biochem. Biophys. Res. Commun. 319 801–809 [DOI] [PubMed] [Google Scholar]
- 56.Holmgren, A., and Luthman, M. (1978) Biochemistry 17 4071–4077 [DOI] [PubMed] [Google Scholar]
- 57.Eriksson, S. (1975) Eur. J. Biochem. 56 289–294 [DOI] [PubMed] [Google Scholar]
- 58.Chabes, A., and Thelander, L. (2000) J. Biol. Chem. 275 17747–17753 [DOI] [PubMed] [Google Scholar]
- 59.Xue, L., Zhou, B., Liu, X., Qiu, W., Jin, Z., and Yen, Y. (2003) Cancer Res. 63 980–986 [PubMed] [Google Scholar]
- 60.Pontarin, G., Fijolek, A., Pizzo, P., Ferraro, P., Rampazzo, C., Pozzan, T., Thelander, L., Reichard, P. A., and Bianchi, V. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 17801–17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spyrou, G., and Holmgren, A. (1996) Biochem. Biophys. Res. Commun. 220 42–46 [DOI] [PubMed] [Google Scholar]
- 62.Rojas, E., Valverde, M., Kala, S. V., Kala, G., and Lieberman, M. W. (2000) Mutat. Res. 447 305–316 [DOI] [PubMed] [Google Scholar]